Abstract
Aims
Vascular perfusion may be impaired in primary open-angle glaucoma (POAG); thus, we evaluated a panel of markers in vascular tone-regulating genes in relation to POAG.
Methods
We used Illumina 660W-Quad array genotype data and pooled P-values from 3108 POAG cases and 3430 controls from the combined National Eye Institute Glaucoma Human Genetics Collaboration consortium and Glaucoma Genes and Environment studies. Using information from previous literature and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, we compiled single-nucleotide polymorphisms (SNPs) in 186 vascular tone-regulating genes. We used the ‘Pathway Analysis by Randomization Incorporating Structure' analysis software, which performed 1000 permutations to compare the overall pathway and selected genes with comparable randomly generated pathways and genes in their association with POAG.
Results
The vascular tone pathway was not associated with POAG overall or POAG subtypes, defined by the type of visual field loss (early paracentral loss (n=224 cases) or only peripheral loss (n=993 cases)) (permuted P≥0.20). In gene-based analyses, eight were associated with POAG overall at permuted P<0.001: PRKAA1, CAV1, ITPR3, EDNRB, GNB2, DNM2, HFE, and MYL9. Notably, six of these eight (the first six listed) code for factors involved in the endothelial nitric oxide synthase activity, and three of these six (CAV1, ITPR3, and EDNRB) were also associated with early paracentral loss at P<0.001, whereas none of the six genes reached P<0.001 for peripheral loss only.
Discussion
Although the assembled vascular tone SNP set was not associated with POAG, genes that code for local factors involved in setting vascular tone were associated with POAG.
Introduction
Primary open-angle glaucoma (POAG), a leading cause of blindness, is characterized by visual field (VF) loss due to retinal ganglion cell death.1 It is believed that there are multifactorial contributors to POAG aetiology in addition to elevated intraocular pressure (IOP), an established risk factor. One major contributor may be regulation of vascular tone.2, 3, 4, 5, 6 For example, glaucoma patients have impaired blood flow and vascular autoregulation in eye tissues,7, 8, 9, 10, 11 and exhibit impaired flow- 12 or acetylcholine-mediated13 vasodilation in the brachial artery and reduced blood flow in finger nailfold capillaries,14 suggesting systemic vascular abnormalities. Of note, POAG patients with initial paracentral VF loss are particularly more likely to have systemic vascular risk factors, such as hypotension and migraines.15, 16, 17
Many studies support the role of vascular factors in glaucoma. Large epidemiologic studies have observed adverse associations with low ocular perfusion pressure18 or low systemic blood pressure,19 and open-angle glaucoma frequency or risk.20, 21 In addition, the Early Manifest Glaucoma Trial22 identified predictors of POAG disease progression that included lower systolic blood and perfusion pressure.
Recent large POAG genome-wide association studies (GWAS) have identified significant associations for single-nucleotide polymorphism (SNP) in the genomic region containing CAV1,23, 24, 25 which codes for caveolins that are involved in vascular regulation, thereby supporting the role of vascular dysregulation in POAG. Because biologic pathway analyses (where SNP sets from functionally related genes are collectively evaluated in relation to a disease of interest) can enhance the power to discover aetiologically relevant pathways and networks, we tested the hypothesis of whether a collection of SNPs in genes functionally involved in vascular tone regulation and identified as significant hits in GWAS of blood pressure are possibly associated with POAG.
In this study, we used high-throughput genotyping SNP data from the Glaucoma Genes and Environment (GLAUGEN) study and the National Eye Institute Glaucoma Human Genetics Collaboration (NEIGHBOR) consortium to assemble a vascular tone SNP panel (8671 SNPs in 186 genes) and used the Pathway Analysis by Randomization Incorporating Structure (PARIS) analysis software package (freeware developed by Dr Marilyn Ritchie, Pennsylvania State University, University Park, State College, PA, USA)26 for pathway analysis. In exploratory analyses, we assessed whether collections of SNPs within genes that comprise the pathway are associated with POAG.
Material and methods
Study population
We used meta-analysed GWAS data from two studies in the United States: GLAUGEN and NEIGHBOR. The GLAUGEN study included nested case–control samples (976 POAG cases and 1140 controls) from the population-based cohorts of the Nurses' Health Study (NHS) and Health Professionals Follow-up Study (HPFS), as well as a clinical case–control sample from the Massachusetts Eye and Ear (MEE). The data from GLAUGEN have been evaluated in investigations of genetic factors and gene–environment interactions in POAG.27, 28 A recent publication describes the criteria for inclusion and exclusion for GLAUGEN,23 and the details are also available at dbGaP, accession number phs000308.v1.p1. The NEIGHBOR study included 2132 POAG cases and 2290 controls from 12 institutions and also provided data for genetic investigations of central corneal thickness,29, 30 IOP,31 and POAG.28 The descriptive information about the design, inclusion/exclusion criteria, study sites, and collected data in the NEIGHBOR study was recently published.32
POAG case and control definitions
All POAG cases were required to have at least one eye with either (1) a cup-to-disc ratio (CDR)>0.7 and VF loss consistent with glaucoma on one reliable VF test or (2) the same VF loss evident on at least two reliable VF tests (reliability of a VF test was defined as fixation loss ≤33%, false positives ≤20%, and false negatives ≤20%). The reproducible VF defect(s) evident on reliable tests were compatible with glaucomatous patterns of nerve fibre layer loss. The type of VF test was not specified, although most cases had threshold VF tests on the Humphrey perimeter. On the basis of slit-lamp biomicroscopy, none of the cases had evidence of exfoliation syndrome, pigment dispersion syndrome, or other glaucoma diagnoses that could produce secondary elevation of IOP. In addition, all cases had documentation of open iridocorneal filtration angles in both eyes. POAG cases were included at the full spectrum of IOP at diagnosis, including both normal tension glaucoma cases with IOP ≤21 mm Hg and high tension glaucoma cases with IOP >21 mm Hg. Controls identified in clinical settings underwent eye exams that showed IOP ≤21 mm Hg and CDR ≤0.6 in both eyes. For nested case–control study controls (from NHS and HPFS), controls reported having had an eye exam at the same time period as the index cases and did not report a glaucoma diagnosis.
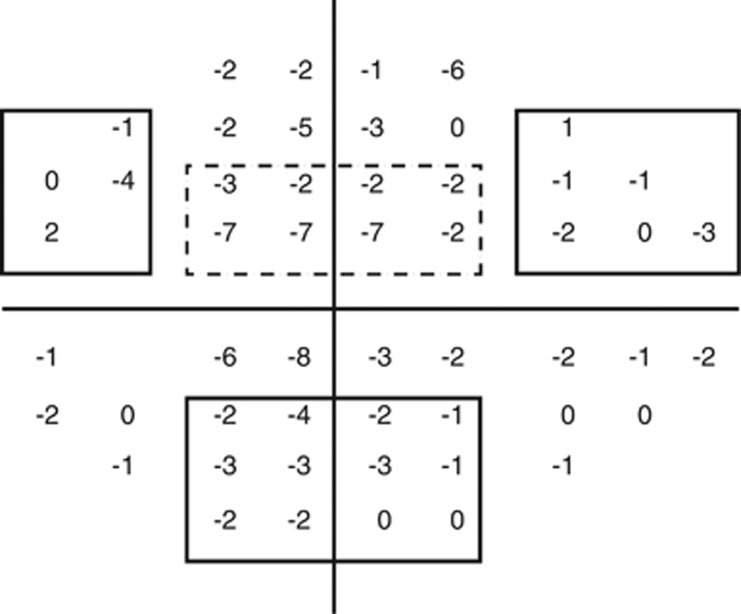
Each case was categorized into POAG subtypes defined by type of VF loss: early paracentral VF loss (with or without peripheral VF loss) or peripheral VF loss only (without any paracentral loss). The earliest VF report for each case was systematically reviewed such that pattern deviation plots were subdivided into paracentral, Bjerrum, nasal step, and temporal wedge zones above and below the horizontal meridian (Figure 1). Two reviewers who were masked to genotype status assessed VFs, and any differences in VF loss-type designation were adjudicated for establishing a consensus designation. The presence of clusters of three or more contiguous points with reduced retinal sensitivity of one-half log unit (−5 dB) relative to age-matched controls were considered to show VF loss; the location of such loss was determined for each of the four regions. Cases were defined as having POAG with ‘early paracentral loss' in the two studies differently: in the GLAUGEN data set, where cases were incident cases from population-based studies with early VF loss, cases had VF loss that occurred in the paracentral zone with or without loss in other peripheral zones, whereas in the NEIGHBOR data set, where cases were prevalent cases from tertiary clinic-based case–control studies with advanced disease, to capture ‘early paracentral loss', cases had to have VF loss that occurred in the paracentral zone only without any loss in other peripheral zones. Cases were defined as having POAG with ‘peripheral loss only' in both GLAUGEN and NEIGHBOR data sets if cases had VF loss in any of the other peripheral zones without loss in the paracentral zone.
Figure 1.
Paracentral and peripheral VF loss definitions. Representative VF pattern deviation plot used for defining the type of VF loss. The box with a dashed line indicates the superior paracentral region. The boxes with solid lines represent the superior nasal step and temporal wedge regions, as well as the inferior Bjerrum region, which are all peripheral regions. For a case to be defined as having early paracentral loss, VF loss in the superior or inferior paracentral zones had to be present; in the GLAUGEN data set, VF loss in other peripheral zones were allowed, whereas in the NEIGHBOR data set, VF loss in other peripheral zones were not allowed. For a case to be defined as having peripheral loss, VF loss in the superior and/or inferior nasal step, temporal wedge, or Bjerrum regions had to be present, with no loss in the paracentral regions.
Genotyping and analyses of genotype associations
Genotyping with the Illumina 660W-Quad array (Illumina, San Diego, CA, USA) was conducted at the Broad Institute (Cambridge, MA, USA) for GLAUGEN and at the Center for Inherited Disease Research (Baltimore, MD, USA) for NEIGHBOR. The details of the methods used for DNA collection, extraction and plating, the algorithm for calling genotypes, the SNP quality control (QC) filters used, and the preliminary analyses undertaken to discover independent predictors of genotype calling rates adjusted for in this paper have been provided in previous publications for both GLAUGEN and NEIGHBOR.23, 28 For analyses of genotype associations with POAG, PLINK v.1.0733 was used with SNPs that passed the QC filters (484 419 SNPs in GLAUGEN and 521 683 SNPs in NEIGHBOR). Analyses were conducted among self-reported Caucasians and were adjusted for population structure, age, gender, and study site in both study groups, and analyses in GLAUGEN were additionally adjusted for the three different DNA extraction methods used and DNA source (blood or cheek cell sample).23, 28 Using the METAL software,34 the GLAUGEN and NEIGHBOR data were meta-analysed, and pooled P-values were obtained for the 480 335 overlapping SNPs in the combined data set.
Defining the vascular tone pathway and analysis of the whole pathway and of specific genes
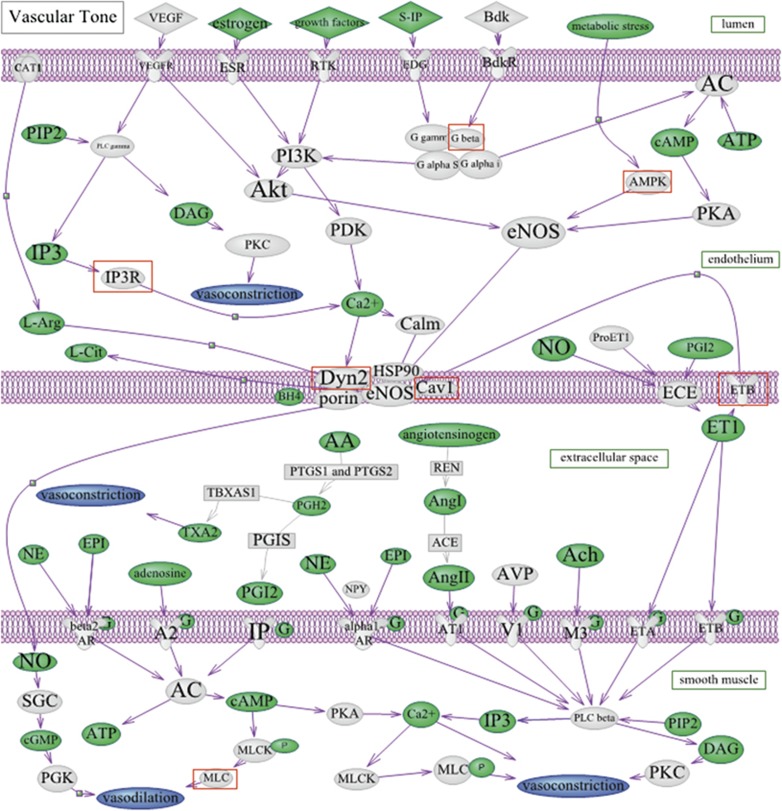
In this paper, we used the P-values for the association with POAG for 8671 unique SNPs in the vascular tone pathway for the analysis that is discussed below. We generated a custom list comprising the vascular tone pathway (see Figure 2) of 8671 SNPs in 186 genes (Supplementary Table 1) across all autosomes. We used the Kyoto Encyclopedia of Genes and Genomes (KEGG)35 online database and other academic sources,36, 37 as well as a recent review38 of the GWAS findings for blood pressure.
Figure 2.
Vascular tone pathway. This pathway was custom-generated by using sources from the Kyoto Encyclopedia of Genes and Genomes35 online database and other academic sources,36, 37 as well as a recent review38 of the GWAS findings for blood pressure. Proteins coded by genes included in the analysis are in grey. Substrates and reaction products are in green. Overall, the vascular tone pathway was not significantly associated with POAG permuted P=0.87. In exploratory analyses, of the eight proteins of genes with permuted P-values <0.001, seven are indicated with red squares, with the remaining being the HFE gene, which was included in the analysis as part of genes identified from a review38 of known blood pressure SNPs.
We uploaded the meta-analysed P-values of the selected SNPs within these genes residing in the EMSEMBL genomic interval±a 50 kB window to PARIS software from the combined GLAUGEN–NEIGHBOR data set. The analytic approach with the PARIS software has been described.26 Briefly, for the assembled pathway, PARIS first determined important parameters such as the number of linkage disequilibrium (LD) blocks, the number of SNPs per block, and the number of SNPs not in any LD block. The vascular tone pathway contained 1405 complex features (LD blocks with two or more SNPs) and 1779 simple features (single SNPs not in any LD block). Then, PARIS conducts a comparison of the overall association of the vascular tone pathway with POAG to the associations of 1000 randomly generated pathways with genetic architectures similar to the vascular tone pathway with POAG; in this comparison, PARIS provides a permuted P-value, which helps determine whether the assembly of SNPs in the pathway of interest as a whole may be significantly associated with POAG. We chose to produce 1000 random pathways, as generating another order of magnitude more pathways with similar genetic architecture would have been prohibitive computationally. To illustrate the methodology of how the random SNP sets are used to assess statistical significance for SNP sets of interest, we, as described previously,39 evaluated an oestrogen pathway among women using the same data set. The results indicated 31 significant features in this pathway, and PARIS generated a significant permuted P-value=0.007. This indicated that only 7 of 1000 random pathways with a genetic architecture similar to the oestrogen pathway had more features that were statistically significant at P<0.05 level in relation to POAG than the oestrogen pathway (i.e., >31 significant features with P<0.05). This overall significant permuted P-value of 0.007 indicated that the assembly of SNPs in the oestrogen pathway was significantly associated with POAG, supporting the role of oestrogen in the aetiology of POAG.39 We used a similar approach in this study to evaluate the vascular pathway in relation to POAG. Because POAG characterized by VF loss in the paracentral region is considered to be more strongly associated with vascular abnormalities,15, 16, 17 in addition to overall POAG, we evaluated subtypes of POAG defined by the type of VF loss separately (POAG with early VF loss in the paracentral region and POAG with VF loss in the peripheral region only), to investigate differences in association.
In subsequent analyses, to determine whether some genes or SNPs in the vascular tone pathway were significantly associated with POAG, we used the ‘-I' (Investigate) option within PARIS, which reports the P-values of genes and SNPs within the pathway. In an approach that was similar to the overall pathway approach, we assessed the association with POAG of the aggregate SNPs within specific genes in the vascular tone pathway to gain insight into which aspects of the pathway may be aetiologically important. Because we evaluated 186 genes in this pathway in relation to three outcomes, there was the potential for significant findings arising owing to chance because of multiple testing. In the PARIS methodology, the genome is permuted rather than applying a Bonferroni correction for multiple testing (to avoid the risk of penalizing larger genes as might happen with Bonferroni correction).26 To minimize false-positive findings, we focused on genes that showed permuted P<0.001 (which represented the lowest possible permuted P-value, as such P-values are based on counts of 1000 random pathways per genes; that is, a count of 1 randomly generated pathway that shows more significant features than the vascular tone pathway out of 1000 random pathways yields a permuted P=0.001, and 0 out of 1000 random pathways leads to a permuted P-value of P<0.001).
We certify that all applicable institutional and governmental regulations concerning the ethical use of data from human volunteers were followed during this research, and the Institutional Review Boards of the participating institutions of GLAUGEN and NEIGHBOR approved this study.
Results
A total of 3430 controls (2290 from NEIGHBOR and 1140 from GLAUGEN) and 3108 cases (2132 from NEIGHBOR and 976 from GLAUGEN) and were analysed in the study (Table 1). Of these 3108 cases, 224 had POAG with early paracentral VF loss, whereas 993 had POAG with peripheral VF loss only.
Table 1. Pathway analysis results using data from the combined GLAUGEN and NEIGHBOR: significance of the association of the overall vascular tone pathway in relation to POAG, POAG with early paracentral visual field loss, and POAG with peripheral visual field loss only.
| Outcome | N |
Mean age (years) |
P-valuea | ||
|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | ||
| POAG | 3108 | 3430 | 65.7 | 67.7 | 0.87 |
| POAG with early paracentral visual field loss | 224 | 3430 | 64.8 | 67.7 | 0.20 |
| POAG with peripheral visual field loss only | 993 | 3430 | 64.8 | 67.7 | 0.39 |
Abbreviations: GLAUGEN, the Glaucoma Genes and Environment study; NEIGHBOR, National Eye Institute Glaucoma Human Genetics Collaboration; POAG, primary open-angle glaucoma.
P-values are permuted P-values.
The vascular tone pathway was not significantly associated with POAG overall (permuted P=0.87); the results were similarly null (permuted P≥0.81) in men and women analysed separately (data not shown). In addition, the vascular tone pathway was not associated with POAG subtypes defined by VF loss: the permuted P-value in relation to early paracentral VF loss was 0.20, whereas the corresponding P-value for peripheral VF loss only was 0.39.
In gene-based analyses, eight genes were associated with overall POAG at permuted P-value <0.001 (Table 2), and the red-boxed items in Figure 2 are the corresponding proteins for seven of the eight (HFE is not included; HFE was a gene with loci that were significantly associated with blood pressure in a recent review38 of the GWAS studies of blood pressure). Of these eight genes, six were those coding for proteins/enzymes that interact with endothelial nitric oxide synthase (eNOS): PRKAA1, CAV1, ITPR3, EDNRB, GNB2, and DNM2. Supplementary Table 2 shows the association between individual SNPs within these eight genes (residing in the EMSEMBL genomic interval±a 50 kB window) and POAG.
Table 2. Gene analysis results for the vascular tone pathway using data from the combined GLAUGEN and NEIGHBOR.
| Protein | Label in Figure 2. | Gene (Ensembl ID) | Chr | No. of simplea features | No. of complexa features |
POAG |
POAG with early paracentral visual field loss |
POAG with peripheral visual field loss only |
|---|---|---|---|---|---|---|---|---|
| P-valueb | P-valueb | P-valueb | ||||||
| 5′ AMP-activated protein kinase | AMPK | PRKAA1c | 5 | 0 | 2 | <0.001 | 0.05 | 0.05 |
| Inositol 1,4,5-trisphosphate receptor, type 3 | IP3R | ITPR3c | 6 | 14 | 9 | <0.001 | <0.001 | >0.99 |
| Hemochromatosis protein | — | HFE | 6 | 0 | 2 | <0.001 | <0.001 | 0.07 |
| Caveolin-1/caveolin 2 | CAV1 | CAV1c | 7 | 2 | 6 | <0.001 | <0.001 | 0.004 |
| G-protein subunit β-2 | G beta | GNB2c | 7 | 0 | 2 | <0.001 | >0.99 | >0.99 |
| Endothelin receptor type B | ETB | EDNRBc | 13 | 0 | 1 | <0.001 | <0.001 | >0.99 |
| Dynamin II | Dyn2 | DNM2c | 19 | 2 | 7 | <0.001 | 0.06 | >0.99 |
| Myosin regulatory light polypeptide 9 | MLC | MYL9 | 20 | 0 | 1 | <0.001 | >0.99 | >0.99 |
Abbreviations: Chr, chromosome; GLAUGEN, the Glaucoma Genes and Environment study; LD, linkage disequilibrium; NEIGHBOR, National Eye Institute Glaucoma Human Genetics Collaboration; POAG, primary open-angle glaucoma; SNP, single-nucleotide polymorphism.
Simple features refer to single SNPs not in any LD block; complex features refer to LD blocks with two or more SNPs.
P-values are permuted P-values.
Genes for proteins that interact with endothelial nitric oxide synthase; see Figure 2.
In addition, three of these (CAV1, ITPR3, and EDNRB), along with HFE, were also associated with the subtype of POAG with early paracentral VF loss with the permuted P-value of <0.001, and PRKAA1 showed borderline significance with the permuted P-value of 0.05. In contrast, none of these reached permuted P-value <0.001 for the subtype of POAG with peripheral VF loss only. However, as observed for POAG with early paracentral VF loss, nominally significant permuted P-values were observed for CAV1 (P-value=0.004) and PRKAA1 (P-value=0.05) in relation to POAG with peripheral VF loss only.
For the subtype of POAG with early paracentral VF loss, one other gene, ADCY7, which was not associated with POAG overall, was found to be associated with this subtype with permuted P<0.001 (data not shown). Notably, ADCY7 codes for a protein that influences eNOS activity. For the subtype of POAG with peripheral VF loss only and not overall POAG or the subtype with early paracentral VF loss (data not shown), there were eight different genes that were associated only with this subtype with permuted P<0.001: PRKAA2, PRKAB2, GNB5, PIK3CG, WNK1, PRKCA, ADORA2A, and AGTR1. Of note, four of these 8 (PRKAA2, PRKAB2, GNB5, and PIK3CG) also coded for proteins that interact with eNOS.
Discussion
In this first study of the collective SNPs in genes involved in the vascular tone pathway and POAG, we observed that the assembled vascular tone pathway SNP panel was not related to the main outcome of overall POAG diagnosis or the subtypes of POAG with early VF loss in the paracentral region or POAG with VF loss in the peripheral region only. However, in gene-based analysis, eight genes were associated with POAG with permuted P<0.001, and of these eight genes, six coded for proteins that interact with eNOS. In particular, three of these six were also significantly related (permuted P<0.001) to the POAG subtype that is characterized by early paracentral VF loss. Thus, although this study did not find associations with the vascular tone panel as a whole, it generated further support for the more specific role of eNOS-related activity in POAG.
Because a myriad of extrinsic (factors that originate from outside the local tissue that affect the arterial blood pressure) and intrinsic factors (local factors that regulate local blood flow) affect vascular tone,3 the panel tested included a comprehensive set of 186 genes (8671 unique SNPs) based on the KEGG pathways and GWAS significant associations for blood pressure based on a recent review.38 The overall null association may have been due to the inclusion of SNPs from gene-regulating factors that may affect blood pressure or regulate vascular tone systemically but are not relevant for blood flow regulation, especially to the optic nerve head (ONH). For example, hormones such as epinephrine, vasopressin, and natriuretic peptides40, 41, 42 or neurogenic factors43 are unlikely to affect the regulation of blood flow to the retina and the ONH (e.g., blood vessels in the retina and the ONH are not innervated),40 whereas intrinsic factors such as vascular endothelial vasoactive agents have an important role in influencing blood flow autoregulation and vascular tone in the ONH.3, 44, 45 Of these agents, NO, generated constitutively by eNOS, has a key role in maintaining the basal vasodilator tone in ocular arteries.40, 46, 47, 48, 49, 50, 51, 52 The major contribution of NO to vascular tone in human ocular vasculature is demonstrated by the observation that systemic administration of an NOS inhibitor reduced blood flow in the ONH53, 54, 55 and by the involvement of NO in the POAG-related vascular conditions, such as hypertension, hypotension, and vasospasms.56
Evidence of abnormal NO production in glaucoma is also provided by genetic studies. Although genetic studies have evaluated SNPs of factors related to the renin–angiotensin system57, 58, 59 and the β-adrenoreceptors60, 61 in relation to POAG, they have been small and conflicting. However, relatively more consistent associations are observed with genetic studies of eNOS-related factors and POAG.62, 63, 64, 65, 66, 67 Although not all studies have found associations with variants in the NOS3 gene that codes for eNOS,68, 69 variants or haplotypes of SNPs in NOS3 have been reported to be associated with POAG overall,62, 65 POAG with migraines,66 POAG or high tension POAG among women only,63, 64 and familial POAG.67 Furthermore, the CAV1/CAV2 gene region, which codes for caveolins that control eNOS activity, contain variants associated with POAG in one GWAS,24 and this finding was replicated in another large candidate gene study.23 The caveolin/eNOS interaction is critical to prevent inadequate NO production under basal conditions and to the translation of extracellular stimuli to intracellular NO signals.70
Consistent with this previous literature, it is suggestive that six of the eight most significant associations in the gene-based analyses were genes encoding proteins that interact with eNOS; based on information from large databases of gene expression in various human tissues, all eight genes showed expression in human retinal tissue.71, 72 ITPR3 codes for inositol triphosphate receptor (IP3R); when IP3 binds to IP3R, calcium levels increase, activating calmodulin, and causing dissociation of caveolin from eNOS. The increased calcium levels directly or indirectly via dynamin 2 (Dyn2), coded by DNM2, activate eNOS. EDNRB codes for endothelin receptor type B (ETB, which is particularly highly expressed in retina71, 72) and PRKAA1 codes for AMP kinase (AMPK); both are additional activators of eNOS. Finally, GNB2 codes for G-protein subunit-β (G beta) that activates PI3K, which in turn activates Akt; Akt then activates eNOS. A limitation of this study was that variants in NOS3, which codes for eNOS, were not significantly associated with POAG, although this could be due to the low coverage (only two non-functional SNPs rs7830 and rs3918188 were included) of the NOS3 SNPs on the Illumina 660W quad platform. This might also be observed if NOS3 effects derive primarily from multiple different rare variants rather than from effects of the kinds of common variants used in this study.
Out of the eight genes showing significant association with POAG, only MYL9, coding for myosin regulatory light polypeptide 9 (MLC) and HFE, which codes for hemochromatosis protein involved in iron metabolism, are not directly involved in eNOS activation. Interestingly, HFE was associated with both POAG overall and the POAG subtype with early paracentral VF loss. Although there are no previous reports of an association between hemochromatosis and glaucoma, iron supplementation has been linked to POAG;73 thus, the association between POAG and iron levels or hemochromatosis warrants further study.
Of the eight genes with P<0.001 associated with POAG overall, three (CAV1, ITPR3, and EDNRB) genes involved in eNOS activity were also associated with the subtype of POAG with early paracentral VF loss, even though we included only 224 such cases. In addition, ADCY7, which codes for adenylate cyclase that activates eNOS, was also significantly related to this subtype. In contrast, none of the genes significantly associated with POAG overall or the POAG subtype with early paracentral VF loss were associated with the POAG subtype with only peripheral VF loss. This is consistent with previous findings showing associations between the POAG subtype showing early paracentral VF loss with systemic vascular dysregulation.15, 17
Among the limitations of this study, first, we lacked replication of the gene-based analyses as there is currently no other data set of comparable size for replication. Thus, although suggestive, these gene-based exploratory analyses, especially in relation to the subtype of POAG with early paracentral VF loss, should be interpreted with caution and replicated. Second, this study was restricted to Caucasians and, therefore, the generalizability of these results to other populations, especially East Asian populations with generally lower IOP,15, 17, 57 is limited.
In this large study to evaluate the collective influence of genes coding for proteins influencing vascular tone, we did not observe associations with POAG. However, in gene-based analysis, we observed confirmatory results supporting the role of the dysregulation of eNOS activity in POAG aetiology.
Acknowledgments
A Horizon Grant to MEEI from Allergan (Irvine, CA, USA) supported the collection of some glaucoma feature data. The Harvard Glaucoma Center of Excellence, a Harvard Department of Ophthalmology Scholar Award, and Margolis fund (Boston, MA) support LRP and JLW. LRP, JER, and JLW are also supported by Research to Prevent Blindness Inc. (New York, NY, USA). The Arthur Ashley Foundation also supports Dr Pasquale. The Glaucoma Research Foundation (San Francisco, CA, USA), American Health Assistance Foundation (Clarksburg, MD, USA), and the Glaucoma Foundation (New York, NY, USA) support YL. EB is supported by NIH Grant R01EY022746. The following National Institutes of Health grants support the maintenance of the NHS and Health Professionals Follow-up, allowing these health professionals to contribute to this analysis: P01 CA87969, CA49449, UM1 CA167552, and HL35464. The following grants from the National Human Genome Research Institute (Bethesda, MD, USA) supported GLAUGEN: HG004728 (LRP), HG004424 (Broad Institute to support genotyping), and HG004446 (C Laurie, U Washington, to support genotype data cleaning and analysis). Genotyping services for the NEIGHBOR study were provided by the Center for Inherited Disease Research (CIDR) and were supported by the National Eye Institute through Grant HG005259-01 (JLW). In addition, CIDR is funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. The National Eye Institute (Bethesda, MD) through ARRA Grants 3R01EY015872-05S1 (JLW) and 3R01EY019126-02S1 (MAH) supported the collection and processing of samples for the NEIGHBOR data set. Funding for the collection of cases and controls was provided by National Institutes of Health (Bethesda, MD) Grants: EY015543 (RRA), EY006827 (DG), HL073389 (E Hauser); EY13315 (MAH); EY09611 (S Hankinson), EY015473 (LRP), EY009149 (PRL), HG004608 (CAM), EY008208 (P Medeiros), EY012118 (MAP-V), EY015682 (TR), EY011671 (JER), EY09580 (JER), EY013178 (JSS), EY010886 (JLW), EY009847 (JLW), EY011008 (L Zangwill), EY144428 (KZ), EY144448 (KZ), EY18660 (KZ), UL1TR000427 (MHB) and U01HG006389 (CAM), P30 EY014104, R01 EY022305, and R21 EY022766.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Eye website (http://www.nature.com/eye)
Supplementary Material
References
- Leske MC. Open-angle glaucoma—an epidemiologic overview. Ophthal Epidemiol. 2007;14 (4:166–172. doi: 10.1080/09286580701501931. [DOI] [PubMed] [Google Scholar]
- Flammer J, Pache M, Resink T. Vasospasm, its role in the pathogenesis of diseases with particular reference to the eye. Prog Retin Eye Res. 2001;20 (3:319–349. doi: 10.1016/s1350-9462(00)00028-8. [DOI] [PubMed] [Google Scholar]
- Hayreh SS. Blood flow in the optic nerve head and factors that may influence it. Prog Retin Eye Res. 2001;20 (5:595–624. doi: 10.1016/s1350-9462(01)00005-2. [DOI] [PubMed] [Google Scholar]
- Hayreh SS, Revie IH, Edwards J. Vasogenic origin of visual field defects and optic nerve changes in glaucoma. Br J Ophthalmol. 1970;54 (7:461–472. doi: 10.1136/bjo.54.7.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Graefe A. Uber die iridectomie bei glaukom und uber den glaucomatosen prozess. Von Graefe's Arch Ophthalmol. 1857;3:456–555. [Google Scholar]
- Moore D, Harris A, Wudunn D, Kheradiya N, Siesky B. Dysfunctional regulation of ocular blood flow: a risk factor for glaucoma. Clin Ophthalmol. 2008;2 (4:849–861. doi: 10.2147/opth.s2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl D, Garhofer G, Schmetterer L. The complex interaction between ocular perfusion pressure and ocular blood flow—relevance for glaucoma. Exp Eye Res. 2011;93 (2:141–155. doi: 10.1016/j.exer.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Flammer J, Orgul S, Costa VP, Orzalesi N, Krieglstein GK, Serra LM, et al. The impact of ocular blood flow in glaucoma. Prog Retin Eye Res. 2002;21 (4:359–393. doi: 10.1016/s1350-9462(02)00008-3. [DOI] [PubMed] [Google Scholar]
- Flammer J, Orgul S. Optic nerve blood-flow abnormalities in glaucoma. Prog Retin Eye Res. 1998;17 (2:267–289. doi: 10.1016/s1350-9462(97)00006-2. [DOI] [PubMed] [Google Scholar]
- Feke GT, Pasquale LR. Retinal blood flow response to posture change in glaucoma patients compared with healthy subjects. Ophthalmology. 2008;115 (2:246–252. doi: 10.1016/j.ophtha.2007.04.055. [DOI] [PubMed] [Google Scholar]
- Feke GT, Hazin R, Grosskreutz CL, Pasquale LR. Effect of brimonidine on retinal blood flow autoregulation in primary open-angle glaucoma. J Ocular Pharmacol Therap. 2011;27 (4:347–352. doi: 10.1089/jop.2011.0014. [DOI] [PubMed] [Google Scholar]
- Su WW, Cheng ST, Ho WJ, Tsay PK, Wu SC, Chang SH. Glaucoma is associated with peripheral vascular endothelial dysfunction. Ophthalmology. 2008;115 (7:1173–1178 e1171. doi: 10.1016/j.ophtha.2007.10.026. [DOI] [PubMed] [Google Scholar]
- Henry E, Newby DE, Webb DJ, O'Brien C. Peripheral endothelial dysfunction in normal pressure glaucoma. Invest Ophthalmol Vis Sci. 1999;40 (8:1710–1714. [PubMed] [Google Scholar]
- Gasser P, Flammer J. Blood-cell velocity in the nailfold capillaries of patients with normal-tension and high-tension glaucoma. Am J Ophthalmol. 1991;111 (5:585–588. doi: 10.1016/s0002-9394(14)73703-1. [DOI] [PubMed] [Google Scholar]
- Park HY, Jung KI, Na KS, Park SH, Park CK. Visual field characteristics in normal-tension glaucoma patients with autonomic dysfunction and abnormal peripheral microcirculation. Am J Ophthalmol. 2012;154 (3:466–475 e461. doi: 10.1016/j.ajo.2012.03.028. [DOI] [PubMed] [Google Scholar]
- Hitchings RA, Anderton SA. A comparative study of visual field defects seen in patients with low-tension glaucoma and chronic simple glaucoma. Br J Ophthalmol. 1983;67 (12:818–821. doi: 10.1136/bjo.67.12.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SC, De Moraes CG, Teng CC, Tello C, Liebmann JM, Ritch R. Initial parafoveal versus peripheral scotomas in glaucoma: risk factors and visual field characteristics. Ophthalmology. 2011;118 (9:1782–1789. doi: 10.1016/j.ophtha.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hulsman CA, Vingerling JR, Hofman A, Witteman JC, de Jong PT. Blood pressure, arterial stiffness, and open-angle glaucoma: the Rotterdam study. Arch Ophthalmol. 2007;125 (6:805–812. doi: 10.1001/archopht.125.6.805. [DOI] [PubMed] [Google Scholar]
- Bonomi L, Marchini G, Marraffa M, Bernardi P, Morbio R, Varotto A. Vascular risk factors for primary open angle glaucoma: the Egna-Neumarkt Study. Ophthalmology. 2000;107 (7:1287–1293. doi: 10.1016/s0161-6420(00)00138-x. [DOI] [PubMed] [Google Scholar]
- Cherecheanu AP, Garhofer G, Schmidl D, Werkmeister R, Schmetterer L. Ocular perfusion pressure and ocular blood flow in glaucoma. Curr Opin Pharmacol. 2013;13 (1:36–42. doi: 10.1016/j.coph.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leske MC. Ocular perfusion pressure and glaucoma: clinical trial and epidemiologic findings. Curr Opin Ophthalmol. 2009;20 (2:73–78. doi: 10.1097/ICU.0b013e32831eef82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leske MC, Heijl A, Hyman L, Bengtsson B, Dong L, Yang Z. Predictors of long-term progression in the early manifest glaucoma trial. Ophthalmology. 2007;114 (11:1965–1972. doi: 10.1016/j.ophtha.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Wiggs JL, Kang JH, Yaspan BL, Mirel DB, Laurie C, Crenshaw A, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet. 2011;20 (23:4707–4713. doi: 10.1093/hmg/ddr382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorleifsson G, Walters GB, Hewitt AW, Masson G, Helgason A, DeWan A, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010;42 (10:906–909. doi: 10.1038/ng.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis SJ, Kang JH, Weinreb RN, Yaspan BL, Cooke Bailey JN, Gaasterland D, et al. Association of CAV1/CAV2 genomic variants with primary open angle glaucoma overall and by gender and pattern of visual field loss. Ophthalmology. 2014;121 (2:508–516. doi: 10.1016/j.ophtha.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaspan BL, Bush WS, Torstenson ES, Ma D, Pericak-Vance MA, Ritchie MD, et al. Genetic analysis of biological pathway data through genomic randomization. Hum Genet. 2011;129 (5:563–571. doi: 10.1007/s00439-011-0956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquale LR, Loomis SJ, Kang JH, Yaspan BL, Abdrabou W, Budenz DL, et al. CDKN2B-AS1 genotype–glaucoma feature correlations in primary open-angle glaucoma patients from the United States. Am J Ophthalmol. 2013;155 (2:342–353 e345. doi: 10.1016/j.ajo.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggs J, Yaspan B, Hauser M, Kang J, Allingham R, Abdrabou W, et al. Common variants in the CDKN2BAS region on 9p21 and a novel region on 8q23 are associated with the normal tension form of open-angle glaucoma with increased susceptibility to optic nerve degeneration. PLoS Genet. 2012;8 (4:e1002654. doi: 10.1371/journal.pgen.1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Vitart V, Burdon KP, Khor CC, Bykhovskaya Y, Mirshahi A, et al. Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat Genet. 2013;45 (2:155–163. doi: 10.1038/ng.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer M, Li J, Yaspan BL, Ozel AB, Richards JE, Moroi SE, et al. Genome-wide analysis of central corneal thickness in primary open-angle glaucoma cases in the NEIGHBOR and GLAUGEN consortia. Invest Ophthalmol Vis Sci. 2012;53 (8:4468–4474. doi: 10.1167/iovs.12-9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozel AB, Moroi SE, Reed DM, Nika M, Schmidt CM, Akbari S, et al. Genome-wide association study and meta-analysis of intraocular pressure. Hum Genet. 2013;133 (1:41–57. doi: 10.1007/s00439-013-1349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggs J, Hauser M, Abdrabou W, Allingham R, Budenz D, DelBono E, et al. The NEIGHBOR Consortium Primary Open Angle Glaucoma Genome-wide Association Study: rationale, study design and clinical variables. J Glaucoma. 2012;22 (7:517–525. doi: 10.1097/IJG.0b013e31824d4fd8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81 (3:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics (Oxford, England) 2010;26 (17:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28 (1:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35 (Database issue:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GS, Miller FW, Pandey JP. The role of genetic factors in autoimmune disease: implications for environmental research. Environ Health Perspect. 1999;107 (Suppl 5:693–700. doi: 10.1289/ehp.99107s5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan S, Newton-Cheh C, Dominiczak AF. Genetic basis of blood pressure and hypertension. Trends Genet. 2012;28 (8:397–408. doi: 10.1016/j.tig.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Pasquale LR, Loomis SJ, Weinreb RN, Kang JH, Yaspan BL, Bailey JC, et al. Estrogen pathway polymorphisms in relation to primary open angle glaucoma: an analysis accounting for gender from the United States. Mol Vis. 2013;19:1471–1481. [PMC free article] [PubMed] [Google Scholar]
- Venkataraman ST, Flanagan JG, Hudson C. Vascular reactivity of optic nerve head and retinal blood vessels in glaucoma—a review. Microcirculation. 2010;17 (7:568–581. doi: 10.1111/j.1549-8719.2010.00045.x. [DOI] [PubMed] [Google Scholar]
- Orgul S, Gugleta K, Flammer J. Physiology of perfusion as it relates to the optic nerve head. Surv Ophthalmol. 1999;43 (Suppl 1:S17–S26. doi: 10.1016/s0039-6257(99)00009-0. [DOI] [PubMed] [Google Scholar]
- Jandrasits K, Luksch A, Soregi G, Dorner GT, Polak K, Schmetterer L. Effect of noradrenaline on retinal blood flow in healthy subjects. Ophthalmology. 2002;109 (2:291–295. doi: 10.1016/s0161-6420(01)00880-6. [DOI] [PubMed] [Google Scholar]
- Harris A, Ciulla TA, Chung HS, Martin B. Regulation of retinal and optic nerve blood flow. Arch Ophthalmol. 1998;116 (11:1491–1495. doi: 10.1001/archopht.116.11.1491. [DOI] [PubMed] [Google Scholar]
- Schmetterer L, Polak K. Role of nitric oxide in the control of ocular blood flow. Prog Retin Eye Res. 2001;20 (6:823–847. doi: 10.1016/s1350-9462(01)00014-3. [DOI] [PubMed] [Google Scholar]
- Haefliger IO, Dettmann E, Liu R, Meyer P, Prunte C, Messerli J, et al. Potential role of nitric oxide and endothelin in the pathogenesis of glaucoma. Surv Ophthalmol. 1999;43:S51–S58. doi: 10.1016/s0039-6257(99)00026-0. [DOI] [PubMed] [Google Scholar]
- Becquet F, Courtois Y, Goureau O. Nitric oxide in the eye: multifaceted roles and diverse outcomes. Surv Ophthalmol. 1997;42 (1:71–82. doi: 10.1016/s0039-6257(97)84043-x. [DOI] [PubMed] [Google Scholar]
- Nathanson JA. Nitric oxide and nitrovasodilators in the eye: implications for ocular physiology and glaucoma. J Glaucoma. 1993;2 (3:206–210. [PubMed] [Google Scholar]
- Wizemann AJ, Wizemann V. Organic nitrate therapy in glaucoma. Am J Ophthalmol. 1980;90 (1:106–109. doi: 10.1016/s0002-9394(14)75085-8. [DOI] [PubMed] [Google Scholar]
- Nathanson JA, McKee M. Alterations of ocular nitric oxide synthase in human glaucoma. Invest Ophthalmol Vis Sci. 1995;36 (9:1774–1784. [PubMed] [Google Scholar]
- Chuman H, Chuman T, Nao-i N, Sawada A. The effect of L-arginine on intraocular pressure in the human eye. Curr Eye Res. 2000;20 (6:511–516. [PubMed] [Google Scholar]
- Behar-Cohen FF, Goureau O, D'Hermies F, Courtois Y. Decreased intraocular pressure induced by nitric oxide donors is correlated to nitrite production in the rabbit eye. Invest Ophthalmol Vis Sci. 1996;37 (8:1711–1715. [PubMed] [Google Scholar]
- Stamer WD, Lei Y, Boussommier-Calleja A, Overby DR, Ethier CR. eNOS, a pressure-dependent regulator of intraocular pressure. Invest Ophthalmol Vis Sci. 2011;52 (13:9438–9444. doi: 10.1167/iovs.11-7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner GT, Garhofer G, Kiss B, Polska E, Polak K, Riva CE, et al. Nitric oxide regulates retinal vascular tone in humans. Am J Physiol-Heart Circul Physiol. 2003;285 (2:H631–H636. doi: 10.1152/ajpheart.00111.2003. [DOI] [PubMed] [Google Scholar]
- Luksch A, Polak K, Beier C, Polska E, Wolzt M, Dorner GT, et al. Effects of systemic NO synthase inhibition on choroidal and optic nerve head blood flow in healthy subjects. Invest Ophthalmol Vis Sci. 2000;41 (10:3080–3084. [PubMed] [Google Scholar]
- Schmetterer L, Krejcy K, Kastner J, Wolzt M, Gouya G, Findl O, et al. The effect of systemic nitric oxide synthase inhibition on ocular fundus pulsations in man. Exp Eye Res. 1997;64 (3:305–312. doi: 10.1006/exer.1996.0213. [DOI] [PubMed] [Google Scholar]
- Resch H, Garhofer G, Fuchsjager-Mayrl G, Hommer A, Schmetterer L. Endothelial dysfunction in glaucoma. Acta Ophthalmol. 2009;87 (1:4–12. doi: 10.1111/j.1755-3768.2007.01167.x. [DOI] [PubMed] [Google Scholar]
- Hashizume K, Mashima Y, Fumayama T, Ohtake Y, Kimura I, Yoshida K, et al. Genetic polymorphisms in the angiotensin II receptor gene and their association with open-angle glaucoma in a Japanese population. Invest Ophthalmol Vis Sci. 2005;46 (6:1993–2001. doi: 10.1167/iovs.04-1100. [DOI] [PubMed] [Google Scholar]
- Bunce C, Hitchings RA, Van Duijn CM, De Jong P, Vingerling JR. Associations between the deletion polymorphism of the angiotensin 1-converting enzyme gene and ocular signs of primary open-angle glaucoma. Graefes Arch Clin Exp Ophthalmol. 2005;243 (4:294–299. doi: 10.1007/s00417-004-1025-5. [DOI] [PubMed] [Google Scholar]
- Ozkur M, Erbagci B, Gungor K, Nacak M, Aynacioglu S, Bekir NA. Angiotensin-converting enzyme insertion–deletion polymorphism in primary open-angle glaucoma. Ophthalmologica. 2004;218 (6:415–418. doi: 10.1159/000080946. [DOI] [PubMed] [Google Scholar]
- Inagaki Y, Mashima Y, Fuse N, Funayama T, Ohtake Y, Yasuda N, et al. Polymorphism of beta-adrenergic receptors and susceptibility to open-angle glaucoma. Mol Vis. 2006;12 (73–75:673–680. [PubMed] [Google Scholar]
- Gungor K, Ozkur M, Cascorbi I, Brockmoller J, Bekir N, Roots I, et al. Beta 2-adrenergic receptor polymorphism and susceptibility to primary congenital and primary open angle glaucoma. Eur J Clin Pharmacol. 2003;59 (7:527–531. doi: 10.1007/s00228-003-0658-3. [DOI] [PubMed] [Google Scholar]
- Ayub H, Khan MI, Micheal S, Akhtar F, Ajmal M, Shafique S, et al. Association of eNOS and HSP70 gene polymorphisms with glaucoma in Pakistani cohorts. Mol Vis. 2010;16 (2–3:18–25. [PMC free article] [PubMed] [Google Scholar]
- da Silva TM, Rocha AV, Lacchini R, Marques CR, Silva ES, Tanus-Santos JE, et al. Association of polymorphisms of endothelial nitric oxide synthase (eNOS) gene with the risk of primary open angle glaucoma in a Brazilian population. Gene. 2012;502 (2:142–146. doi: 10.1016/j.gene.2012.04.047. [DOI] [PubMed] [Google Scholar]
- Kang JH, Wiggs JL, Rosner BA, Hankinson SE, Abdrabou W, Fan BJ, et al. The relation between endothelial nitric oxide synthase gene variants and primary open-angle glaucoma: interactions with gender and postmenopausal hormone use. Invest Ophthalmol Vis Sci. 2010;51:971–979. doi: 10.1167/iovs.09-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao QO, Wang DH, Sun HJ. Association of genetic polymorphisms of eNOS with glaucoma. Mol Vis. 2011;17 (17-20:153–158. [PMC free article] [PubMed] [Google Scholar]
- Logan JF, Chakravarthy U, Hughes AE, Patterson CC, Jackson JA, Rankin SJ. Evidence for association of endothelial nitric oxide synthase gene in subjects with glaucoma and a history of migraine. Invest Ophthalmol Vis Sci. 2005;46 (9:3221–3226. doi: 10.1167/iovs.05-0368. [DOI] [PubMed] [Google Scholar]
- Tunny TJ, Richardson KA, Clark CV. Association study of the 5′ flanking regions of endothelial-nitric oxide synthase and endothelin-1 genes in familial primary open-angle glaucoma. Clin Exp Pharmacol Physiol. 1998;25 (1:26–29. doi: 10.1111/j.1440-1681.1998.tb02138.x. [DOI] [PubMed] [Google Scholar]
- Weiss J, Frankl SA, Flammer J, Grieshaber MC, Hollo G, Teuchner B, et al. No difference in genotype frequencies of polymorphisms of the nitric oxide pathway between Caucasian normal and high tension glaucoma patients. Mol Vis. 2012;18 (229–31:2174–2181. [PMC free article] [PubMed] [Google Scholar]
- Fan BJ, Liu K, Wang DY, Tham CCY, Tam POS, Lam DSC, et al. Association of polymorphisms of tumor necrosis factor and tumor protein p53 with primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2010;51 (8:4110–4116. doi: 10.1167/iovs.09-4974. [DOI] [PubMed] [Google Scholar]
- Rath G, Dessy C, Feron O. Caveolae, caveolin and control of vascular tone: nitric oxide (NO) and endothelium derived hyperpolarizing factor (EDHF) regulation. J Physiol Pharmacol. 2009;60 (Suppl 4:105–109. [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10 (11:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101 (16:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SY, Singh K, Lin SC. The association between glaucoma prevalence and supplementation with the oxidants calcium and iron. Invest Ophthalmol Vis Sci. 2012;53 (2:725–731. doi: 10.1167/iovs.11-9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.