Abstract
In the last decade, atypical Listeria monocytogenes and L. innocua strains have been detected in food and the environment. Because of mutations in the major virulence genes, these strains have different virulence intensities in eukaryotic cells. In this study, we performed phenotypic and genotypic characterization of atypical L. monocytogenes and L. innocua isolates obtained from swine slaughterhouses and meat markets. Forty strains were studied, including isolates of L. monocytogenes and L. innocua with low-hemolytic activity. The isolates were characterized using conventional phenotypic Listeria identification tests and by the detection and analysis of L. monocytogenes-specific genes. Analysis of 16S rRNA was used for the molecular identification of the Listeria species. The L. monocytogenes isolates were positive for all of the virulence genes studied. The atypical L. innocua strains were positive for hly, plcA, and inlC. Mutations in the InlC, InlB, InlA, PI-PLC, PC-PLC, and PrfA proteins were detected in the atypical isolates. Further in vitro and transcriptomic studies are being developed to confirm the role of these mutations in Listeria virulence.
1. Introduction
Listeria monocytogenes and L. innocua are closely related species of the Gram-positive genus Listeria. They are widely distributed in the environment and frequently isolated from food. L. monocytogenes is the causative agent of listeriosis, a foodborne disease with a high fatality rate (20–30%) that mostly affects the elderly, neonates, and immunocompromised individuals [1, 2]. L. monocytogenes cannot be distinguished from other Listeria species using conventional isolation methods. Standard biochemical methods and selective and differential media are used for the identification of L. monocytogenes [3, 4]; however, some L. ivanovii, L. innocua, and L. seeligeri strains generate similar results to L. monocytogenes in these tests [5–7]. Therefore, it is necessary to confirm the virulence characteristics of L. monocytogenes to distinguish the Listeria species.
The best-characterized L. monocytogenes virulence factors are listeriolysin O (LLO), phosphatidylinositol phospholipase C (PI-PLC), and the internalins A and B (InlA and InlB). LLO and PI-PLC are encoded by the hly and plcA genes, respectively, which belong to the virulence gene cluster Listeria pathogenicity island 1 (LIPI-1), which contains the major virulence genes of L. monocytogenes [8]. Few atypical L. innocua strains have been reported to contain L. monocytogenes-specific genes and exhibit phenotypic characteristics similar to L. monocytogenes such as weak hemolysis [6, 7, 9]. Furthermore, certain low-hemolytic L. monocytogenes strains retain their virulence despite the presence of mutations in major virulence genes [10–12]. The existence of these atypical strains indicates that traditional phenotypic and genotypic characterization methods must be used with care and that further studies are required to improve the identification of Listeria isolates.
This study used phenotypic and genotypic methods to characterize atypical L. monocytogenes and L. innocua isolates obtained from swine slaughterhouses and meat markets in Sao Paulo State, Brazil.
2. Material and Methods
2.1. Bacterial Strains and Culture Conditions
Forty Listeria sp. isolates were studied. Of these, 25 were isolated from pork, slaughterhouses, and markets (15 isolates of L. monocytogenes and 10 of L. innocua), 11 isolates of L. monocytogenes were obtained from human infections, and four were control strains (L. monocytogenes ATCC 19115 and ATCC 19111 and L. innocua ATCC 33090 and CLIP 12612) (Table 1). The environmental and pork isolates were isolated as described by Moreno et al. [13]; the clinical strains and Listeria controls were obtained from the Public Health Laboratory (School of Public Health, University of Sao Paulo) and Laboratory of Swine Health (School of Veterinary Medicine and Animal Science, University of Sao Paulo) collections. The environmental and pork isolates were obtained from different swab samples taken from the slaughterhouses environment and carcasses from Sao Paulo State; the clinical isolates were obtained from the blood, placenta, and cerebrospinal fluid samples of different patients from different Brazilian states (Tables 1 and 2).
Table 1.
Sources and phenotypic and genotypic characteristics of the Listeria monocytogenes isolates used in this study.
| Isolate | Species | Serotype | Origin | Site | Year | ALOA | Hemolysis | inlA | inlB | inlC | inlJ* | plcA | plcB | prfA | hly |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lm1 | L. monocytogenes | 1/2a | Slaught 1 | Floor | 2008 | Halo | Positive | + | + | + | + | + | + | + | + |
| Lm2 | L. monocytogenes | 1/2b | Slaught 1 | Floor | 2008 | Halo | Positive | + | + | + | + | + | + | + | + |
| Lm3 | L. monocytogenes | 4b | Market 1 | Floor | 2008 | Halo | Positive | + | + | + | + | + | + | + | + |
| Lm21 | L. monocytogenes | 1/2a | Slaught 1 | Floor | 2008 | Halo | Positive | + | + | + | + | + | + | + | + |
| Lm22 | L. monocytogenes | 1/2a | Slaught 1 | Floor | 2008 | Halo | Positive | + | + | + | + | + | + | + | + |
| Lm23 | L. monocytogenes | 1/2a | Slaught 1 | Floor | 2008 | Halo | Positive | + | + | + | + | + | + | + | + |
| Lm25 | L. monocytogenes | 1/2a | Slaught 1 | Floor | 2008 | Halo | Positive | + | + | + | + | + | + | + | + |
| Lm26 | L. monocytogenes | 1/2a | Slaught 1 | Floor | 2008 | Halo | Positive | + | + | + | + | + | + | + | + |
| Lm27 | L. monocytogenes | 1/2a | Slaught 1 | Floor | 2008 | Halo | Positive | + | + | + | + | + | + | + | + |
| Lm28 | L. monocytogenes | 1/2a | Market 2 | Pork | 2008 | Halo | Weak positive | + | + | + | + | + | + | + | + |
| Lm29 | L. monocytogenes | 1/2a | Market 2 | Pork | 2008 | Halo | Weak positive | + | + | + | + | + | + | + | + |
| Lm30 | L. monocytogenes | 1/2a | Market 2 | Pork | 2008 | Halo | Weak positive | + | + | + | + | + | + | + | + |
| Lm31 | L. monocytogenes | 1/2a | Market 2 | Pork | 2008 | Halo | Weak positive | + | + | + | + | + | + | + | + |
| Lm34 | L. monocytogenes | 1/2a | Human | Blood | 1989 | Halo | Strong positive | + | + | + | + | + | + | + | + |
| Lm35 | L. monocytogenes | 4b | Human | Blood | 2004 | Halo | Strong positive | + | + | + | + | + | + | + | + |
| Lm36 | L. monocytogenes | 4b | Human | Blood | 1977 | Halo | Strong positive | + | + | + | + | + | + | + | + |
| Lm37 | L. monocytogenes | 4b | Human | CSF | 1982 | Halo | Strong positive | + | + | + | + | + | + | + | + |
| Lm38 | L. monocytogenes | 1/2b | Human | CSF | 1983 | Halo | Strong positive | + | + | + | + | + | + | + | + |
| Lm39 | L. monocytogenes | 1/2a | Human | Placenta | 1978 | Halo | Strong positive | + | + | + | + | + | + | + | + |
| Lm39a | L. monocytogenes | 1/2a | Human | Placenta | 1978 | Halo | Positive | + | + | + | + | + | + | + | + |
| Lm40 | L. monocytogenes | 1/2a | Human | Blood | 1985 | Halo | Strong positive | + | + | + | + | + | + | + | + |
| Lm41 | L. monocytogenes | 4b | Human | CSF | 1997 | Halo | Strong positive | + | + | + | + | + | + | + | + |
| Lm42 | L. monocytogenes | 4b | Human | CSF | 1997 | Halo | Positive | + | + | + | + | + | + | + | + |
| Lm43 | L. monocytogenes | 1/2a | Human | CSF | 1983 | Halo | Positive | + | + | + | + | + | + | + | + |
| Lm4 | L. monocytogenes | 1/2a | Market 2 | Floor | 2008 | Halo | Weak positive | + | + | + | + | + | + | + | + |
| Lm33 | L. monocytogenes | 1/2a | Market 2 | Floor | 2008 | Halo | Weak positive | + | + | + | + | + | + | + | + |
| Lm10 | L. monocytogenes | 4b | ATCC 19115 | — | — | Halo | Strong positive | + | + | + | + | + | + | + | + |
| Lm15 | L. monocytogenes | 1/2a | ATCC 19111 | — | — | Halo | Strong positive | + | + | + | + | + | + | + | + |
Slaught 1: slaughterhouse 1. CSF: cerebrospinal fluid. *All isolates were positive for fragments of inlJ but presented variable results for hole gene amplification (see Table 4).
Table 2.
Sources and phenotypic and genotypic characteristics of the Listeria innocua isolates used in this study.
| Isolate | Species | Serotype | Origin | Site | Year | ALOA | Hemolysis | inlA x | inlB | inlC | inlJ | plcA | plcB | prfA | hly x |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lin5 | L. innocua | 6a | Market 1 | Floor | 2008 | Halo* | Weak positive** | − | − | + | − | + | − | − | + |
| Lin6 | L. innocua | 6a | Slaught 2 | Floor | 2008 | Halo* | Weak positive** | − | − | + | − | + | − | − | + |
| Lin7 | L. innocua | 6a | Slaught 2 | Floor | 2008 | Halo* | Weak positive** | − | − | + | − | + | − | − | + |
| Lin8 | L. innocua | 6a | Slaught 2 | Floor | 2008 | Halo* | Weak positive** | − | − | + | − | + | − | − | + |
| Lin9 | L. innocua | 6a | Slaught 2 | Floor | 2008 | Halo* | Weak positive** | − | − | + | − | + | − | − | + |
| Lin16 | L. innocua | 6a | Slaught 1 | Floor | 2006 | Negative | Negative | − | − | − | − | − | − | − | − |
| Lin17 | L. innocua | 6a | Slaught 1 | Floor | 2006 | Negative | Negative | − | − | − | − | − | − | − | − |
| Lin18 | L. innocua | 6a | Slaught 1 | Floor | 2006 | Negative | Negative | − | − | − | − | − | − | − | − |
| Lin19 | L. innocua | 6a | Slaught 1 | Floor | 2006 | Negative | Negative | − | − | − | − | − | − | − | − |
| Lin20 | L. innocua | 6a | Slaught 1 | Floor | 2006 | Negative | Negative | − | − | − | − | − | − | − | − |
| Lin11 | L. innocua | 6a | ATCC 33090 | — | — | Negative | Negative | − | − | − | − | − | − | − | − |
| Lin46 | L. innocua | 6a | CLIP 12612 | — | — | Negative | Negative | − | − | − | − | − | − | − | − |
Slaught 1: slaughterhouse 1; Slaught 2: slaughterhouse 2. *Subtle halo. ** Very weak positive hemolysis. xAtypical isolates were positive for fragments of inlAB operon and hly but presented variable results for inlA and hly complete amplification (see Table 4).
The isolates were maintained in a stock medium containing glycerol at −80°C. The isolates were reactivated in brain-heart infusion (BHI) medium (Difco, Sparks, MD, USA) and plated on tryptone soy agar supplemented with yeast (TSAYE) (Oxoid, Lenexa, USA) to isolate pure colonies before use.
2.2. Conventional Listeria Identification Tests
The isolates were serotyped using polyclonal antisera produced against Listeria somatic and flagellar antigens in rabbits, according to the method described by Seeliger and Höhne [14]. The isolates were also characterized by catalase, motility, and biochemical tests including acid production from D-xylose, D-mannitol, L-rhamnose, and α-methyl-D-mannoside. Cultivation in selective agar Listeria according to Ottaviani and Agosti (ALOA) (Biolife, Milan, Italy) was used to identify L. monocytogenes isolates, and β-hemolysis was assessed by sting inoculation on 5% sheep blood agar.
2.3. Detection of L. monocytogenes-Specific Genes
Genomic DNA extraction was performed as described by Ausubel et al. [15]. All isolates were screened for the inlA, inlB, inlC, inlJ, hly, prfA, plcA, and plcB genes. The primers described by Johnson et al. [6], Liu et al. [16], and Jung et al. [17] were used for detection of prfA, inlC and inlJ, and inlA, respectively. Specific primers were designed for the complete amplification of the virulence genes (Table 3). The PCRs were performed using an Eppendorf Mastercycler gradient thermal cycler. Each reaction (25 μL) contained 5 μL of genomic DNA, MilliQ water, 10X PCR buffer, 1.5 mM MgCl2, 200 μM of dNTPs (Fermentas, Burlington, Canada), 200 μM of each primer, and 1.25 U of Taq DNA polymerase (Promega). The PCR programs were as follows: 30 cycles of denaturation at 94°C for 1 min, annealing at primer-specific temperature for 1–1.5 min, elongation at 72°C for 1 min per 1 Kb, and final extension at 72°C for 10 min. The amplified products were separated by electrophoresis on 1.5% agarose gels and stained with ethidium bromide (1 μg/mL). The molecular weights of the products were determined using the 1 Kb Plus DNA Ladder (Fermentas, Burlington, Canada).
Table 3.
Primers designed in this study for the amplification of the L. monocytogenes virulence genes.
| Primer | Sequence 5′-3′ | Target | Product (bp) |
|---|---|---|---|
| inlA ext Fw | CGGCTCCGTAGACAGATTAG | inlA | 2884 |
| inlA ext Rv | GTGATAGTCTCCGCTTGTAC | ||
| inlA In 1 -Fw | GTGAGAAGAAAACGA | 1200 | |
| inlA Detec-Rv | TGGTGTAAGATCGCT | ||
| inlA Detec-Fw | AAGTGATATAACTCC | — | |
|
| |||
| inlB ext Fw | GCTAGATGTGGTTTTCGGACT | inlB | 2146 |
| inlB ext Rv | TAAGCAGCGCAAAGGTGATTCCTAC | ||
| inlB In-Fw | GTGAAAGAAAAGCAC | 1227 | |
| inlB Seq 3 -Rv | ATTCCCGCGAATATA | ||
| inlB Seq 2 -Fw | TGATGGAACGGTAAT | 900 | |
| inlB End 3 -Rv | TNATTTCTGTGCCCT | ||
|
| |||
| plcB ext Fw | CCATACGACGTTAATTCTTGCAATG | plcB | 1039 |
| plcB ext Rv | TATCCACCCGCTAACGAGTG | ||
|
| |||
| plcA ext Fw | GAGGTTGCTCGGAGATATAC | plcA | 1100 |
| plcA ext Rv | CTGCTGTCCCTTTATCGTCG | ||
| plcA Detec-Fw | AACCATTATTATGCG | 396 | |
| plcA Detec-Rv | TGCAGCATACTGACG | ||
|
| |||
| hly ext Fw | CGATAAAGGGACAGCAGGACT | hly | 1796 |
| hly ext Rv | AGCCTGTTTCTACATTCTTCACAA | ||
| hly Detec-Fw | TAACAACGCAGTAAA | 566 | |
| hly Detec-Rv | CGTAAGTCTCCGAGG | ||
| hly End-Fw | CCTCCTGCATATATC | 725 | |
| hly End-Rv | TTATTCGATTGGATT | ||
|
| |||
| inlC In 1 -Fw | ATGCTAGTNTTAATTGTA | inlC | 852 |
| inlC End 2 -Rv | CTAATTCTTGATAGGTTGTG | ||
|
| |||
| prfA Detec-Fw | CTGCTAACAGCTGAGCTATG | prfA | 404 |
| prfA Detec-Rv | GCTACCGCATACGTTATC | ||
| prfA End Rv | ATGAACGCTCAAGCA | — | |
In: primers corresponding to the beginning of the gene; End: primers corresponding to the end of the gene; Detec: internal primers designed for gene detection; ext: external primers; Seq: internal primers designed for sequencing.
2.4. DNA Sequencing
The amplified fragments were purified using the Illustra GFX PCR DNA and Gel Band Purification kit (GE Healthcare) according to the manufacturer's protocol and sequenced directly at Genomic (Genomic Engenharia Molecular, Sao Paulo, Brazil). DNA sequencing was performed on an Applied Biosystems 3130xl DNA analyzer using the BigDye Terminator v3.1 cycle sequencing kit.
2.5. Detection of Mutations in L. monocytogenes Virulence Genes
Sequence analysis was performed using the BIOEDIT Sequence Alignment Editor 7.0.9 [18]. The obtained sequences of the virulence genes were compared to previously published L. monocytogenes sequence accessions from GenBank (NCBI, Bethesda, USA). The sequencing products were edited and compared with the sequences available in the GenBank database by manual alignment and using the ClustalW application. The nucleotide sequences obtained were translated into their corresponding amino acid sequences by the Nucleotide Translate application. Subsequently, the amino acid sequences were analyzed to identify changes in the compositions of their respective proteins, which might modify or eliminate protein functions.
2.6. Identification of Protein Domains and Prediction of Secondary Structures
The domains of InlC, InlB, InlA, PI-PLC, PC-PLC, and Hly from reference strain L. monocytogenes EGD-e were determined using the PROSITE database [19] of the ExPASy server (SIB, Swiss Institute of Bioinformatics). The locations of these domains were compared to the mutations identified in the studied isolates.
2.7. Species-Level Identification by 16S rRNA Amplification and Phylogenetic Analysis
Species identity was confirmed using 16S rRNA analysis. The primers and amplification protocol described by Thompson et al. [20] were used to amplify complete 16S rRNA genes. The fragments were sequenced and phylogenetic analysis was performed using the Mega 5.10 software [21]. The dendrogram was constructed using the maximum-likelihood method with the Tamura-3-parameter model.
2.8. Nucleotide Sequence Accession Numbers
All DNA sequences from this study were deposited in GenBank under the accession numbers KC618415-KC618420, KC666995-KC667019, KC808518-KC808549, and KC808567-KC808583.
3. Results
3.1. Conventional Listeria Identification Tests
The phenotypic characterization of Listeria sp. isolates is shown in Tables 1 and 2. Five atypical L. innocua isolates (Lin5–9) and six low-hemolytic L. monocytogenes (Lm4, Lm33, and Lm28–31) isolates were observed. The atypical L. innocua isolates exhibited phenotypic characteristics similar to L. monocytogenes with weak hemolysis and subtle halo in ALOA cultivation. These isolates could be distinguished only by serotyping, which revealed that the atypical isolates were L. innocua serotype 6a.
3.2. Detection and Analysis of L. monocytogenes Virulence Genes
The detection and complete amplification of the inlB, inlC, plcA, plcB, hly, and prfA genes were performed using previously published primers and primers designed in this study. The inlA and inlJ genes were only partially amplified using the primers inlA In-Fw/inlA Detec-Rv, designed in this study,and inlJ-Fw/inlJ-Rv, which were described by Liu et al. [16]. All L. monocytogenes isolates including the six low-hemolytic isolates (Lm4, Lm33, and Lm28–31) contained the studied genes. The five atypical L. innocua isolates (Lin5–9) contained inlC and plcA and fragments of the hly gene (Table 4).
Table 4.
Distribution of the results of the virulence gene amplification from Listeria species.
| Primer | Species | Positive | Negative |
|---|---|---|---|
| N (%) | N (%) | ||
| inlC_Liu 1 | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 0 | 12 (100.0) | |
| inlC In–End | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 5 (41.7) | 7 (58.3) | |
| prfA Johnson–End | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 0 | 12 (100.0) | |
| prfA_Johnson2 | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 0 | 12 (100.0) | |
| prfA Detec | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 0 | 12 (100.0) | |
| plcA ext | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 5 (41.7) | 7 (58.3) | |
| plcA Detec | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 5 (41.7) | 7 (58.3) | |
| plcB ext | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 0 | 12 (100.0) | |
| inlB In– Seq 3 | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 0 | 12 (100.0) | |
| inlB Seq 2 | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 0 | 12 (100.0) | |
| inlA In–Detec | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 0 | 12 (100.0) | |
| inlAB_Jung 3 | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 5 (41.7) | 7 (58.3) | |
| hly ext | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 0 | 12 (100.0) | |
| hly End | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 5 (41.7) | 7 (58.3) | |
| hly_Border 4 | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 5 (41.7) | 7 (58.3) | |
| hly Detec | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 5 (41.7) | 7 (58.3) | |
| inlJ_Liu 1 | L. monocytogenes | 28 (100.0) | 0 |
| L. innocua | 0 | 12 (100.0) | |
| inlJ ext | L. monocytogenes | 23 (82.1) | 5 (17.9) |
| L. innocua | 0 | 12 (100.0) |
Nucleotide substitutions were detected in inlC, inlB, inlA, plcA, plcB, and prfA, only in the six low-hemolytic L. monocytogenes isolates (Lm4, Lm33, and Lm28–31). Seven substitutions were detected in the inlC gene; however, only the transition of adenine to cytosine and the inversion of thiamine to adenine at codon 10 led to the mutation Ile10His, and the transition of thiamine to cytosine at codon 12 resulted in the mutation Met12Thr. Ten substitutions were detected in plcA, leading to the mutations Ile17Val and Phe119Tyr in the PI-PLC. In the plcB sequence, only two transitions of thiamine to cytosine were identified at codon 13, which resulted in the mutation Ile13Thr. Seven substitutions were detected in inlB; however, only the transitions of adenine to guanine at codons 117 and 132 resulted in the mutations Ala117Thr and Val132Ile (Figures 1 and 2).
Figure 1.
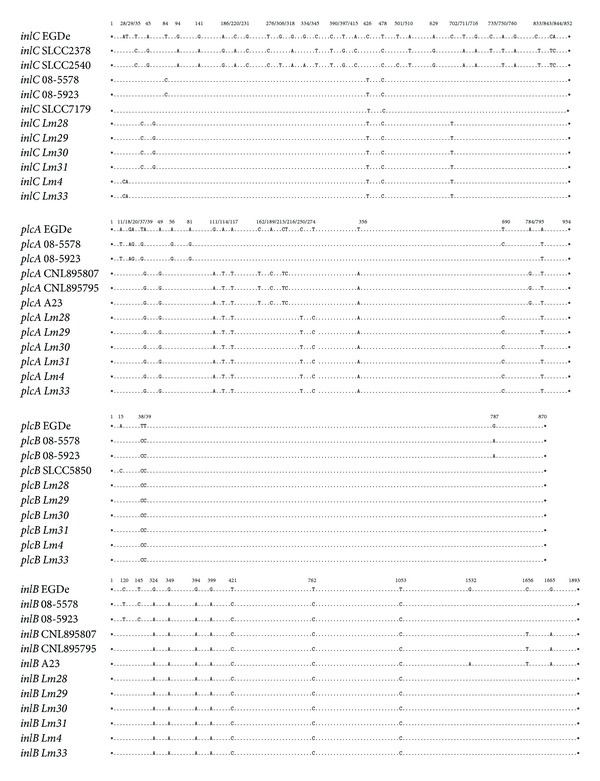
Nucleotide substitutions detected in the inlC, plcA, plcB, and inlB genes. The Lm28–31, Lm4, and Lm33 isolates were aligned with L. monocytogenes EGDe and the previously described mutant strains. Asterisks indicate the start and stop codons, dots represent identical nucleotides, and numbers indicate the positions of the substitutions.
Figure 2.
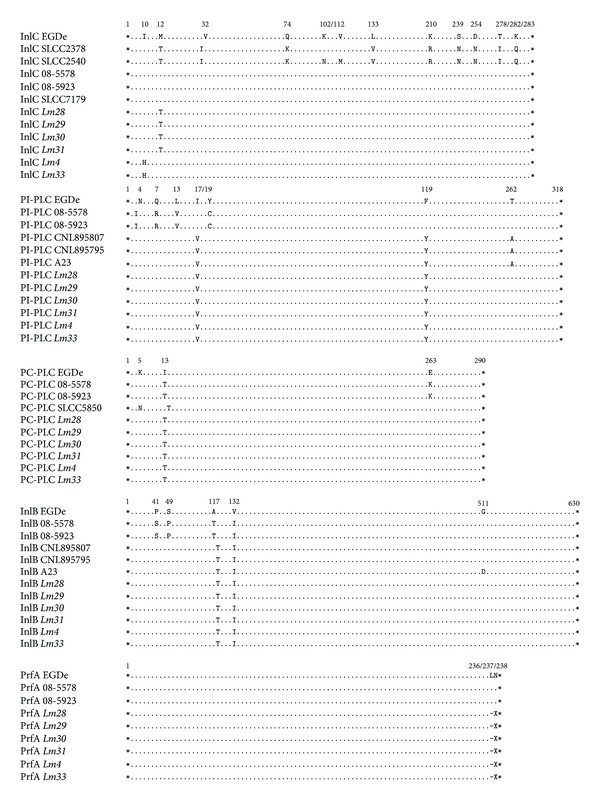
Amino acids substitutions in the InlC, PI-PLC, PC-PLC, InlB, and PrfA proteins. The Lm28–31, Lm4, and Lm33 isolates were aligned with L. monocytogenes EGDe and the previously described mutant strains. Asterisks indicate the start and stop codons, dots represent identical amino acids, and numbers indicate the positions of the substitutions.
A deletion of five nucleotides was also detected in the prfA sequence, leading to the deletion of codons 236 and 237 in the Lm4, Lm33, and Lm28–31 isolates. Eight substitutions were detected in the inlA fragments of the low-hemolytic L. monocytogenes isolates, resulting in the mutations Thr51Ala and Ile157Leu (Figure 3). The Lm4, Lm33, and Lm28–31 isolates also contained 15 substitutions in the hly sequence, whereas the Lin5 and Lin6–9 isolates only contained 14 and 13 of these substitutions, respectively. However, all these atypical isolates contained only the mutations Val438Ile and Lys523Ser (Figure 3).
Figure 3.
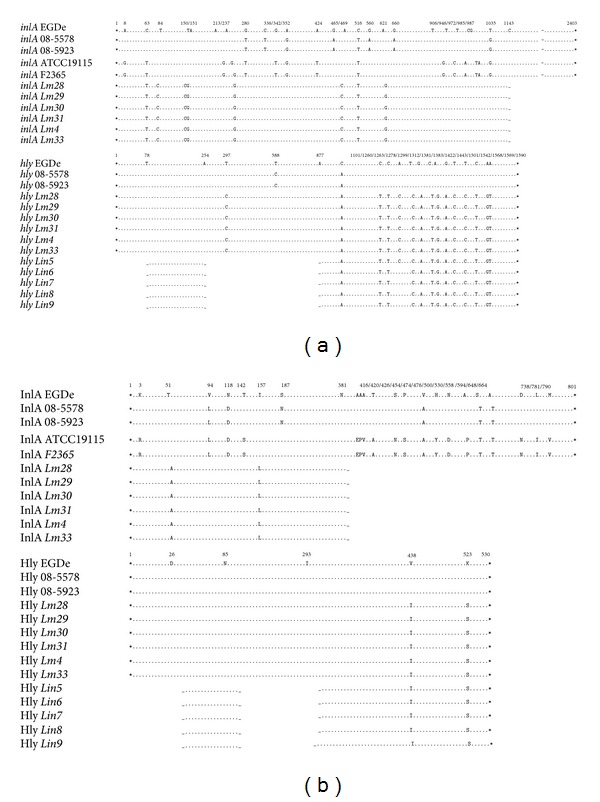
Nucleotide substitutions detected in inlA and hly (a) and mutations identified in InlA and Hly (b). The Lm28–31, Lm4, Lm33, and Lin5–9 isolates were aligned with L. monocytogenes EGDe and previously described mutant strains. Asterisks indicate the start and stop codons, dots represent identical amino acids, and numbers indicate the positions of the substitutions. Gaps represent the regions that were not amplified.
3.3. Identification of Protein Domains
Of the identified mutations, only Ala117Thr and Val132Ile in InlB and Ile157Leu in InlA were located in the leucine-rich repeat (LRR) domains that are characteristic of these proteins. The Phe119Tyr mutation in PI-PLC was also located in the PI-PLC X-box domain. The thiol-activated cytolysin signature motifs in Hly and the zinc-dependent phospholipase C domain in PC-PLC presented distinct locations of the mutations identified in the respective proteins.
3.4. Species Confirmation by 16S rRNA Phylogenetic Analysis
From the amplification and analysis of the 16S rRNA genes, a dendrogram was constructed, which allowed the distinction of L. monocytogenes and L. innocua species. The dendrogram contained three major groups; the first group consisted of L. grayi and L. murrayi, the second group contained L. rocourtiae, and the third group consisted of clusters of L. monocytogenes and L. marthii, L. innocua, L. welshimeri, L. seeligeri, and L. ivanovii (Figure 4). The isolates Lin5 – 9 and Lin11 were grouped with the standard strains of L. innocua, whereas the isolates Lm28–31, Lm4, and Lm33 were grouped together with the standard strains of L. monocytogenes.
Figure 4.
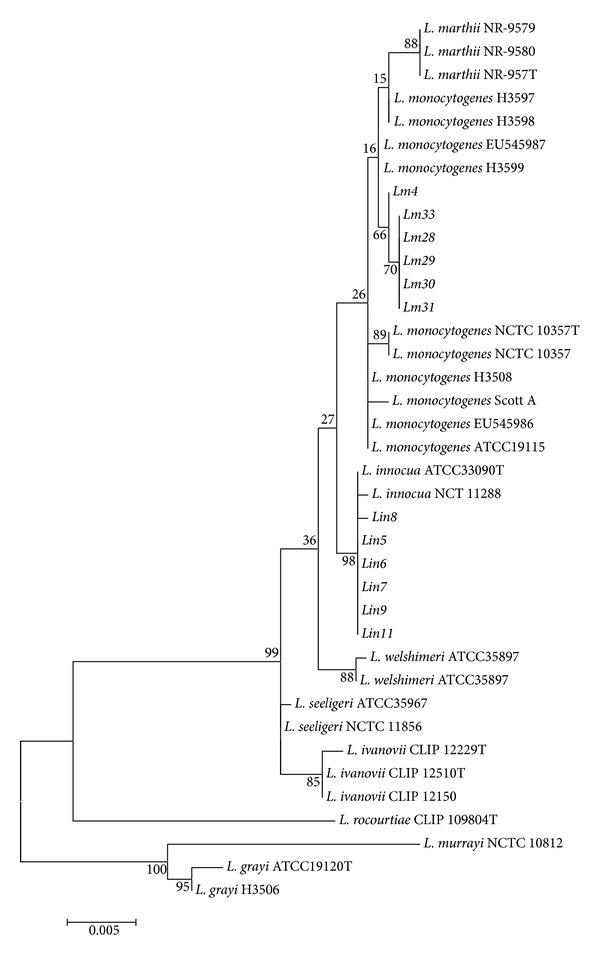
Dendrogram showing the evolutionary relationships among the Listeria isolates based on the 16S rRNA nucleotide sequences. The dendrogram was constructed using the maximum-likelihood method (Tamura-3-parameter model) with the MEGA 5.10 software. The bootstrap values presented at corresponding branches were evaluated using 500 replicates.
4. Discussion
Studies on Listeria virulence mechanisms have become important in recent decades because this microorganism is used as a model of intracellular infection. L. monocytogenes virulence factors have been described, and their mechanisms of action and respective genes have been studied using distinct molecular techniques and in vivo and in vitro experiments. In addition to the use of Listeria as a model organism, there is great interest in studying this organism because of the increasing incidence of listeriosis in the United States of America (USA) and Europe [23, 24].
Our results using conventional Listeria identification tests are consistent with the subjectivity and ambiguity of phenotypic tests that have been discussed in the last decade [6, 7]. Although these conventional methods are still utilized, biochemical and phenotypic tests yield variable results during the identification of Listeria species and serotypes, and the emergence of atypical isolates has further increased the uncertainty of the application of these tests. From a public health perspective, a drastic measure could be adopted to classify all isolates with doubtful hemolytic status as L. monocytogenes or as isolates with pathogenic risk without major efforts to identify the species and serovars. However, for better epidemiological, microbiological, and evolutionary understanding, it is important to identify and characterize the phenotypes and molecular features of these atypical isolates.
This study aimed to detect the hly, plcA, plcB, prfA, inlA, inlB, inlC, and inlJ genes in L. monocytogenes and L. innocua isolates. These genes are characteristic of L. monocytogenes and are essential for intracellular infection. The presence of these genes in isolates from meat and the environment suggests the pathogenic potential of these isolates and a risk to human health. We detected these virulence genes in all L. monocytogenes isolates including the six low-hemolytic isolates (Lm4, Lm33, and Lm28–31); additionally, the five atypical L. innocua isolates (Lin5–9) contained the inlC, hly, and plcA genes.
Our results are consistent with the data of Johnson et al. [6] and Volokhov et al. [7], who identified some L. monocytogenes virulence genes in L. innocua strains with atypical hemolysis. Therefore, the use of traditional PCR methods based mostly on the detection of hly and plcA for the distinction of Listeria pathogenic species should be reconsidered because these methods do not enable the distinction of atypical isolates. Accurate identification of Listeria species was possible only by the complete sequencing and phylogenetic analysis of the 16S rRNA gene (Figure 4). We propose that the detection of prfA, plcB, and inlB might be a better and reliable alternative to enable the rapid distinction of L. monocytogenes and L. innocua. We also suggest that analysis of the complete 16S rRNA gene sequences is important for the accurate identification of Listeria species.
The inlC and plcA genes from the atypical L. innocua isolates did not contain nucleotide substitutions and mutations in their respective proteins. The only mutations identified in these isolates were the Val438Ile and Lys523Ser in Hly. The hly gene could not be completely amplified, but this might be due to insertions or deletions between the detected fragments. However, the hemolytic phenotypes of these atypical isolates suggest that despite the difficulty in amplifying this locus there were no gross alterations in Hly function. Further studies will be carried out to confirm and quantify hly expression.
Because the atypical L. innocua isolates presented the low-hemolytic phenotype and halo in ALOA cultivation, we concluded that these isolates produce at least Hly and PI-PLC. Since the only detected mutations were not located in the thiol-activated cytolysin signature motifs in Hly, the low expression of the hly and plcA genes might be due to altered promoter activity. As the prfA gene was also not detected in these isolates, we suggest that a secondary promoter might activate the expression of hly and plcA and originate the observed phenotype. However, further in vitro and proteomic studies are necessary to verify the activity and integrity of these virulence factors.
The mutations detected in InlB and PI-PLC in the low-hemolytic L. monocytogenes isolates (Lm4, Lm33, and Lm28–31) are consistent with results from previous studies on low-virulent L. monocytogenes field strains [10–12]. The mutations Ala117Thr and Val132Ile in InlB are located in the LRR domains of this protein, which are directly related to the interaction of this internalin with the Met cellular receptor and might compromise the adhesin function of InlB [11, 12]. The Ile17Val and Phe119Tyr mutations in PI-PLC are located in the signal sequence and the X-box domain, respectively, whereas the Thr262Ala mutation causes the introduction of an amino acid with different physicochemical properties, which might inhibit PI-PLC activity [12].
The mutations identified in PC-PLC, InlC, InlA, PrfA, and Hly are novel. The Ile13Thr mutation in PC-PLC is not located at the zinc-dependent phospholipase C domain of this protein, and the Ile10His and Met12Thr mutations in InlC are not located in the LRR domains of this internalin. The Thr51Ala and Ile157Leu mutations in InlA are also novel, and although they do not cause the truncation of InlA [11, 12], they are located in the LRR domains; therefore, these mutations might compromise the internalization of L. monocytogenes in epithelial cells. Further expression studies are required to confirm whether these mutations affect the expression and function of these virulence factors.
The low-hemolytic L. monocytogenes isolates contained the same Hly mutations as the atypical L. innocua; theconsequence of this observation is unclear. The deletion in prfA in the low-hemolytic L. monocytogenes isolates might underlie the reduced hemolytic activity in these strains because PrfA is the activator of the LIPI-1 cluster. However, the impairment of prfA would result in the reduced expression of all LIPI-1 genes. Therefore, further transcriptomic studies are required to completely characterize these atypical isolates, enhance our knowledge of their evolution and impact on public health, and develop more efficient methods for the identification and distinction of Listeria species.
Acknowledgments
This study was sponsored by FAPESP-São Paulo Research Foundation (Process nos. 06/55501-0; 10/19005-4; 10/13511-5) and CAPES-Coordenação de Aperfeiçoamento de Pessoal de NívelSuperior.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Farber JM, Peterkin PI. Listeria monocytogenes, a food-borne pathogen. Microbiological Reviews. 1991;55(3):476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mead PS, Slutsker L, Dietz V, et al. Food-related illness and death in the United States. Emerging Infectious Diseases. 1999;5(5):607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rocourt J, Schrettenbrunner A, Seeliger HPR. Biochemical differenciation of the Listeria monocytogenes (sensu lato) Annales de l’Institut Pasteur. Microbiologie. 1983;134(1):65–71. [PubMed] [Google Scholar]
- 4.Lemes-Marques EG, Yano T. Influence of environmental conditions on the expression of virulence factors by Listeria monocytogenes and their use in species identification. FEMS Microbiology Letters. 2004;239(1):63–70. doi: 10.1016/j.femsle.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 5.Gouin E, Mengaud J, Cossart P. The virulence gene cluster of Listeria monocytogenes is also present in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, a nonpathogenic species. Infection and Immunity. 1994;62(8):3550–3553. doi: 10.1128/iai.62.8.3550-3553.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson J, Jinneman K, Stelma G, et al. Natural atypical Listeria innocua strains with Listeria monocytogenes pathogenicity island 1 genes. Applied and Environmental Microbiology. 2004;70(7):4256–4266. doi: 10.1128/AEM.70.7.4256-4266.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volokhov DV, Duperrier S, Neverov AA, George J, Buchrieser C, Hitchins AD. The presence of the internalin gene in natural atypically hemolytic Listeria innocua strains suggests descent from L. monocytogenes . Applied and Environmental Microbiology. 2007;73(6):1928–1939. doi: 10.1128/AEM.01796-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cossart P, Lecuit M. Interactions of Listeria monocytogenes with mammalian cells during entry and actin-based movement: bacterial factors, cellular ligands and signaling. EMBO Journal. 1998;17(14):3797–3806. doi: 10.1093/emboj/17.14.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perrin M, Bemer M, Delamare C. Fatal case of Listeria innocua bacteremia. Journal of Clinical Microbiology. 2003;41(11):5308–5309. doi: 10.1128/JCM.41.11.5308-5309.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roche SM, Gracieux P, Milohanic E, et al. Investigation of specific substitutions in virulence genes characterizing phenotypic groups of low-virulence field strains of Listeria monocytogenes . Applied and Environmental Microbiology. 2005;71(10):6039–6048. doi: 10.1128/AEM.71.10.6039-6048.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roche SM, Grépinet O, Corde Y, et al. A Listeria monocytogenes strain is still virulent despite nonfunctional major virulence genes. Journal of Infectious Diseases. 2009;200(12):1944–1948. doi: 10.1086/648402. [DOI] [PubMed] [Google Scholar]
- 12.Témoin S, Roche SM, Grépinet O, Fardini Y, Velge P. Multiple point mutations in virulence genes explain the low virulence of Listeria monocytogenes field strains. Microbiology. 2008;154(3):939–948. doi: 10.1099/mic.0.2007/011106-0. [DOI] [PubMed] [Google Scholar]
- 13.Moreno LZ, Paixão R, Gobbi DD, et al. Characterization of atypical Listeria innocua isolated from swine slaughterhouses and meat markets. Research in Microbiology. 2012;163(4):268–271. doi: 10.1016/j.resmic.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Seeliger HPR, Höhne K. Serotyping of Listeria monocytogenes and related species. Methods in Microbiology. 1979;13:31–49. [Google Scholar]
- 15.Ausubel FM, Brent R, Kingston RE. Short Protocols in Molecular Biology. New York, NY, USA: John Wiley & Sons; 1995. [Google Scholar]
- 16.Liu D, Lawrence ML, Ainsworth AJ, Austin FW. Toward an improved laboratory definition of Listeria monocytogenes virulence. International Journal of Food Microbiology. 2007;118(2):101–115. doi: 10.1016/j.ijfoodmicro.2007.07.045. [DOI] [PubMed] [Google Scholar]
- 17.Jung YS, Frank JF, Brackett RE, Chen J. Polymerase chain reaction detection of Listeria monocytogenes on frankfurters using oligonucleotide primers targeting the genes encoding internalin AB. Journal of Food Protection. 2003;66(2):237–241. doi: 10.4315/0362-028x-66.2.237. [DOI] [PubMed] [Google Scholar]
- 18.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 19.Sigrist CJA, Cerutti L, de Castro E, et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Research. 2009;38(1):D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson FL, Hoste B, Vandemeulebroecke K, Swings J. Genomic diversity amongst Vibrio isolates from different sources determined by fluorescent amplified fragment length polymorphism. Systematic and Applied Microbiology. 2001;24(4):520–538. doi: 10.1078/0723-2020-00067. [DOI] [PubMed] [Google Scholar]
- 21.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Border PM, Howard JJ, Plastow GS, Siggens KW. Detection of Listeria species and Listeria monocytogenes using polymerase chain reaction. Letters in Applied Microbiology. 1990;11(3):158–162. doi: 10.1111/j.1472-765x.1990.tb00149.x. [DOI] [PubMed] [Google Scholar]
- 23.Angulo FJ. Listeriosis in the United States. FSIS, USDA, 2009, http://www.fsis.usda.gov/PDF/Lm_Angulo_062309.pdf.
- 24.Allerberger F, Wagner M. Listeriosis: a resurgent foodborne infection. Clinical Microbiology and Infection. 2010;16(1):16–23. doi: 10.1111/j.1469-0691.2009.03109.x. [DOI] [PubMed] [Google Scholar]


