Abstract
Background
The management of genitourinary malignancies requires a multidisciplinary care team composed of urologists, medical oncologists and radiation oncologists. A genitourinary (GU) oncology clinical database is an invaluable resource for patient care and research. Although electronic medical records provide a single web-based record used for clinical care, billing and scheduling, information is typically stored in a discipline-specific manner and data extraction is often not applicable to a research setting. A GU oncology database may be used for the development of multidisciplinary treatment plans, analysis of disease-specific practice patterns, and identification of patients for research studies. Despite the potential utility, there are many important considerations that must be addressed when developing and implementing a discipline-specific database.
Methods and Materials
The creation of the GU oncology database including prostate, bladder and kidney cancers with the identification of necessary variables was facilitated by meetings of stakeholders in medical oncology, urology, and radiation oncology at the University of North Carolina (UNC) at Chapel Hill with a template data dictionary provided by the Department of Urologic Surgery at Vanderbilt University Medical Center. Utilizing Research Electronic Data Capture (REDCap, version 4.14.5), the UNC Genitourinary OncoLogy Database (UNC GOLD) was designed and implemented.
Results
The process of designing and implementing a discipline-specific clinical database requires many important considerations. The primary consideration is determining the relationship between the database and the Institutional Review Board (IRB) given the potential applications for both clinical and research uses. Several other necessary steps include ensuring information technology security and federal regulation compliance; determination of a core complete data set; creation of standard operating procedures; standardizing entry of free text fields; use of data exports, queries, and de-identification strategies; inclusion of individual investigators’ data; and strategies for prioritizing specific projects and data entry.
Conclusions
A discipline-specific database requires a buy-in from all stakeholders, meticulous development, and data entry resources in order to generate a unique platform for housing information that may be used for clinical care and research with IRB approval. The steps and issues identified in the development of UNC GOLD provide a process map for others interested in developing a GU oncology database.
Keywords: Urologic Oncology, Clinical Database, Genitourinary Oncology, REDCap, Oncology Database
INTRODUCTION
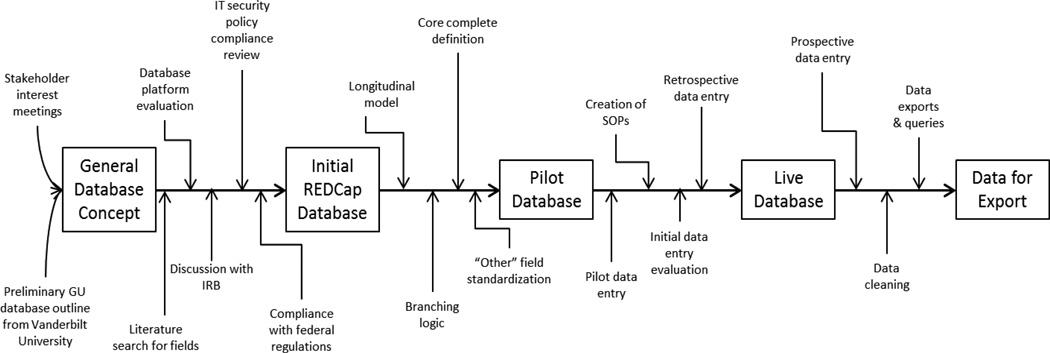
Clinical databases are an integral part of patient care and research, providing a clear format for reviewing both aggregate and individual patient information. To date, utilization of medical databases has included translational bioinformatics [1], tissue banks [2], quality control and monitoring [3], as well as others. Although databases are associated with many advantages, their development and implementation may be challenging. For this reason, we describe the successful initiation, design, and execution of the University of North Carolina Genitourinary OncoLogy Database (UNC GOLD), in order to provide others with a process map (Figure 1).
Figure 1.
Database development process map
The electronic medical record (EMR) provides a single web-based patient record, which has become an invaluable component of modern clinical care. Unfortunately, its potential use as a research tool is hampered by difficulty extracting data efficiently and in a format that is applicable to the research setting [4,5]. The capacity for medical records to be accessible over the internet allows for greater data portability [6], however, because they are so large, and their uses so diverse, they lack many options for discipline-specific customization. Furthermore, EMRs are designed to provide a patient-specific picture for clinical care, billing, and scheduling. While this is practical in a clinic setting, the narrative organizational structure does not allow querying of data across multiple patients, outputs information in only a limited number of formats, and lacks options for de-identification [7]. Furthermore, EMRs often heterogeneously capture treatment mapping, functional status, clinical stages, or other relevant factors that may be used during routine care, despite their patient-based approach. Finally, while access to clinic notes from the entire hospital system may provide interdisciplinary insights, there is often limited information from referring physicians for discipline-specific care, especially with respect to patients seen outside of the hospital system. Because of the complex nature of genitourinary malignancies, a multidisciplinary team of urologists, medical oncologists, and radiation oncologists share in the care of patients. This leads to patient information stored in a discipline-centered manner with limited ability to integrate and analyze the information originating from different providers.
A disease group-specific clinical database may help to compensate for the limitations of the electronic medical record as well as provide a more readily-accessible means of analyzing patient data [8]. The clinical database may be used to generate patient lists for tumor board meetings, analyze disease-specific practice patterns, and identify patients for clinical trials and other correlative or translational research studies. It may also help to determine the feasibility of conducting clinical trials in a specific patient population. Furthermore, the prospective completion of disease-specific data entry forms at predetermined time points can track disease progression, survival, and other outcomes while avoiding issues such as recall bias. Finally, the availability of these data to researchers within the disease group offers an invaluable resource for the conception and development of research projects with IRB approval.
DATABASE PLANNING AND IMPLEMENTATION
Planning and Input
Although there are clear advantages to clinical databases, the vast amount of information to be collected renders database development and upkeep a substantial undertaking. For this reason, cooperation from members of the disease group team is crucial [9]. A buy-in from these stakeholders in the form of providing input related to the important data points for each subspecialty increases the robustness of the database and promotes continued data quality over time. In the development of the current database, UNC GOLD members with leadership roles in this project (Steering Committee) conferred with other stakeholders including urologists, radiation oncologists, and medical oncologists at the University of North Carolina at Chapel Hill to gather input on what information to collect. Three disease-specific meetings for prostate, kidney and bladder cancer were held to determine the group and individual goals for the database and to review the necessary data points. This feedback was then integrated into a genitourinary oncology database backbone provided by colleagues at Vanderbilt University (D.A.B., C.Y.).
In addition to individual physician involvement, it was necessary to review the literature regarding clinical databases in general and those specifically designed for kidney, prostate, and bladder cancer. To that end, PubMed literature searches for “clinical database,” “REDCap,” “urology database,” and “oncology database” were performed. Articles specifically discussing databases (as opposed to those citing the database as a data source) were selected for evaluation. Citations within those articles were also reviewed. This research yielded recommendations from relevant organizations regarding the variables to include and specifications about how to record this information [10]. There was also a wealth of information discussing the merits of a variety of specific database programs in a clinical context [11,12].
These publications were beneficial in outlining some of the specific concerns that accompany clinical databases. Specifically, Smith et al. discuss the importance of developing a strong but flexible data dictionary and presenting a variety of design considerations to ease data entry [13]. Others have reported on the necessity of ensuring the quality and security of database data and methodologies for doing so [14]. Finally, discussions of information technology considerations [15], and other factors [16,17] in the literature enabled the UNC GOLD group to successfully integrate the developing database into existing infrastructure and navigate issues associated with web-based database platforms.
Database Features
There were several features that the UNC GOLD team identified as critical (Table 1). The most important feature was compliance with the university data security policy for patient data. Additionally, it was necessary that both the initial setup as well as the routine data entry were efficient, error-free, and customizable. Because many of the data points were defined by stakeholders, the database had to be flexible with respect to both the type and number of fields. Additionally, since UNC GOLD’s goal is to capture baseline data for every patient seen in the clinic, as well as periodic updates, it was critical to have a mechanism to reduce opportunities for data entry errors. Research Electronic Data Capture (REDCap) [17,18] is fast to navigate, allows for the use of branching logic to hide fields that are unused for a given patient (e.g. prostate cancer forms for female patients), and runs real-time validation to ensure that the type of information entered is correct within user-defined limits (e.g. verifies that all zip codes have 5 numbers and no letters). Within REDCap, another way to verify data consistency is through the use of “required” fields. When these fields are not completed, data entry personnel are given an error message and asked to provide a response so that a minimum dataset is available for all patients. Finally, one of REDCap’s greatest merits is the speed with which a database can be built; development does not require any programming knowledge or special training, but forms can be created in a matter of minutes. Thus, REDCap (current version, 4.14.5) was selected as the platform for UNC GOLD.
Table 1.
Critical Database Features
| Feature | Research Electronic Data Capture (REDCap) |
|---|---|
| Turn-around time | Database development time dependent on amount of information to collect and amount of branching logic necessary; single form creation and set up time <30 minutes in most cases |
| Ease of use | User-friendly interface for both design and data entry |
| Flexibility | Completely user-defined database, within calculation and logical limits of program |
| Speed | Fast; dependent on server and amount of branching logic on page |
| Availability | Supported by the NC TraCS Institute19 at UNC-Chapel Hill |
| Ease of customization | All fields and logic user-defined; specific special fields/calculations available; user interface standard |
| Data entry & data validation options | Branching logic hides fields that should be blank; specifications for data format/type; set ranges for date and numeric fields; double data entry options; definition of “required” fields |
| Linkage to other databases in health care system | Currently, no linkage is available |
| Querying | User-defined queries at both form- and field-level available; options to remove identifiers and shift date; real-time results |
| Data importing and exporting | Available for use with Excel, SAS, STATA, SPSS, and R statistical software packages |
| Security | Compliant with UNC data security and HIPAA requirements |
| User access rights | Can be defined for individuals or sites to prevent unauthorized viewing and editing of data |
| Additional features | Scheduling; option for multiple arms; option for longitudinal structure; file dropbox; integrated survey tool; data imports |
NC TraCS: North Carolina Translational & Clinical Sciences Institute
UNC: University of North Carolina
HIPAA: Health Insurance Portability and Accountability Act
Database Setup
Starting with the template in use at Vanderbilt University, the data fields from the stakeholders and recommendations from the literature were integrated. This preliminary version was then modified by the addition of extensive branching logic. Because the UNC GOLD database includes bladder, ureter, renal pelvis, kidney, and prostate cancers, branching logic allowed data entry personnel to select only the applicable primary cancer site, while hiding those remaining, thus avoiding any unwarranted data entry. This modification also provided for a more streamlined, universal demographics page and condensed large pages of possible fields to shorter forms that would automatically show only the relevant fields. With respect to flexibility, UNC GOLD was developed to account for cases in which a patient develops a second genitourinary primary malignancy, or a tumor believed to originate from one site is determined to have a different primary (e.g. a kidney tumor derived from an upper tract urothelial primary).
Development, Database Structure, and Modification
As the development progressed, it became evident that due to the desired data points such as progression of disease and survival outcomes, entry would need to occur at multiple time points. To capture this information, it was necessary to institute a longitudinal data collection model. REDCap has this capacity, thus the only change was to consider whether individual forms should be made available at all, or only some, time points. Although similar databases specify preoperative, operative, and postoperative dates, consideration of the possibility for treatment variability in these malignancies ultimately led UNC GOLD to simply number “visits” (dates when the EMR was abstracted). For example, because many patients do not ultimately undergo surgical intervention as primary therapy, surgically-defined forms may fail to collect information about other therapies. Additionally, for cases that require more than one surgery, there are also several preoperative and postoperative periods. Therefore, rather than disabling forms, information may be entered at any time point. This setup allows for the potential for multiple operative procedures. To further increase the capacity of the database, forms for questionnaires relevant to each cancer were also created.
To ensure the accuracy of UNC GOLD, the database coordinators and Steering Committee met regularly to discuss possible modifications. Because of the growing size of the project, and the large number of data entry spaces, the team also defined a core complete set of variables that would be generally applicable in the context of both clinical quality improvement, as well as potential research projects (Table 2). By defining this set, the number of individual data points to enter was reduced, but still captured the information deemed highest priority. Rather than eliminating the fields that were not on this list, database coordinators annotated the database by marking those fields that were core complete. Retaining the unmarked fields was done both to give the database the capacity to grow over time, as well as to provide the structure for smaller projects looking at those particular variables.
Table 2.
Selected UNC GOLD forms with variables
| Preoperative - Bladder Ureter and Renal Pelvis |
Operative – Kidney | Pathology - Prostate |
|---|---|---|
| When was bladder cancer initially diagnosed? Referring physician last name Referring physician first name Prior Pelvic Radiation? Prior Abdominal/Pelvic Surgery? Has patient had a TURBTa or biopsy? Date of TURBT/biopsy leading to current treatment Number of prior TURBTs or Biopsies Histology Specify benign histology Specify histology other Specify other benign histology TURBT biopsy grade TURBT Stage Clinical N Stage Clinical M Stage TURBT muscle present? TURBT multifocal? Lymphovascular invasion on pathology Did patient have a restaging TURBT (within 3 months)? Did pathology change? What is new pathology? High or low grade? Was muscle present? N Stage M Stage Lymphovascular invasion on pathology Prognostic Factors Risk Factors Intravesical chemotherapy use Intravesical chemotherapy type How many induction courses has patient had? Preop METSb Preoperative comments Date of data entry: Data entry person: |
Was surgery performed? Date of surgery Attending surgeon Laterality Procedure on right Procedure on left Surgical approach Was surgery retroperitoneal? ASAc score Crystalloid Colloids Packed red blood cells Fresh Frozen Plasma Platelets Estimated blood loss Was the surgery cytoreductive? Tumor thrombus present? What is the level of thrombus? Was cardiac bypass used? Was lymph node dissection performed? Was clamping used on the right? Was ischemia warm or cold? What was the right side clamp time? Was early unclamping performed? Was only the artery clamped? Was clamping used on the left? Was ischemia warm or cold? What was the left side clamp time? Was early unclamping performed? Was only the artery clamped? What was the total clamp time? Was fluorescence with ICGd used? What was the time of incision? What was the time of closure? Operative comments Were there any operative complications? (List of complications) Specify complication grade Specify other operative complications Data entry date: Data entry person: |
Was tumor tissue sent to pathology? Histology Specify other histology Percent tumor Prostate volume Calculated tumor volume Gleason primary postop Gleason secondary postop Gleason sum postop Gleason tertiary postop Margin status Margin location Specify other margin location Side of margin Aggregate length of margin Pathologic tumor stage Pathologic tumor stage Extracapsular extension Extracapsular extension location Extracapsular extension location, specify Seminal vesicle invasion Seminal vesicle invasion location Perineural invasion? Lymph nodes sampled Lymph tissue in specimen Sampled as a group? Total number positive Total number sampled Right lymph nodes positive? Left lymph nodes positive? Pathology comments Data entry date: Data entry person: |
TURBT: Transurethral resection of bladder tumor
MET: Metabolic equivalent of task
ASA score: American Society of Anesthesiologists physical status classification system
ICG: Indocyanine green
Standardization of User-Defined Fields
To account for individual differences while maintaining a database that could be queried for relevant variables, the UNC GOLD team instituted methodologies to standardize the entry of information into user-defined text boxes. By keeping a list of all “other” entries which may be copied and pasted as needed, data abstraction personnel verify that information is entered in a consistent manner. Copying and pasting this information directly out of the “other” entry list reduces the likelihood of typos while also avoiding multiple records referring to the same finding with different terminology, for example “kidney stones,” “renal calculi,” and “nephrolithiasis.” Standardizing in this manner limits the number of queries to a single term.
Database Testing
Once UNC GOLD had been created and refined, it was necessary to ensure that data defined by the stakeholders was available in the medical record, and able to be accurately recorded. To do this, the database was piloted using a small set of patients with each malignancy identified from existing Institutional Review Board (IRB) approved clinical databases. These records were each abstracted by UNC GOLD personnel and compared to the data contained within the existing databases. This process illustrated several technical glitches (since rectified) and allowed for analysis of abstraction methodologies. Furthermore, several disconnects regarding the availability of some of the data points were identified. In some cases, the hospital or clinic notes did not consistently report specific variables, including smoking status and the extent of lymph node dissection. Additionally, it was noted that many older records did not contain information about intraoperative fluids or surgery start and stop times. Because this information is readily available intra-operatively, a form to prospectively collect this information specifically for UNC GOLD was created. This form included all of the operative questions, and could be filled out manually by a surgeon. To further ease the integration of this surgical data and to reduce the amount of time spent on each chart abstraction, options for direct data entry were evaluated. One option included Teleform, (Cardiff Software, Hewlett Packard, Palo Alto, CA) a service that reads scanned forms and generates raw data that can then be imported into REDCap. Although this feature could directly read the paper forms, it raised several concerns regarding data quality and security. Another alternative that was discussed was the use of the integrated REDCap Survey Tool. This feature allowed specific fields from UNC GOLD to be compiled and emailed as a survey to the surgeon, rather than as a paper questionnaire. The provider could complete the form online for each patient treated, using the same data security as the database, and the data would automatically populate database fields as if a data entry person had abstracted the information. One limitation of this methodology was the restriction that each database could only be connected to a single survey; a problem for a database encompassing several malignancies with distinct surgical treatments.
Production Mode
Because database development allows for the creation and deletion of fields, it is possible to unintentionally delete or overwrite data. To mitigate that problem, UNC GOLD has been put into “production mode” wherein modifications to the database structure may not be made by UNC GOLD personnel. Instead, all proposed changes are provided to the UNC Clinical Data Manager (C.W.P.) who updates the database and verifies that all data are retained.
ISSUES IDENTIFIED
Although database creation is associated with technical hurdles, more substantive considerations may also be revealed in the process. These matters require careful attention prior to data entry. Because clinical databases do not follow the same well-defined set of requirements as research studies or hospital-level electronic medical records, it is necessary to search a variety of resources and policies to ensure compliance.
Regulatory Considerations and Project Prioritization
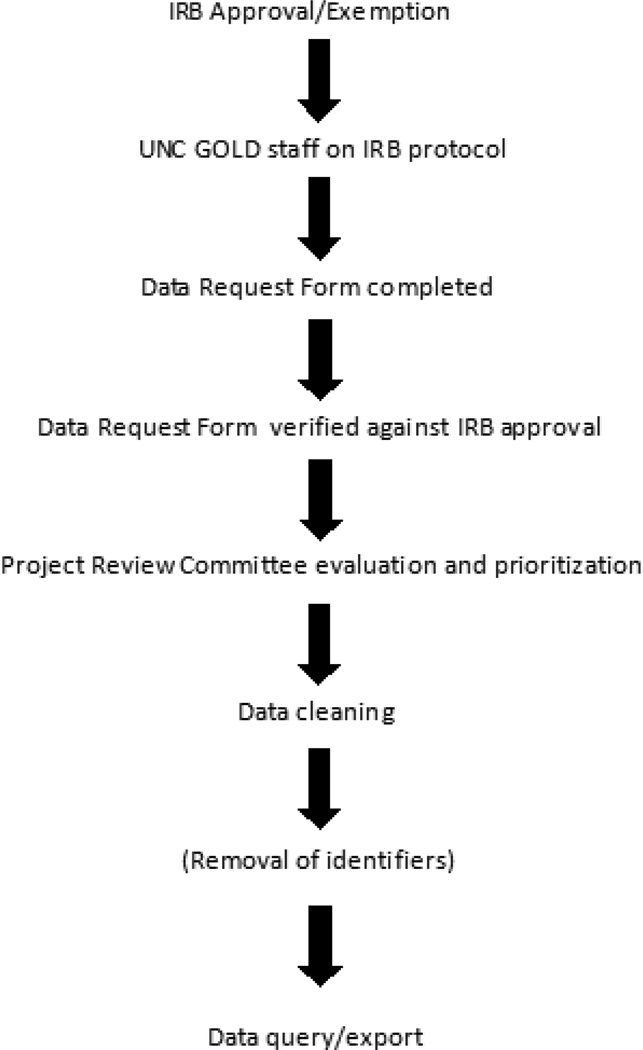
One of the first issues to be addressed was the involvement of the IRB in the project. Although the database was created to store clinical information, it may also serve as a primary data source for research projects. In its capacity as a clinical storehouse, the IRB does not consider UNC GOLD to be research, and no specific approvals are required for its creation or for abstraction of patient records. However, if information contained within the database is used for research purposes, IRB protocol submission and approval are required (although in many cases these submissions qualify for a waiver of informed consent). In some cases, physicians requesting de-identified data are exempt from IRB approval, however, it is the policy of the UNC GOLD group that all studies using this data apply for IRB approval as part of the data request process (Figure 2). Additionally, because the research requires review of the medical records, querying of data, and exporting data to researchers, all IRB applications must include UNC GOLD staff members working with these data. To simplify the process, researchers requesting access to data must complete a data request form, which specifies the desired forms or variables. This request is then reviewed against the IRB protocol to ensure appropriate approval. Once appropriate IRB approval or exemption has been established, the proposed project is reviewed by the UNC GOLD Project Review Committee. This committee is composed of individual users of the database and has a rotating membership roster. The committee determines the priority of each research project based on the status of ongoing projects as well as available resources. Finally, database coordinators query the database for the exact information listed on the request form. In order to maintain the integrity of UNC GOLD, individual investigators do not receive access to the database or the primary data therein. Instead, only Steering Committee members and the database coordinators have access to the primary data, and the coordinators generate reports specific to the data that has been requested. REDCap contains features to randomly shift dates while maintaining the time between the dates within an individual record, as well as an option to remove fields marked as identifiers. In addition to verifying the completeness, accuracy, and regulatory compliance of all data queries, the data request form also provides a record for auditing purposes.
Figure 2.
Data request and release
Information Security Compliance
Similar to determining the relationship between the department’s database and the IRB, compliance with the institutional data security policy as well as the FDA’s regulations on electronic records [19] is critical. Although REDCap is designed to be in accord with national research and clinical standards so as to address federal regulations, UNC has additional password and access requirements. Particularly with clinical databases that contain identifiable information and for which patients have not provided consent, it is important to verify that all aspects of data security are regularly monitored and compared against any relevant information security policies. It is also the duty of all involved with the database to use the same precautions as they would for the EMR when reviewing any information from REDCap. At UNC, the system Clinical Data Manager is responsible for the day-to-day compliance monitoring.
Funding
Funding for a database is critical to support data management and abstraction staff as well as IT resources including server space. Possible funding mechanisms include institutional, cancer center, or department support; grant funding; philanthropy; or industry sponsorship. The UNC GOLD project is supported by departmental resources.
External Data Sources
Apart from regulatory matters, there are considerations regarding the data itself. Specifically, information on cause and date of death is very limited in the EMR. Because a large component of a clinical database involves the tracking of outcomes and survival, it is important to have high quality information. To supplement the medical record, while still maintaining a clinical database that does not require IRB approval, it is necessary to acquire this information from a publically available source. As an example, the North Carolina State Center for Health Statistics provides vital record information that is available to anyone and can provide a report of deaths in the state. Another possible source of this data is the Social Security Death Index (SSDI).
Independent Provider Databases
Another major point of interest in developing UNC GOLD is the ability to integrate IRB approved patient databases that have been created and maintained by individual providers. Although these databases do not include the entire core complete dataset, they are a significant resource for retrospective analyses. Additionally, because that data has heretofore been limited to a single physician, inclusion of that data into UNC GOLD with IRB approval allows for greater data sharing and larger patient populations. Despite these advantages, it is important to note that these records are often antiquated in their formatting, limited in scope to the specific interests of a single provider, and may not have undergone data entry and cleaning with the level of rigor of data entered prospectively into the disease group database. For that reason, identifying and separating the retrospective data from prospective data (in this case by patient ID numbers) is essential. Review of the data, formatting for upload to REDCap, and supplementation of retrospective records with information from the EMR to fulfill the core complete dataset may be utilized to improve the quality of these resources.
DATA ENTRY
After responding to the issues arising out of database creation, it is necessary to devise methods for prospective data collection. Principal in this endeavor is the prioritization of patients for entry into the database. Although the goal is to ultimately capture the entire clinic population, initially this is not feasible. One option is to limit data collection to patients with a specific primary cancer site. Alternatively, it is possible to begin data entry with all new patients seen in clinic. Due to time and resource limitations, the initial goal selected is prospective data collection for new patients seen in clinic after the date that the database “went live.” This restriction helps to standardize the way in which data is collected and reported by reducing the amount of older or archived records, which may be unavailable. After their initial abstraction, all records in UNC GOLD are updated biannually. This length of time was chosen to limit the redundancy of information entered for patients seen frequently (e.g. patients regularly receiving weekly systemic chemotherapy), while still capturing information for people with longer follow-up schedules.
Development of Standard Operating Procedures
In addition to setting the frequency of abstraction and updating, it is important to outline standard operating procedures (SOPs) for data collection. These SOPs indicate where information for each field may be found within the medical record, provide clarification about staging and procedures, and define dummy dates for cases in which a specific date is unavailable. Additionally, the operating procedures provide guidance with respect to recording information listed inconsistently in the medical record. All SOPs are updated regularly based on physician input and issues identified during abstraction.
Data Quality and Staff Abstraction Training
Referring to the SOPs, the database coordinators began record abstraction. Once each had reviewed and input information from 10–20 patients, the other coordinator then reviewed the same medical records, noting discrepancies or questions. This check was critical in identifying fields requiring more explanation, making staff members comfortable with record abstraction, and providing insight into the inter-rater reliability and data quality that could be expected from UNC GOLD. Notably, this process illustrated several fields for which coordinators had different responses supported by information in the medical record. Discussing among themselves and with relevant clinicians, these conflicts were resolved, and explanations appended to the abstraction protocol for future reference. This continued until over 50 patients representing the three genitourinary sites, all treatment modalities, and a variety of stages had been abstracted.
Duplicate data entry represents a significant resource burden [20]. Because error rates from the initial 50 patients were low, rather than reviewing each record, bulk data exports are periodically analyzed for any aberrant, missing, or otherwise problematic data according to guidelines in the literature [21]. Using this methodology, all records containing a possible error are reviewed, and changes made if necessary. Data are monitored and cleaned in this fashion immediately prior to any analysis.
Retrieving, Querying, and Exporting Data
After ensuring sufficient data quality, it was necessary to consider strategies for pulling project-specific and user-defined datasets. REDCap includes two strategies for compiling data for analysis: reports and exports. Queries to generate reports may be designed with fields in any order that the user defines. These queries provide data for view within REDCap prior to downloading to Microsoft Excel (Microsoft Corporation, Redmond, WA) and can be saved and reused. Alternatively, exports allow specification of data at the field, form, and database level, but the ordering of the data maps exactly to that within the database. Exports also allow for downloading of data in a format compatible with Microsoft Excel as well as a variety of statistical software packages, with additional options for removing identifiers and shifting dates within records to comply with date de-identification. Typically, reports and queries are more useful for data monitoring tasks such as verifying the absence of duplicated records, while the de-identification features in the export tool better meet the needs of researchers requesting data.
Rules for Dissemination of UNC GOLD Related Research
UNC GOLD may be used to support individual research and group projects. In the case of these larger, collaborative group projects, no individual may publish results, design future, apply for a patent, or otherwise use data or information from UNC GOLD without the prior authorization of the Project Review Committee. For collaborative projects, the Project Review Committee working with members of the specific group is tasked with determining primary and corresponding authors in such a manner to foster continued collaboration. Finally, all individual and group investigations should acknowledge UNC GOLD in related publications.
CONCLUSION
The development of a genitourinary clinical database within a large research institution provides substantial benefit to clinical and research goals. This project, however, requires a significant buy-in from all stakeholders, careful development, consideration of a variety of regulatory and technical issues, and support for ongoing abstraction, updating, and data cleaning. Although UNC GOLD represents a widely-applicable case study in this process, individual departments and institutions must consider their own needs.
Acknowledgments
The North Carolina Clinical and Translational Sciences Institute (NC TraCS) at the University of North Carolina at Chapel Hill. NIH CTSA program of the division of Research Resources, National Institutes of Health. Supported by Grant Award Number UL1RR025747; REDCap (Research Electronic Data Capture). Supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health. Supported by Grant Award Number UL1TR000083. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. UNC Lineberger Comprehensive Cancer Center Office of Clinical and Translational Research; Department of Urologic Surgery, Vanderbilt University Medical Center; University Cancer Research Fund.
REFERENCES
- 1.Butte AJ, Ito S. Translational bioinformatics: data-driven drug discovery and development. Clinical Pharmacology & Therapeutics. 2012;91(6):949–952. doi: 10.1038/clpt.2012.55. [DOI] [PubMed] [Google Scholar]
- 2.Dhir R, Patel AA, Winters S, et al. A multidisciplinary approach to honest broker services for tissue banks and clinical data: a pragmatic and practical model. Cancer. 2008;113(7):1705–1715. doi: 10.1002/cncr.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson JG. Clearing the way for physicians’ use of clinical information systems. Communications of the ACM. 1997;40(8):83–90. [Google Scholar]
- 4.Furukawa MF, Poon E. Meaningful use of health information technology: evidence suggests benefits and challenges lie ahead. American Journal of Managed Care. 2011;17(12 Spec No) SP76a-SP. [PubMed] [Google Scholar]
- 5.MacKenzie SL, Wyatt MC, Schuff R, Tenenbaum JD, Anderson N. Practices and perspectives on building integrated data repositories: results from a 2010 CTSA survey. Journal of the American Medical Informatics Association. 2012;19:119–124. doi: 10.1136/amiajnl-2011-000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall WW, Haley RW. Use of a secure internet web site for collaborative medical research. JAMA. 2000;284(14):1843–1849. doi: 10.1001/jama.284.14.1843. [DOI] [PubMed] [Google Scholar]
- 7.West SL, Blake C, Zhiwen Liu, McKoy JN, Oertel MD, Carey TS. Reflections on the use of electronic health record data for clinical research. Health Informatics Journal. 2009;15(2):108–121. doi: 10.1177/1460458209102972. [DOI] [PubMed] [Google Scholar]
- 8.Prokosch HU, Ganslandt T. Perspectives for medical informatics. Reusing the electronic medical record for clinical research. Methods of Information in Medicine. 2009;48(1):38–44. [PubMed] [Google Scholar]
- 9.Yackanicz L, Kerr R, Levick D. Physician buy-in for EMRs. Journal of Healthcare Information Management. 2010;24(2):41–44. [PubMed] [Google Scholar]
- 10.Lotan Y, Amiel G, Boorjian SA, et al. on Behalf of the Bladder Cancer Think Tank and Bladder Cancer Advocacy Network. Comprehensive handbook for developing a bladder cancer cystectomy database. Urologic Oncology. 2011 Nov 4; doi: 10.1016/j.urolonc.2011.09.004. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 11.Deo S. Computerized clinical database development in oncology. Indian Journal of Palliative Care. 2011;17(Suppl):S2–S3. doi: 10.4103/0973-1075.76229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin JD, Guidry A, Brinkley JF. A partnership approach for Electronic Data Capture in small-scale clinical trials. Journal of Biomedical Informatics. 2011;44(Suppl 1):S103–S108. doi: 10.1016/j.jbi.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith JP, Elliott AC, Hynan LS, Reisch JS, Waddell SA. Creation and use of a database in clinical and translational research. Journal of Investigative Medicine. 2010;58(3):544–553. doi: 10.231/JIM.0b013e3181c9f668. [DOI] [PubMed] [Google Scholar]
- 14.Krishnankutty B, Bellary S, Kumar NB, Moodahadu LS. Data management in clinical research: An overview. Indian Journal of Pharmacology. 2012;44(2):168–172. doi: 10.4103/0253-7613.93842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohmann C, Kuchinke W. Future developments of medical informatics from the viewpoint of networked clinical research. Interoperability and integration. Methods of Information in Medicine. 2009;48(1):45–54. [PubMed] [Google Scholar]
- 16.Payne PR, Greaves AW, Kipps TJ. CRC clinical trials management system (CTMS): an integrated information management solution for collaborative clinical research. AMIA Annual Symposium Proceedings. 2003:967. [PMC free article] [PubMed] [Google Scholar]
- 17.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. Journal of Biomedical Informatics. 2009 Apr;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Supported by Grant Award Number UL1RR025747 from the Clinical and Translational Science Award program of the Division of Research Resources, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
- 19.CFR – Code of federal regulations title 21. [Accessed August 27, 2012]; Available at: http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfCFR/CFRSearch.cfm?CFRPart=11.
- 20.Gibson D, Harvey AJ, Everett V, Parmar MK. Is double data entry necessary? The CHART trials. CHART steering committee. Continuous, hyperfractioned, accelerated radiotherapy. Controlled Clinical Trials. 1994;15(6):482–488. doi: 10.1016/0197-2456(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 21.Day S, Fayers P, Harvey D. Double data entry: what value, what price? Controlled Clinical Trials. 1998;19(1):15–24. doi: 10.1016/s0197-2456(97)00096-2. [DOI] [PubMed] [Google Scholar]