Abstract
Cancer research has seen tremendous changes over the past decade. Fast progress in sequencing technology has afforded us with landmark genetic alterations, which had immediate impact on clinical science and practice by pointing to new kinase targets, such as PI 3-Kinase, the EGF receptor or BRAF. The PI 3-Kinase pathway for growth control has emerged as a prime example for both oncogene activation and tumor suppressor loss in cancer.
Here, we discuss how therapy using PI 3-kinase pathway inhibitors could benefit from information on specific phosphatases, which naturally antagonize the kinase targets. This PI 3-Kinase pathway is found mutated in most cancer types, including prostate, breast, colon and brain tumors. The tumor suppressing phosphatases operate at two levels. Lipid level phosphatases, such as PTEN and INPP4b revert PI 3-kinase activity to keep the lipid second messengers inactive. At the protein level, PHLPP1/2 protein phosphatases inactivate AKT kinase, thus antagonizing mTOR complex 2 activity. However, in contrast to their kinase counterparts the phosphatases are unlikely drug targets. They would need to be stimulated by therapy and are commonly deleted and mutated in cancer. Yet, since they occupy critical nodes in preventing cancer initiation and progression, the information on their status has tremendous potential in outcome prediction, and in matching the available kinase inhibitor repertoire with the right patients.
1. Background
1.1. The PTEN/ PI 3-Kinase pathway
Phosphatase and Tensin homologue deleted on chromosome Ten (PTEN) was discovered in 1997 as the result of a chase for the candidate tumor suppressor in the frequently deleted chromosome 10q23 region (1, 2). The two teams immediately saw that the gene encodes a phosphatase, which launched a flurry of investigations for its substrate. In spite of the logical appeal for a phosphatase tumor suppressor to reverse the action of an oncogenic protein kinase, a landmark study identified the PTEN substrate to be the membrane phospholipid Phosphatidylinositol 3,4,5 trisphosphate, PI(3,4,5)P3 (3). Since PTEN showed specificity for removing the phosphate at the 3-position of the inositol ring (creating PI(4,5)P2) it immediately became clear that its activity antagonizes the previously identified class I PI 3-Kinases, which conversely phosphorylate the PI(4,5)P2 lipid at that position (4), (reviewed in (5)). These results gave birth to our current concept of PTEN and the class I PI 3-Kinases as top level communicators of growth control in cancer (see Figure). Today we know that this pathway constitutes the major oncogenic signaling axis next to the RAS/ MAP Kinase pathway.
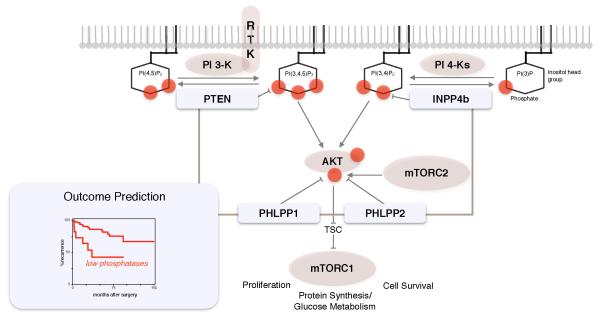
Figure 1.
Core phosphatases of the PI 3-Kinase pathway. PTEN and INPP4b phosphatases inactivate PIP-lipid second messengers to prevent AKT activation. Functionally, they both antagonize class I PI 3-Kinase dependent membrane recruitment of AKT. PHLPP1 and PHLPP2 revert AKT activation by dephosphorylation at Serine 473 to antagonize the phosphorylation that the mTOR Complex 2 carries out on this site. AKT activation signals mTORC1 activation via inhibition of the TSC tumor suppressor complex.
Outcome prediction. Phosphatase status at the DNA, RNA or protein level can be used to predict disease outcome.
In this Review, we discuss how the lipid and protein level phosphatases cooperate to protect from cancer and how their use as biomarkers could assist outcome prediction and therapy approach.
Phosphoinositide 3-kinases (PI3Ks) constitute a conserved family of lipid kinases that phosphorylate phosphoinositides (PIs) at the 3-position of their inositol head group (5). The family is classified into several subtypes depending on the substrate PIs that they can phosphorylate, yet the class I PI3Ks are unique: only they can create the master growth control second messenger, PI(3,4,5)P3 (below termed PIP3 ). The class IA PI3Ks relay extracellular growth and survival signals into the cell by producing PIP3 after activation by ligand bound receptor tyrosine kinases (RTKs). The PIP3 lipid then attracts proteins such as AKT kinase and its activating kinase 3-Phosphoinositide-dependent protein kinase 1 (PDK1) via their Pleckstrin Homology (PH) domains, thus converting the lipid-phosphorylation code into protein signaling cascades (see Figure). Accordingly, tumor suppression by the pathway’s phosphatases occurs at two fundamentally different levels: lipid level phosphatases convert the actively signaling PIP3 lipids to their inactive isoforms, and protein level phosphatases inactivate the downstream phosphorylated proteins back to the non-phosphorylated state. Below, we discuss the functions and interactions among the pathway’s major phosphatases, and their potential in predicting disease outcomes and therapy response.
1. 2. The lipid level phosphatases
The phosphatidylinositol membrane lipids (PIPs) constitute only a few percent of total membrane lipid mass, consistent with the notion that they do not define physical membrane properties but instead serve as top level intracellular second messengers for signaling (6). Of the seven naturally occurring PIPn phospho-isoforms, PIP3 executes the major known signaling function in cancer. However, PIP3 is only present at very low levels in cell membranes (6) reflecting the transient nature of signaling at the level of the lipids. Keeping these second messenger levels low is the major known function of the phosphatase PTEN (7).
PTEN
PTEN occupies a unique position in antagonizing PI-3 Kinases (see Figure) by dephosphorylating PI(3,4,5)P3 to PI(4,5)P2. To date, it represents the most efficient suppressor of the PI 3-Kinase pathway. In vitro, it inhibits cell growth, survival and proliferation so efficiently that stable expression in cancer cells is very hard to achieve. The PTEN gene has been mutated in heritable cancer syndromes, which are now collectively referred to as the PTEN Hamartoma Tumor Syndromes (PTHS), with Cowden Disease, Proteus-like- and Bannayan-Riley-Ruvalcaba syndromes showing the highest PTEN germline mutation frequency (80%, 60% and 60%, respectively, (8). Somatic PTEN-alterations are seen in such a high number of human epithelial cancers that they rival those of p53 (9), see also COSMIC database, http://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=PTEN). However, deregulation of PTEN that does not involve gene alterations is also of critical importance in cancer (10, 11). Primary human prostate cancers for example, frequently present with partial reduction of the PTEN protein but rarely with complete gene inactivation (12-14). Animal models have shown that Pten is haploinsufficient in prostate and other cancer types (15-18). Given the prominent role of PTEN in cancer signaling, this led to the realization that the classical ‘two-hit-hypothesis’ for tumor suppression (19) needed to be expanded (reviewed in (20)). Analysis of PTEN alteration in surgically removed prostate cancers has revealed that 86% of tumors retain the gene while protein loss or strong reduction is found at 75% percent frequency (13). Similar observations were made in colorectal and lung cancer (21, 22) where PTEN protein loss is far more frequent than gene/ RNA loss, and in thyroid cancer, endocrine pancreatic tumors, and melanoma where PTEN is often lost from cell nuclei (23-25) (reviewed in (26). These cases mostly exhibit normal RNA levels, suggesting that PTEN protein degradation is a common cause of cancer formation (reviewed in (26). The finding that homozygous PTEN-loss triggers p53-mediated senescence explained why the partial loss is widespread in cancer cells with functional p53 (27). In prostate for example, p53-deletion with concurrent deletion of PTEN is frequently observed in metastasis, but not in primary disease (12, 13).
INPP4b
The inositol polyphosphate 4-phosphatase (INPP4) activity was first identified in brain lysates (28) and cloning of the human cDNA revealed that it was widely expressed and responsible for most of the cell’s phosphatase activity towards the 4’ position on PIPs (29). A second human gene sharing 42% identity at the protein level was soon identified and classified as Type II phosphatase (30) with the gene name INPP4b. In vitro analysis showed that the enzyme primarily hydrolyzes PI(3,4)P2 at the 4-position through its C-terminal catalytic domain to generate PI(3)P (29), a result that was confirmed in cells (31). Through this action, INPP4B suppresses P1(3,4)P2, which can still serve as a platform for AKT recruitment and activation (31). In agreement, knockdown of INPP4b was found to phenocopy hallmarks of PTEN-suppression, such as increased AKT signaling triggered by insulin (31, 32) and showed an increased p53-dependent cellular senescence response upon co-suppression with PTEN. Taken together, PTEN and INPP4b have emerged as the most strongly cooperating lipid level PI 3-Kinase pathway phosphatases.
1. 3. The Protein level phosphatases
PHLPP1
The Pleckstrin Homology domain Leucine-rich repeat Protein Phosphatase 1 (PHLPP1) was discovered in a logical search for AKT antagonists that link a phosphatase- to a PH-domain (33). PHLPP1, previously implicated in circadian rhythms (34), fulfilled these criteria and was confirmed to directly dephosphorylate and inactive AKT at the serine 473 activation site (33) in addition to other targets (reviewed in (35)). The gene locates to chromosome 18q21 and expresses two alternatively spliced isoforms. PHLPP1b differs from PHLPP1a by a 50Kd N-terminal extension, containing a RAS-association domain of still unclear function. Both isoforms share the same features: the PH domain, leucine-rich repeats (LRR), a PP2C catalytic domain and a PDZ-ligand domain (reviewed in (35)). PHLPP1 has been well-studied in cancer models (13, 36-38), development and function of T cells (39), cardiac cell survival (40), and circadian rhythms (41). In the mouse prostate, Phlpp1-loss leads to Akt-driven neoplasia and synergizes with partial Pten-loss to accelerate tumor proliferation, onset and incidence. Importantly, loss of Phlpp1, just like loss of Pten, triggers activation of p53 which causes cellular senescence. This response acts as a barrier against disease progression and is spontaneously overcome by p53-inactivation in the mutant mice (13).
PHLPP2
PHLPP2 (on chromosome 16q21) shares its domain structure with the longer PHLPP1b splice isoform (42). Just like PHLPP1, PHLPP2 is also ubiquitously expressed in most tissues and shows highest abundance in the brain. PHLPP2 also acts predominantly on the serine 473 site of AKT and differential specificities of PHLPP1/2 for AKT1/2/3 have been reported in vitro (43, 44) and are being investigated further in animal knockout models. One critical distinction between Phlpp1 and Phlpp2 is their differential response to PI 3-Kinase pathway activation in genetically controlled experiments: while Phlpp1 levels remain constant, Phlpp2 is surging to antagonize Akt (13). Thus, after Pten-loss, Phlpp2 is critically attenuating pathway output. It remains to be seen if the two PHLPP isoforms show differential tissue specific-roles for this function. Mechanistically, the PHLPP protein levels are regulated downstream of mTORC1, thus linking levels and activity to PI 3-kinase pathway output in a negative feedback (13, 45) (reviewed in (46)). This role of PHLPP2 in partially substituting for PTEN may explain the frequent co-deletion of PTEN and PHLPP2 in lethal prostate cancers.
2. Clinical-Translational Advances
2.1 Phosphatases as prognostic biomarkers
Single gene associations
PTEN has been extensively investigated as biomarker for prediction of disease outcome across many cancer types (47). As a single factor, low PTEN gene expression is associated with prostate metastasis (48), faster rising PSA levels after surgery, (13, 49), and with castration-resistance (50). In breast cancer, signatures of PTEN-loss have been associated with poor prognosis (51), similar to findings in colon (52). Several studies showed the correlation between disease progression and/ or relapse after intervention, when low or absent PTEN protein levels were detected in a prostate tumor (53-55). Similarly, PTEN protein status has been associated with better response to HER2 target therapy in breast cancer (56-59), although the straightforward correlation has been called into question by a recent large-scale study (60), which highlighted a major issue with pathway biomarkers. How much reduction of a tumor suppressor is called to be functionally relevant ((61), see also Conclusions)?
Gene loss and reduction of INPP4b protein has been frequently seen in breast and ovarian cancer and correlated with increased progression of disease and shorter overall survival (31, 32). In basal-like breast cancer furthermore, loss of INPP4b (and PTEN) strongly correlated with PI 3-Kinase pathway activation as confirmed through the TCGA consortium ((32, 62) reviewed in (63)). INPP4b protein is also frequently lowered in prostate cancer, an event that was associated with shorter times to biochemical recurrence ((64), reviewed in (65)). Intriguingly, the study found that the androgen receptor (AR) positively regulated INPP4b transcription and protein levels, consistent with an emerging pattern of AR-mediated suppression of AKT: two recent studies demonstrated that AR also positively regulates PHLPP1 to suppress AKT signaling (37, 38). These findings suggest that anti-hormone therapy could come at the price of increased AKT activity, when INPP4b (and PHLPP1) are intact. Decreased INPP4b expression was furthermore found to correlate with disease progression in melanocytic tumors (66). Collectively, these results confirm the key pathway position of INPP4b and point to its usefulness as pathway biomarker.
The PHLPP phosphatases have quickly moved into the spotlight of tumor suppressor studies by virtue of their ability to directly dephosphorylate AKT kinase. Strong evidence to confirm the mouse tumor suppressor function of Phlpp1 in human has come from studies on prostate cancer where the gene is frequently deleted (12). The expression analysis of clinically annotated patient samples from this study revealed significant association of low PHLPP1/2 expression with disease recurrence after surgery (13). In colon cancer, reduced protein levels have been described (67) and PHLPP1-status has also been linked to treatment response after chemo- and hormone therapy (37, 38, 68). A cancer associated polymorphism in the PHLPP2 gene that reduces AKT-suppressing activity has been identified (69) and validated in breast and ovarian cancer (70, 71). Furthermore, several studies have shown a compensatory role for PHLPP proteins after pathway activation through e.g. PTEN-loss. This failsafe response is triggered by aberrant mTOR activation and serves to limit cell proliferation (13, 45). Thus, on the one hand, the PHLPP phosphatases are emerging as critical pathway breaks at the protein level, and on the other hand they serve as rheostats that actively dampen the malfunction of lipid level phosphatases.
2.2 Recovering phosphatase function through target therapy
At the lipid level, several candidate drugs against PI 3-Kinases have shown success and are currently in advanced clinical trials (see Figure, Lipid kinase inhibitors). Functionally, this approach supports or recovers PTEN or INPP4b activity, or it reverts activity of mutant PI 3-Kinase. The BKM120 inhibitor (Buparlisib), which has activity against all four isoforms of the class I PI 3-Kinase catalytic subunit (p110 alpha, beta, gamma delta), is currently in a Phase III trial for metastatic (HR+, HER2−) breast cancer. Other approaches include isoform specific inhibitors, such as CAL101 (Idelalisib), a p110-delta inhibitor, which Phase III trials showed a significant improvement in overall survival for Chronic Lymphocytic Leukemia (CLL) (72), in this type of cancer PTEN LOH has been observed at 20% frequency (73).
The PI 3-kinase pathway has been successfully targeted at the protein level after the discovery of the naturally occurring mTORC1 inhibitor rapamycin and its derivatives, the rapalogs. They are used as immunosuppressants after organ transplantation as they inhibit T-cell activation (74). The RAD001 derivative (Everolimus) has been FDA approved in 2010 for the treatment of Tuberous Sclerosis (TSC) syndrome, which predis-poses patients with inherited TSC mutations (see Figure) to precancerous lesions. In cancer, the drug has been approved for advanced kidney cancer, a TSC associated astrocytoma, a HR+, HER2− breast cancer subtype and for treatment of pancreatic neuroendocrine tumors (PNETs) (reviewed in (75)). AKT inhibitors are targeting the protein by two mechanisms. Allosteric inhibitors, such as MK2206 or Perifosine prevent translocation of AKT to the plasma membrane, thus it cannot be activated by phosphorylation. The ATP-competitive inhibitor (e.g GSK2110183, Afuresertib) in contrast targets the AKT active site, which results in hyperphosphorylation of the kinase. Both MK2206 and GSK2110183 are currently in Phase II trials against blood and solid cancers (see Figure), while the Phase III trials of Perifosine in colon cancer and multiple myeloma have not shown significant results.
Finally, several promising compounds exhibit so-called dual specificity, due to the close evolutionary relationship between the kinase domains of PI 3-kinases and mTOR– in spite of the diversification into lipid and protein specific kinases (76). Although no dual specificity inhibitor has so far been FDA approved, several of them are currently used in Phase 2 trials (see Figure) against PNET and other advanced cancers.
3. Conclusions
The lipid and protein level phosphatases of the PI 3-Kinase pathway form a tight natural network against cancer, which should be routinely monitored at the genetic and protein level to assist outcome prediction. In addition, successful drug discovery programs have afforded us with many compounds that can support or replace core functions of these phosphatases when they are lost. The challenge now consists in matching molecular genetic events on the phosphatase side with therapeutic strategies on the inhibitor side. It remains unclear if alterations at a specific level of the pathway sensitize cancers to drugs that inhibit at the same exact level. For example, one could envision PTEN-mutant tumors to be more sensitive to PI 3-kinase than Akt- or mTORC-inhibitors. Such linkages can be established in controlled, primary model systems using pharmacological and genetic tools. However, it is also expected that these linkages are perturbed by the context of spontaneous aberrations and feedbacks that arise in a tumor (77). Furthermore, functional readouts for the relevance of phosphatase alterations are needed to prevent false status calls, as recently suggested by comparing different studies on PTEN status in trastuzumab therapy of breast cancer (reviewed in (61)). Yet in spite of this complexity, there is hope for discovering distinct linkages between alterations and therapeutic effects. Patients harboring TSC germline mutations that activate mTORC1 clearly benefit from targeting of mTORC1 with rapalogs. Thus it can be envisioned, that the precise matching of drugs with predetermined pathway defects and known resistance routes of a tumor may provide a winning anti-cancer strategy.
Table 1. Clinical trial relevance.
Phosphatase status could be used for patient stratification into matching clinical trials. Current Phase II/III trials against pathway targets at the lipid and/ or protein level are listed. Phosphatases which are functionally supported by each approach are indicated in the Relevant Phosphatase column.
| Kinase target(s) | Antagonistic phosphatase |
Drug | Trial phase |
Cancer | Trial ID | ||
|---|---|---|---|---|---|---|---|
|
Protein
kinase in- hibitors |
AKT | GSK2110183 | Phase 2 | Solid tumors, hemato- logic malignancies |
NCT01531894 | ||
| AKT | MK2206 | Phase 2 | Relapsed or refractory acute myeloid leukemia |
NCT01253447 | |||
| mTORC1 | Everolimus | Phase 2 | Melanoma | NCT01960829 | |||
| mTORC1 | Sirolimus | Phase 2 | Hepatocellular carci- noma |
NCT01374750 | |||
| mTORC1 | Temsirolimus | Phase 1/2 | Advanced cancers | NCT00877773 | |||
| mTORC1/2 | INK128 | Phase 1 | Advanced non- hematologic malignan-cies |
NCT01899053 | |||
| mTORC1/2 | PHLPP1/2 | OSI-027 | Phase 1 | Solid tumor, lymphoma | NCT00698253 | ||
| mTORC1/2 | AZD8055 | Phase 1 | Glioblastoma multiforme, other brain tumors |
NCT01316809 | |||
|
Lipid ki-
nase in- hibitors |
PI3K | BAY80-6946 | Phase 2 | Non-Hodgkin lympho- ma |
NCT01660451 | ||
| PI3K | BKM120 | Phase 3 | Metastatic breast cancer HR+, HER2- |
NCT01633060 | |||
| PI3K | PTEN | CAL101 | Phase 3 | Chronic lymphocytic leukemia |
NCT01659021 | ||
| PI3K | GDC0941 | Phase 2 | Non-small cell lung cancer |
NCT01493843 | |||
| PI3K | IPI145 | Phase 2 | Indolent non-Hodgkin lymphoma |
NCT01882803 | |||
| PI3K | XL147 | Phase 1/2 | Breast cancer, breast neoplasms |
NCT01042925 | |||
|
Dual spec-
ificity in- hibitors |
PI3K | mTORC1/2 | BEZ235 | Phase 2 | Pancreatic neuroendo- crine tumors (pNET) |
NCT01628913 | |
| PI3K | mTORC1/2 | PTEN | BGT226 | Phase 1/2 | Advanced breast cancer | NCT00600275 | |
| PI3K | mTORC1/2 | PF04691502 | Phase 2 | Endometrial neoplasms | NCT01420081 | ||
| PI3K | mTORC1/2 | PHLPP1/2 | PF05212384 | Phase 2 | Metastatic colorectal cancer |
NCT01925274 | |
| PI3K | mTORC1/2 | XL765 | Phase 1 | Glioblastoma, astrocy- toma |
NCT01240460 | ||
Acknowledgments
We thank members of the Trotman Lab and D. Tsang for valuable discussion and help with the manuscript. This work was supported by grants to L.C. Trotman from the NIH (CA137050), the Department of the Army (W81XWH-09-1-0557), the STARR foundation, and the Robertson Research Fund of Cold Spring Harbor Laboratory.
Abbreviations
- LOH
Loss of Heterozygosity
- PIPs
phosphatidylinositol membrane lipids
- PIs
phosphoinositides
- PI(3,4,5)P3
Phosphatidylinositol 3,4,5 trisphosphate
- PI(4,5)P2
Phosphatidylinositol 4,5 bisphosphate
- PI3Ks
Phosphoinositide 3-kinases
- RTKs
receptor tyrosine kinases
- PDK1
3-Phosphoinositide-dependent protein kinase 1
- INPP4
Inositol polyphosphate 4-phosphatase
- INPP4B
Inositol polyphosphate 4-phosphatase type II
- PHLPP1
Pleckstrin Homology domain Leucine-rich repeat Protein Phosphatase 1
- PHLPP2
Pleckstrin Homology domain Leucine-rich repeat Protein Phosphatase 2
- Trp53
Transformation Related Protein 53 gene, p53-gene
Footnotes
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
Note: We aim to consistently use the convention for nomenclature rules of human genes (e.g. PTEN), proteins (PTEN), and mouse gene and protein homologues (Pten, Pten, respectively) wherever possible.
References
- 1.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 2.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–62. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 3.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–8. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 4.Whitman M, Downes CP, Keeler M, Keller T, Cantley L. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature. 1988;332:644–6. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- 5.Vanhaesebroeck B, Stephens L, Hawkins P. PI3K signalling: the path to discov ery and understanding. Nat Rev Mol Cell Biol. 2012;13:195–203. doi: 10.1038/nrm3290. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Bankaitis VA. Phosphoinositide phosphatases in cell biology and disease. Prog Lipid Res. 2010;49:201–17. doi: 10.1016/j.plipres.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–8. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 8.Mester J, Eng C. When overgrowth bumps into cancer: the PTEN-opathies. Am J Med Genet C Semin Med Genet. 2013;163C:114–21. doi: 10.1002/ajmg.c.31364. [DOI] [PubMed] [Google Scholar]
- 9.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer ge nomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leslie NR, Foti M. Non-genomic loss of PTEN function in cancer: not in my genes. Trends Pharmacol Sci. 2011;32:131–40. doi: 10.1016/j.tips.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–96. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 12.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Inte grative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M, Pratt CP, Zeeman ME, Schultz N, Taylor BS, O’Neill A, et al. Identifica tion of PHLPP1 as a tumor suppressor reveals the role of feedback activation in PTEN-mutant prostate cancer progression. Cancer Cell. 2011;20:173–86. doi: 10.1016/j.ccr.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwabi-Addo B, Giri D, Schmidt K, Podsypanina K, Parsons R, Greenberg N, et al. Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer pro gression. Proc Natl Acad Sci U S A. 2001;98:11563–8. doi: 10.1073/pnas.201167798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–55. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 18.Alimonti A, Carracedo A, Clohessy JG, Trotman LC, Nardella C, Egia A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nature Genet. 2010;42:454–8. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppres sion. Nature. 2011;476:163–9. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naguib A, Cooke JC, Happerfield L, Kerr L, Gay LJ, Luben RN, et al. Alterations in PTEN and PIK3CA in colorectal cancers in the EPIC Norfolk study: associations with clinicopathological and dietary factors. BMC Cancer. 2011;11:123. doi: 10.1186/1471-2407-11-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yanagawa N, Leduc C, Kohler D, Saieg MA, John T, Sykes J, et al. Loss of phosphatase and tensin homolog protein expression is an independent poor prognostic marker in lung adenocarcinoma. J Thorac Oncol. 2012;7:1513–21. doi: 10.1097/JTO.0b013e3182641d4f. [DOI] [PubMed] [Google Scholar]
- 23.Gimm O, Perren A, Weng LP, Marsh DJ, Yeh JJ, Ziebold U, et al. Differential nu clear and cytoplasmic expression of PTEN in normal thyroid tissue, and benign and ma lignant epithelial thyroid tumors. Am J Pathol. 2000;156:1693–700. doi: 10.1016/s0002-9440(10)65040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whiteman DC, Zhou XP, Cummings MC, Pavey S, Hayward NK, Eng C. Nuclear PTEN expression and clinicopathologic features in a population-based series of primary cutaneous melanoma. Int J Cancer. 2002;99:63–7. doi: 10.1002/ijc.10294. [DOI] [PubMed] [Google Scholar]
- 25.Perren A, Komminoth P, Saremaslani P, Matter C, Feurer S, Lees JA, et al. Mu tation and expression analyses reveal differential subcellular compartmentalization of PTEN in endocrine pancreatic tumors compared to normal islet cells. Am J Pathol. 2000;157:1097–103. doi: 10.1016/S0002-9440(10)64624-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naguib A, Trotman LC. PTEN plasticity: how the taming of a lethal gene can go too far. Trends Cell Biol. 2013;23:374–9. doi: 10.1016/j.tcb.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collado M, Serrano M. Senescence in tumours: evidence from mice and hu mans. Nat Rev Cancer. 2010;10:51–7. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norris FA, Majerus PW. Hydrolysis of phosphatidylinositol 3,4-bisphosphate by inositol polyphosphate 4-phosphatase isolated by affinity elution chromatography. J Biol Chem. 1994;269:8716–20. [PubMed] [Google Scholar]
- 29.Norris FA, Auethavekiat V, Majerus PW. The isolation and characterization of cDNA encoding human and rat brain inositol polyphosphate 4-phosphatase. J Biol Chem. 1995;270:16128–33. doi: 10.1074/jbc.270.27.16128. [DOI] [PubMed] [Google Scholar]
- 30.Norris FA, Atkins RC, Majerus PW. The cDNA cloning and characterization of inositol polyphosphate 4-phosphatase type II. Evidence for conserved alternative splic ing in the 4-phosphatase family. J Biol Chem. 1997;272:23859–64. doi: 10.1074/jbc.272.38.23859. [DOI] [PubMed] [Google Scholar]
- 31.Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16:115–25. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fedeale CG, Ooms LM, Ho M, Vieusseux J, O’Toole SA, Millar EK, et al. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human ba sal-like breast cancers. Proc Natl Acad Sci U S A. 2010;107:22231–6. doi: 10.1073/pnas.1015245107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphory lates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Shimizu K, Okada M, Nagai K, Fukada Y. Suprachiasmatic nucleus circadian os cillatory protein, a novel binding partner of K-Ras in the membrane rafts, negatively reg ulates MAPK pathway. J Biol Chem. 2003;278:14920–5. doi: 10.1074/jbc.M213214200. [DOI] [PubMed] [Google Scholar]
- 35.Newton AC, Trotman LC. Turning off AKT: PHLPP as a drug target. Ann Rev Pharmacol Toxicol. 2014;54:537–58. doi: 10.1146/annurev-pharmtox-011112-140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molina JR, Agarwal NK, Morales FC, Hayashi Y, Aldape KD, Cote G, et al. PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene. 2012;31:1264–74. doi: 10.1038/onc.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carver BS, Chapinski CC, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN--deficient prostate cancer. Cancer Cell. 2011;19:575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 2011;19:792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patterson SJ, Han JM, Garcia R, Assi K, Gao T, O’Neill A, et al. Cutting edge: PHLPP regulates the development, function, and molecular signaling pathways of regu latory T cells. J Immunol. 2011;186:5533–7. doi: 10.4049/jimmunol.1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyamoto S, Purcell NH, Smith JM, Gao T, Whittaker R, Huang K, et al. PHLPP-1 negatively regulates Akt activity and survival in the heart. Circ Res. 2010;107:476–84. doi: 10.1161/CIRCRESAHA.109.215020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu K, Okada M, Takano A, Nagai K. SCOP, a novel gene product ex pressed in a circadian manner in rat suprachiasmatic nucleus. FEBS Letters. 1999;458:363–9. doi: 10.1016/s0014-5793(99)01190-4. [DOI] [PubMed] [Google Scholar]
- 42.Brognard J, Newton AC. PHLiPPing the switch on Akt and protein kinase C sig naling. Trends Endocrinol Metabo. 2008;19:223–30. doi: 10.1016/j.tem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007;25:917–31. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 44.Nitsche C, Edderkaoui M, Moore RM, Eibl G, Kasahara N, Treger J, et al. The phosphatase PHLPP1 regulates Akt2, promotes pancreatic cancer cell death, and inhib its tumor formation. Gastroenterology. 2012;142:377–87. e1–5. doi: 10.1053/j.gastro.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J, Stevens PD, Gao T. mTOR-dependent regulation of PHLPP expression controls the rapamycin sensitivity in cancer cells. J Biol Chem. 2011;286:6510–20. doi: 10.1074/jbc.M110.183087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newton AC, Trotman LC. Turning Off AKT: PHLPP as a Drug Target. Annu Rev Pharmacol Toxicol. 2014;54:537–58. doi: 10.1146/annurev-pharmtox-011112-140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of com mon cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koksal IT, Dirice E, Yasar D, Sanlioglu AD, Ciftcioglu A, Gulkesen KH, et al. The assessment of PTEN tumor suppressor gene in combination with Gleason scoring and serum PSA to evaluate progression of prostate carcinoma. Urol Oncol. 2004;22:307–12. doi: 10.1016/j.urolonc.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 49.Yoshimoto M, Cunha IW, Coudry RA, Fonseca FP, Torres CH, Soares FA, et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer. 2007;97:678–85. doi: 10.1038/sj.bjc.6603924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bertram J, Peacock JW, Fazli L, Mui AL, Chung SW, Cox ME, et al. Loss of PTEN is associated with progression to androgen independence. Prostate. 2006;66:895–902. doi: 10.1002/pros.20411. [DOI] [PubMed] [Google Scholar]
- 51.Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She QB, Maurer M, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104:7564–9. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colakoglu T, Yildirim S, Kayaselcuk F, Nursal TZ, Ezer A, Noyan T, et al. Clinicopathological significance of PTEN loss and the phosphoinositide 3-kinase/Akt pathway in sporadic colorectal neoplasms: is PTEN loss predictor of local recurrence? Am J Surg. 2008;195:719–25. doi: 10.1016/j.amjsurg.2007.05.061. [DOI] [PubMed] [Google Scholar]
- 53.Chaux A, Peskoe SB, Gonzalez-Roibon N, Schultz L, Albadine R, Hicks J, et al. Loss of PTEN expression is associated with increased risk of recurrence after prosta tectomy for clinically localized prostate cancer. Mod Pathol. 2012;25:1543–9. doi: 10.1038/modpathol.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmitz M, Grignard G, Margue C, Dippel W, Capesius C, Mossong J, et al. Complete loss of PTEN expression as a possible early prognostic marker for prostate cancer metastasis. Int J Cancer. 2007;120:1284–92. doi: 10.1002/ijc.22359. [DOI] [PubMed] [Google Scholar]
- 55.Lotan TL, Gurel B, Sutcliffe S, Esopi D, Liu W, Xu J, et al. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical co hort of prostate cancer patients. Clin Cancer Res. 2011;17:6563–73. doi: 10.1158/1078-0432.CCR-11-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Razis E, Bobos M, Kotoula V, Eleftheraki AG, Kalofonos HP, Pavlakis K, et al. Evaluation of the association of PIK3CA mutations and PTEN loss with efficacy of trastuzumab therapy in metastatic breast cancer. Breast Cancer Res Treat. 2011;128:447–56. doi: 10.1007/s10549-011-1572-5. [DOI] [PubMed] [Google Scholar]
- 57.Esteva FJ, Guo H, Zhang S, Santa-Maria C, Stone S, Lanchbury JS, et al. PTEN, PIK3CA, p-AKT, and p-p70S6K status: association with trastuzumab response and sur vival in patients with HER2-positive metastatic breast cancer. Am J Pathol. 2010;177:1647–56. doi: 10.2353/ajpath.2010.090885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujita T, Doihara H, Kawasaki K, Takabatake D, Takahashi H, Washio K, et al. PTEN activity could be a predictive marker of trastuzumab efficacy in the treatment of ErbB2-overexpressing breast cancer. Br J Cancer. 2006;94:247–52. doi: 10.1038/sj.bjc.6602926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–27. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 60.Perez EA, Dueck AC, McCullough AE, Chen B, Geiger XJ, Jenkins RB, et al. Im pact of PTEN protein expression on benefit from adjuvant trastuzumab in early-stage human epidermal growth factor receptor 2-positive breast cancer in the North Central Cancer Treatment Group N9831 trial. J Clin Oncol. 2013;31:2115–22. doi: 10.1200/JCO.2012.42.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rexer BN, Shyr Y, Arteaga CL. Phosphatase and tensin homolog deficiency and resistance to trastuzumab and chemotherapy. J Clin Oncol. 2013;31:2073–5. doi: 10.1200/JCO.2012.48.5243. [DOI] [PubMed] [Google Scholar]
- 62.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lehmann BD, Pietenpol JA. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J Pathol. 2014;232:142–50. doi: 10.1002/path.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hodgson MC, Shao LJ, Frolov A, Li R, Peterson LE, Ayala G, et al. Decreased expression and androgen regulation of the tumor suppressor gene INPP4B in prostate cancer. Cancer Res. 2011;71:572–82. doi: 10.1158/0008-5472.CAN-10-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Agoulnik IU, Hodgson MC, Bowden WA, Ittmann MM. INPP4B: the new kid on the PI3K block. Oncotarget. 2011;2:321–8. doi: 10.18632/oncotarget.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perez-Lorenzo R, Gill KZ, Shen CH, Zhao FX, Zheng B, Schulze HJ, et al. A tu mor suppressor function for the lipid phosphatase INPP4B in melanocytic neoplasms. J Invest Dermatol. 2013 Nov 29; doi: 10.1038/jid.2013.511. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 67.Liu J, Weiss HL, Rychahou P, Jackson LN, Evers BM, Gao T. Loss of PHLPP expression in colon cancer: role in proliferation and tumorigenesis. Oncogene. 2009;28:994–1004. doi: 10.1038/onc.2008.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–66. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brognard J, Niederst M, Reyes G, Warfel N, Newton AC. Common polymorphism in the phosphatase PHLPP2 results in reduced regulation of Akt and protein kinase C. J Biol Chem. 2009;284:15215–23. doi: 10.1074/jbc.M901468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Santarpia L, Qi Y, Stemke-Hale K, Wang B, Young EJ, Booser DJ, et al. Mutation profiling identifies numerous rare drug targets and distinct mutation patterns in different clinical subtypes of breast cancers. Breast Cancer Res Treat. 2012;134:333–43. doi: 10.1007/s10549-012-2035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stemke-Hale K, Shipman K, Kitsou-Mylona I, de Castro DG, Hird V, Brown R, et al. Frequency of mutations and polymorphisms in borderline ovarian tumors of known cancer genes. Modern Pathol. 2013;26:544–52. doi: 10.1038/modpathol.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharman JP, Coutre SE, Cheson BD, Pagel JM, Hillmen P, Barrientos JC, et al. A phase 3, randomized, double-blind, placebo-controlled study evaluating the efficacy and safety of idelalisib and rituximab for previously treated patients with chronic lym phocytic leukemia (CLL). 2013 ASH Annual Meeting and Exposition; New Orleans, Lou isiana. [Google Scholar]
- 73.Leupin N, Cenni B, Novak U, Hugli B, Graber HU, Tobler A, et al. Disparate expression of the PTEN gene: a novel finding in B-cell chronic lymphocytic leukaemia (BCLL) Br J Haematol. 2003;121:97–100. doi: 10.1046/j.1365-2141.2003.04227.x. [DOI] [PubMed] [Google Scholar]
- 74.Kahan BD. Fifteen years of clinical studies and clinical practice in renal trans plantation: reviewing outcomes with de novo use of sirolimus in combination with cyclo sporine. Transplant Proc. 2008;40:S17–20. doi: 10.1016/j.transproceed.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 75.Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol. 2013;10:143–53. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- 76.Feldman ME, Shokat KM. New inhibitors of the PI3K-Akt-mTOR pathway: in sights into mTOR signaling from a new generation of Tor Kinase Domain Inhibitors (TORKinibs) Curr Top Microbiol Immunol. 2010;347:241–62. doi: 10.1007/82_2010_64. [DOI] [PubMed] [Google Scholar]
- 77.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]