Abstract
Background
Environmental factors may play a role in colon cancer. In this view, several studies investigated tumor samples for the presence of various viral DNA with conflicting results.
Findings
We undertook a systematic DNA analysis of 44 consecutive, prospectively collected primary tumor samples by real time and qualitative PCR for viruses of known or potential oncogenic role in humans, including polyomavirus (JCV, BKV, Merkel cell polyomavirus), HPV, HTLV, HHV-8 and EBV. Negative controls consisted of surgical resection margins. No evidence of genomic DNA fragments from tested virus were detected, except for EBV, which was found in a significant portion of tumors (23/44, 52%). Real-time PCR showed that EBV DNA was present at a highly variable content (median 258 copies in 105 cells, range 15–4837). Presence of EBV DNA had a trend to be associated with high lymphocyte infiltration (p = 0.06, χ2 test), and in situ hybridization with EBER1-2 probes revealed latency in a fraction of these lymphoid cells, with just a few scattered plasma cells positive for BZLF-1, an immediate early protein expressed during lytic replication. LMP-1 expression was undetectable by immunohistochemistry.
Conclusions
These results argue against a significant involvement of the tested oncogenic viruses in established colon cancer.
Keywords: Colon cancer, Oncogenic viruses, EBV
Findings
It is estimated that 16-18% of the global cancer burden can be associated with oncogenic viruses [1,2]. The DNA viruses of recognized pathogenic role in humans include HCV, HBV, HPV, HHV-8, MCPyV and EBV, while the role of polyomaviruses JCV and BKV is controversial [1,3]. Colon cancer is a leading cause of cancer-related death and morbidity in western countries. The pathogenesis of this cancer is highly complex and it involves sequential genetic and epigenetic mechanisms [4,5]. A possible contribution of environmental agents, including bacteria and viruses, is also considered [6,7]. However, the search for a pathogenic agent generated conflicting results, possibly related to technical reasons or other unknown factors [6].
To contribute to this issue, we prospectively collected a series of consecutive 44 colon cancers to perform a systematic PCR-analysis for human polyomaviruses (JCV, BKV, MCPyV), HPV, HTLV, HHV-8 and EBV. The study was approved by Fondazione IRCCS Policlinico San Matteo Review Board. Fondazione IRCCS Policlinico San Matteo Review Board. Clinical and pathology characteristics are reported in Table 1. The population was representative of colon cancer cases who underwent primary tumor resection (low numbers of metastatic cases, Duke’s stage D). Internal control consisted of surgical resection margin (at least 5 cm distant from the tumor). Quality of extracted DNA samples (QIAmp DNA Mini Kit, Qiagen, Italy) was verified by means of amplification of the housekeeping gene beta-2-microglobulin (DNA amplicons >105 copies). Positive controls included virus infected human samples from various pathologies, including tumor samples, and plasmid DNA. Amplification methods consisted of real time (EBV, JCV, BKV, HTLV and HHV-8) [8-11] and qualitative PCR (MCPyV, HPV) [12,13]. Given the very controversial evidence concerning JCV, which was found from 0% to nearly 80% of the tumor samples tested in the literature [6,14-18], JCV was additionally investigated by the specific qualitative PCR described in positive reports [18] and that employed primers spanning a different portion of the large T antigen. Sensitivity for JCV real-time PCR assay was 1 viral copy in 105 cells. Experiments showed that no genomic DNA fragments from the tested viruses were detectable, with the notable exception of EBV that was positive in a consistent portion of cases (23/44 samples, 52%). Tumors associated with EBV positivity had EBV negative surgical resection margins. EBV DNA content was highly variable in tumors (median 258 viral copies in 105 cells, range 15–4837), and EBV had a trend to be observed in tumors displaying high lymphoid infiltration (p = 0.06, χ2 test). In situ hybridization analyses for the detection of EBER1-2 RNAs (PNA Probe/FITC and ISH detection kit, Dako, Denmark) demonstrated virus latency in a variable fraction of infiltrating non-neoplastic lymphoid cells, which could reach 20% in a few cases (Figure 1), but not in tumor cells. On the same line, immunohistochemistry to EBNA-1 nuclear protein (Fitzgerald Industries International – USA, clone M5042521), an antigen that is expressed during both latent and lytic phases, failed to show positive nuclei in neoplastic cells. Immunohistochemistry for LMP-1, a membrane associated protein involved in activation, was also negative, while lytic cycle was detected via expression of the immediate early protein BZLF1 (ZEBRA antigen, LSBio, clone LS-C102904) in a few scattered plasma cells (Figure 2). These findings essentially confirm latency of EBV in lymphoid infiltrates. The presence of EBV was not associated with the other tumor or clinical parameters studied including age, stage, tumor localization, or the presence of necrosis.
Table 1.
Clinical and pathological features of the colon cancer cases analysed for potential oncogenic viruses
| Colon cancer (n = 44) | No. of cases | % |
|---|---|---|
| Sex |
|
|
| Male |
26 |
59 |
| Female |
18 |
41 |
| Age groups |
|
|
| Median of age = 75 years (range 46–84) |
|
|
| ≤ 65 years |
9 |
20 |
| ≥ 65 years |
35 |
80 |
| Anatomic site |
|
|
| Caecum |
4 |
9 |
| Ascending |
11 |
25 |
| Transverse |
9 |
20 |
| Descending |
6 |
14 |
| Sigmoid |
4 |
9 |
| Sigmoid - Rectal |
10 |
23 |
| Colon dx |
18 |
41 |
| < 65 years |
26 |
59 |
| Staging |
|
|
| B1 |
7 |
16 |
| B2 |
16 |
36 |
| C1 |
2 |
5 |
| C2 |
15 |
34 |
| D |
4 |
9 |
| TNM |
|
|
| T1 |
0 |
0 |
| T2 |
9 |
20 |
| T3 |
29 |
66 |
| T4 |
6 |
14 |
| N0 |
23 |
52 |
| N1 |
11 |
25 |
| N2 |
10 |
23 |
| M0 |
40 |
91 |
| M1 |
4 |
9 |
| Grading |
|
|
| G1 |
0 |
0 |
| G2 |
32 |
73 |
| G3 | 12 | 27 |
Figure 1.
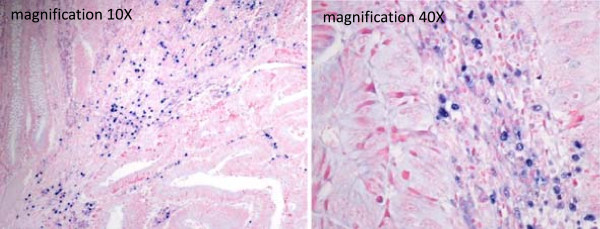
EBV latency is restricted to the inflammatory infiltrate in colon cancer. Tissue RNA in situ hybridization using EBER 1–2 PNA probes to detect EBV latency (Dako, Denmark). A blue nuclear staining indicates a positive reaction. Epithelial and cancer cells are negative, whereas occasionally positivity of non-neoplastic lymphoid cells may be extensive (reaching 20%).
Figure 2.
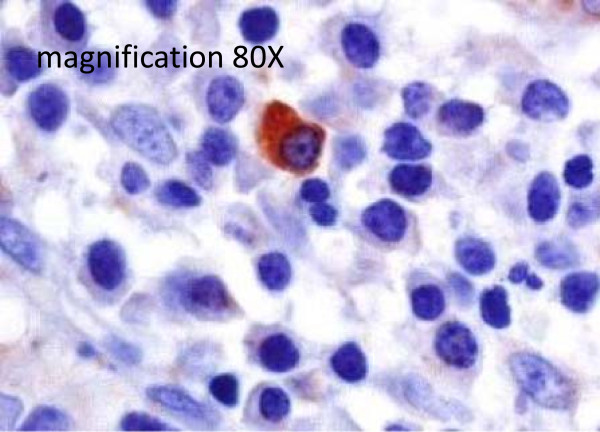
EBV lytic phase was limited to a few scatter plasma cells in the lymphoid infiltrate. Immunohistochemistry with a monoclonal antibody specific for the early immediate protein BZLF-1. These findings confirmed that EBV is essentially in latency phase in the lymphoid infiltrates of colorectal cancers.
In conclusion, we performed a PCR-based systematic analysis for potential oncogenic viruses in clinically established colon cancer and EBV was the only one detected. The viral infection was restricted to latency in the lymphoid infiltrate, in line with the few reports that used in situ hybridization with EBER probes [19,20], while we noted an association with high lymphocyte infiltration, a well-recognized favorable prognostic parameter. EBV positivity in lymphoid infiltrates may occasionally be extensive (Figure 1), much higher than expected on the numbers of circulating EBV positive memory B-lymphocytes in normal individuals, and it might be of interest to studying this phenomenon in specifically designed studies. In summary, the present analysis does not support a significant involvement of the tested viruses in manifest colon cancer, and suggests that new approaches [21] capable of detecting known and unknown non-human sequences should be investigated to study the role of infectious agents in colon cancer.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
VP and FB designed and coordinated the study and wrote the manuscript; MR collected data on patients and contributed to elaboration and interpretation of results; LF and SP performed PCR experiments; ED and RR carried out in situ hybridization and immunohistochemistry analyses; OL performed histological evaluation; AV performed primary tumor collection, histological evaluation and contributed to study design and interpretation of results, reviewed the manuscript; SB collected data on patients and critically reviewed the manuscript; MP critically reviewed the manuscript; PP contributed to study coordination and critically reviewed the manuscript. All authors read and approved the final manuscript.
Contributor Information
Loretta Fiorina, Email: loretta75@libero.it.
Mattia Ricotti, Email: mattiaricotti@alice.it.
Alessandro Vanoli, Email: ale.vanol@virgilio.it.
Ombretta Luinetti, Email: o.luinetti@smatteo.pv.it.
Elena Dallera, Email: e.dallera@smatteo.pv.it.
Roberta Riboni, Email: r.riboni@smatteo.pv.it.
Stefania Paolucci, Email: s.paolucci@smatteo.pv.it.
Silvia Brugnatelli, Email: s.brugnatelli@smatteo.pv.it.
Marco Paulli, Email: m.paulli@smatteo.pv.it.
Paolo Pedrazzoli, Email: p.pedrazzoli@smatteo.pv.it.
Fausto Baldanti, Email: f.baldanti@smatteo.pv.it.
Vittorio Perfetti, Email: v.perfetti@smatteo.pv.it.
Acknowledgements
The study was supported by Fondazione IRCCS Policlinico “San Matteo”, Ricerca Corrente to VP and FB. ED is recipient of a fellowship from Anatomic Pathology Section, IRCCS Fondazione Policlinico “San Matteo,” Pavia, Italy; RR is a recipient of a grant (project #580) from the Italian Ministry of Health (5 per Mille) of IRCCS Fondazione Policlinico “San Matteo”, Pavia, Italy.
References
- Bergonzini V, Salata C, Calistri A, Parolin C, Palù G. View and review on viral oncology research. Infect Agent Cancer. 2010;5:11. doi: 10.1186/1750-9378-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13(6):607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- Perfetti V, Ricotti M, Buonaguro F, Tirelli U, Pedrazzoli P. An overview of viral oncology in Italy - report from the Pavia meeting on solid tumors. Infect Agent Cancer. 2012;7(1):23. doi: 10.1186/1750-9378-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135(4):1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Engeland M, Derks S, Smits KM, Meijer GA, Herman JG. Colorectal cancer epigenetics: complex simplicity. J Clin Oncol. 2011;29(10):1382–1391. doi: 10.1200/JCO.2010.28.2319. [DOI] [PubMed] [Google Scholar]
- Hasan N, Pollack A, Cho I. Infectious causes of colorectal cancer. Infect Dis Clin North Am. 2010;24(4):1019–1039. doi: 10.1016/j.idc.2010.07.009. [DOI] [PubMed] [Google Scholar]
- Travaglione S, Fabbri A, Fiorentini C. The Rho-activating CNF1 toxin from pathogenic E. coli: a risk factor for human cancer development? Infect Agent Cancer. 2008;3:4. doi: 10.1186/1750-9378-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldanti F, Gatti M, Furione M, Paolucci S, Tinelli C, Comoli P, Merli P, Locatelli F. Kinetics of Epstein-Barr virus DNA load in different blood compartments of pediatric recipients of T-cell-depleted HLA-haploidentical stem cell transplantation. J Clin Microbiol. 2008;46(11):3672–3677. doi: 10.1128/JCM.00913-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens B, López G, Adaui V, González E, Kerremans L, Clark D, Verdonck K, Gotuzzo E, Vanham G, Cassar O, Gessain A, Vandamme AM, van Dooren S. Development and validation of a multiplex real-time PCR assay for simultaneous genotyping and human T-lymphotropic virus type 1, 2, and 3 proviral load determination. J Clin Microbiol. 2009;47(11):3682–3691. doi: 10.1128/JCM.00781-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watzinger F, Suda M, Preuner S, Baumgartinger R, Ebner K, Baskova L, Niesters HG, Lawitschka A, Lion T. Real-time quantitative PCR assays for detection and monitoring of pathogenic human viruses in immunosuppressed pediatric patients. J Clin Microbiol. 2004;42(11):5189–5198. doi: 10.1128/JCM.42.11.5189-5198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saundh BK, Tibble S, Baker R, Sasnauskas K, Harris M, Hale A. Different patterns of BK and JC polyomavirus reactivation following renal transplantation. J Clin Pathol. 2010;63(8):714–718. doi: 10.1136/jcp.2009.074864. [DOI] [PubMed] [Google Scholar]
- Pancaldi C, Corazzari V, Maniero S, Mazzoni E, Comar M, Martini F, Tognon M. Merkel cell polyomavirus DNA sequences in the buffy coats of healthy blood donors. Blood. 2011;117(26):7099–7101. doi: 10.1182/blood-2010-09-310557. [DOI] [PubMed] [Google Scholar]
- Qu W, Jiang G, Cruz Y, Chang CJ, Ho GY, Klein RS, Burk RD. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J Clin Microbiol. 1997;35(6):1304–1310. doi: 10.1128/jcm.35.6.1304-1310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb PA, Bush AC, Stoner GL, Lampe JW, Potter JD, Bigler J. No evidence of an association of JC virus and colon neoplasia. Cancer Epidemiol Biomarkers Prev. 2004;13(4):662–666. [PubMed] [Google Scholar]
- Militello V, Trevisan M, Squarzon L, Biasolo MA, Rugge M, Militello C, Palù G, Barzon L. Investigation on the presence of polyomavirus, herpesvirus, and papillomavirus sequences in colorectal neoplasms and their association with cancer. Int J Cancer. 2009;124(10):2501–2503. doi: 10.1002/ijc.24224. [DOI] [PubMed] [Google Scholar]
- Theodoropoulos G, Panoussopoulos D, Papaconstantinou I, Gazouli M, Perdiki M, Bramis J, Lazaris AC. Assessment of JC polyoma virus in colon neoplasms. Dis Colon Rectum. 2005;48(1):86–91. doi: 10.1007/s10350-004-0737-2. [DOI] [PubMed] [Google Scholar]
- Chiaravalli AM, Longhi E, Vigetti D, De Stefano FI, Deleonibus S, Capella C, Solcia E, Parravicini C. Gastrointestinal cancers reactive for the PAb416 antibody against JCV/SV40 T-Ag lack JCV DNA sequences while showing a distinctive pathologic profile. J Clin Pathol. 2013;66(1):44–49. doi: 10.1136/jclinpath-2012-200963. [DOI] [PubMed] [Google Scholar]
- Laghi L, Randolph AE, Chauhan DP, Marra G, Major EO, Neel JV, Boland CR. JC virus DNA is present in the mucosa of the human colon and in colorectal cancers. Proc Natl Acad Sci U S A. 1999;96(13):7484–7489. doi: 10.1073/pnas.96.13.7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Ding YQ, Li X, Yao KT. Investigation of Epstein-Barr virus in Chinese colorectal tumors. World J Gastroenterol. 2003;9(11):2464–2468. doi: 10.3748/wjg.v9.i11.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong NA, Herbst H, Herrmann K, Kirchner T, Krajewski AS, Moorghen M, Niedobitek F, Rooney N, Shepherd NA, Niedobitek G. Epstein-Barr virus infection in colorectal neoplasms associated with inflammatory bowel disease: detection of the virus in lymphomas but not in adenocarcinomas. J Pathol. 2003;201(2):312–318. doi: 10.1002/path.1442. [DOI] [PubMed] [Google Scholar]
- Barzon L, Lavezzo E, Costanzi G, Franchin E, Toppo S, Palù G. Next-generation sequencing technologies in diagnostic virology. J Clin Virol. 2013;58(2):346–350. doi: 10.1016/j.jcv.2013.03.003. [DOI] [PubMed] [Google Scholar]


