Abstract
Knowledge of the genetics of type 2 diabetes mellitus (T2DM) has evolved tremendously over the past few years. Following advances in technology and analytical approaches, collaborative case–control genome-wide association studies have revealed up to 65 loci credibly associated with T2DM. Prospective population studies have demonstrated that aggregated genetic risk scores, so-called because they sum the genetic risk attributed to each locus, can predict incident T2DM among individuals of various age ranges and diverse ethnic backgrounds. With each set of T2DM loci discovered, increasing the number of loci in these scores has improved their predictive ability, although a prediction plateau may already have been reached. The current literature shows that intensive lifestyle interventions are effective for preventing T2DM at any level of genetic risk and might be particularly efficacious among individuals with high genetic susceptibility. By contrast, counselling to inform patients about their personal T2DM genetic risk profiles does not seem to improve motivation or attitudes that lead to positive lifestyle behaviour changes. Future studies should investigate the role of genetics for both T2DM prediction and prevention in young populations in the hope of reducing disease burden for future generations.
Introduction
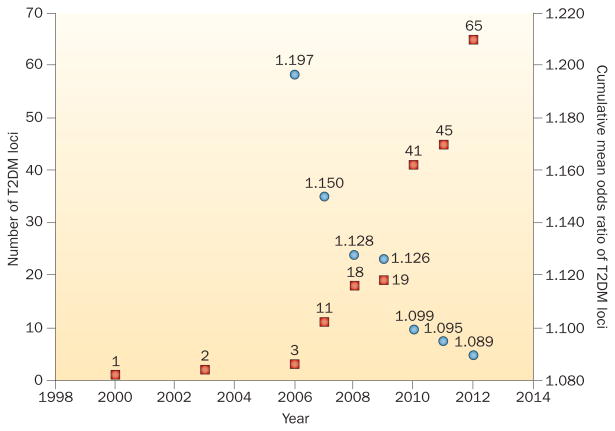
Type 2 diabetes mellitus (T2DM) is the archetype of a complex disease, influenced by both genetic and environmental determinants. Advances in knowledge and technology in the field of genetics have enabled the discovery of T2DM-associated genetic loci at a speed unthinkable just a few years ago. From a handful of genes identified through candidate-gene approaches, this area of investigation has evolved rapidly by pooling the data generated by consortia of multiple individual cohorts in case–control genome-wide associations study (GWAS) analyses, as demonstrated by the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) consortium. This approach has now uncovered at least 65 distinct genetic loci associated with T2DM;1 nonetheless, as the number of loci identified has increased, their individual (per allele) effect size on T2DM risk prediction has decreased. This finding indicates that the common variants with the largest effect size associated with T2DM have already been discovered (Figure 1). Looking at the physiological indices, approximately 10 of the first 37 loci reported by the DIAGRAM consortium seem to be implicated in β-cell function and insulin secretion or processing, whereas only four are thought to be involved in insulin sensitivity.2 Consequently, many loci have no obvious function in glycaemic regulation, and further work is required to elucidate the biology behind the fact that a substantial association with risk of T2DM was detected.2,3 The revelation of new biological pathways implicated in T2DM pathophysiology could be one of the most promising contributions of T2DM genetics to date.
Figure 1.
Cumulative number of T2DM loci discovered and T2DM risk per risk allele, by year, since 2000. The left axis (red squares) indicates the number of replicated, genome-wide significant loci associated with T2DM identified during the period 2000–2012 in populations of European, South Asian and East Asian ancestry. No such loci were discovered before 2000. The right axis (blue circles) indicates the cumulative mean effect size, expressed as the odds ratio, for the index variants at loci discovered from 2006 to 2012.1 As the number of T2DM loci identified has increased over time, the mean odds ratio has fallen from 1.197 for the three variants known in 2006 to 1.089 for the 65 variants known in 2012. These values equate to a fall from 19.7% increased mean odds ratio per additional risk allele in 2006 to 8.9% in 2012. Abbreviation: T2DM, type 2 diabetes mellitus.
Despite the excitement raised as a consequence of these novel findings, there has been scepticism about the clinical usefulness of T2DM genetics. One hope has been to use genetic information to identify individuals at high risk of developing T2DM, with the goal of providing ‘personalized medicine’ to offer targeted T2DM prevention strategies. Indeed, lifestyle intervention can prevent disease onset in individuals at high risk of T2DM.4–7 Nonetheless, interventions tested in T2DM prevention trials are resource-intensive, and current healthcare systems are unable to support such approaches at a population level or even for subgroups of individuals with prediabetes.8 Another hope for T2DM genetics is that personal knowledge of genetic risk would improve individual motivation for healthful behaviour change.9
This Review examines current knowledge about T2DM genetic risk prediction in general populations (with insights from diverse ethnic backgrounds and age ranges), the effect of lifestyle intervention according to gen etic risk and the capacity of genetic counselling to support be haviour modification. Future areas of research linking T2DM genetics to T2DM prevention are also considered.
Genetic prediction of T2DM risk
Genetic risk scores
Essentially all T2DM genetic risk variants identified to date have been found using a cross-sectional, case–control design methodology. From the time that more than a handful of T2DM genetic variants were known, investigators have tested whether aggregating genetic information from multiple variants could prospectively predict new cases of T2DM in unselected general populations where most individuals are at low phenotypic risk. Genetic risk scores (GRS; Box 1) to predict T2DM are built by summing the risk alleles that an individual carries, assuming that model is additive (zero, one or two risk alleles at each locus). These scores can be weighted according to the effect size reported in the initial case– control studies. This approach allows some risk alleles to contribute more risk information to the score than other alleles. For example, the risk allele at TCF7L2 carries an odds ratio (OR) of 1.40 per risk allele (the largest effect size known for a common variant), whereas most other loci have been found to carry ORs of 1.05–1.20 per risk allele.1 However, weighting the effect alleles did not markedly influence the results in most of the studies that performed both weighted and non-weighted analyses.10
Box 1. Glossary of terms.
C-statistic. The value of the area under the receiver operating characteristics (ROC) curve, which is a graphic representation of the ability of a test to distinguish between individuals who have an outcome or disease and those who do not. The ROC curve plots the sensitivity (or true positive rate) on the y axis versus 1 minus the specificity (or true negative rate) on the x axis. If the value is 0.5, the test result is equivalent to pure chance; the higher the value, the more accurate the discrimination.
Genetic risk score (GRS). A score aggregating information from more than one variant at different genetic loci associated with a disease. A GRS is generally constructed according to an additive genetic model, such that one point is accorded for each copy of the risk allele carried by an individual at each loci included. The value of the allele can also be weighted by the disease risk effect size reported in previous association analyses.
Net reclassification improvement index (NRI). An estimate of the ability of a prediction model to correctly reassign individuals to risk categories when compared to another model. For example, one prediction model may classify individuals as having low, intermediate or high risk of an outcome. However, some individuals in the low-risk category will have the outcome and some in the high-risk category will not. If a second model assigns a greater number of the eventual cases to a high-risk category and a greater number of non-cases to a low-risk category, it is considered an improvement over the first model and has a positive NRI value. The NRI is calculated from discrete risk categories or from continuous risk probability estimates. Here, models evaluated with continuous NRI are primarily discussed. A continuous NRI of 0.2, 0.4 or ≥0.6 indicates a weak, modest or strong improvement, respectively.60
One of the first prospective T2DM prediction studies was the Framingham Heart Study, which reported the association of a GRS comprising 18 known T2DM loci with disease incidence over 28 years of follow-up among individuals who were nondiabetic at the beginning of observation.10 On average, individuals who ultimately developed T2DM had approximately one-half of a risk allele more than those who did not develop T2DM (17.7 ± 2.7 versus 17.1 ± 2.6, respectively; P <0.001). The sex-adjusted OR for T2DM was 1.12 per risk allele, indicating that each risk allele had increased the odds of developing T2DM by approximately 12%. Adding the GRS to a T2DM prediction model including sex only (representing the most basic genetic information that can be determined by phenotypic examination at birth) markedly improved the C-statistic (Box 1) from 0.53 to 0.58 (P = 0.01). The GRS remained significantly associated with T2DM risk in a clinical prediction model that included sex, age, family history of T2DM, BMI, fasting plasma glucose level, systolic blood pressure, HDL cholesterol and fasting triglyceride levels.10 However, measurements of model discrimination and reclassification demonstrated that the GRS added only marginal information to the clinical prediction model.
An analysis of the Malmö and Botnia studies, two European prospective population-based studies, with at least 23 years of follow-up, reached a comparable conclusion using an 11-locus GRS.11 The predictive ability was increased over a long duration of follow-up, with a C-statistic of 0.56 in evaluating the GRS in the shortest quintile of follow-up time (16.3 ± 2.5 years) versus a C-statistic of 0.62 in the longest quintile of follow-up time (28.4 ± 0.5 years; P = 0.01). By contrast, the clinical prediction model performed better over the shortest follow-up time, with a C-statistic of 0.75 versus 0.67 for the longest quintile of follow-up time (P = 0.01). This observation is consistent with the static nature of the genotype, whereas any measured clinical factor, such as BMI or blood pressure, is dynamic in nature and varies over an individual's lifespan. This finding also raises the possibility that genetic testing early in life might better discriminate T2DM risk than genetic testing later in life.
Plateau for predictive value
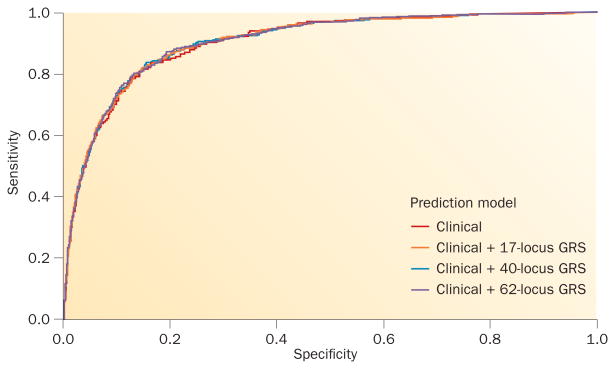
Other researchers have tested the ability of GRS to predict T2DM in prospective studies, mostly finding positive associations (reviewed elsewhere12); however, many investigators highlighted the marginal improvement in predictive ability of the models reported in such studies. One hypothesis suggests that as the number of T2DM-associated loci identified grows, thereby capturing an increased proportion of the heritability of T2DM, generation of more-inclusive GRS would improve the predictive value of genetic testing, similar to observations with the genetic determinants of other metabolic traits.13 Following the second publication of large-scale T2DM GWAS by the DIAGRAM consortium,14 a GRS that included 40 loci was tested and compared to the previously reported 17-locus GRS used in the Framingham Heart Study.15 The mean GRS was approximately one and a half risk alleles higher among individuals who developed T2DM than those who did not (P = 1.7 × 10−10). As shown in Figure 2, the addition of the 40-locus GRS improved the predictive power of the clinical model: the C-statistic was 0.906 with this score and 0.903 without it (P = 0.04). Performance of the 40-locus GRS, therefore, exceeded that of the original 17-locus GRS.15
Figure 2.
Prediction of T2DM in the Framingham Offspring Study adding genetic risk scores to a clinical prediction model. Receiver operating characteristic curves for the prediction of T2DM in the Framingham Offspring Study using a clinical model alone (red) or in combination with a 17-locus (orange), 40-locus (blue) or 62-locus (purple) GRS. The C-statistics for each prediction model were 0.9031, 0.9050, 0.9055 and 0.9058, respectively. Despite being statistically associated with an increased risk of T2DM, each GRS provided only marginal additional information to the prediction model comprising known clinical risk factors (sex, age, family history of T2DM, BMI, fasting plasma glucose levels, systolic blood pressure, HDL cholesterol levels and fasting triglyceride levels). Abbreviations: GRS, genetic risk score; T2DM, type 2 diabetes mellitus.
Nevertheless, other research suggests that using an even greater number of common variants for T2DM prediction might not have additional benefits. One analysis suggests that using a 62-locus GRS does not further improve the predictive value of the 40-locus GRS in a sample of participants enrolled in the Framingham Heart Study.16 The C-statistic was identical (0.906) regardless of which GRS was added to the clinical risk factor model (Figure 2). This observation suggests that a GRS might reach a plateau for its predictive value after a certain number of T2DM loci have been included, particularly because newly identified risk alleles are associated with smaller and smaller effect sizes.1
Influence of age
The timeline for T2DM prediction influences the performance of individual risk factors and prediction models. That is, a model optimized to predict incident T2DM over a 5-year period might perform poorly for predic tion over 30 years; furthermore, this discrepancy might differ between genotype-based and phenotype-based models.11 Unlike clinical risk factors, such as BMI and fasting blood glucose level, genetic coding variants have the rare property of being an exposure that remains static over the life course. In theory, genetic models should outperform those that consider clinical risk factors that change over time in predicting T2DM over long time frames.
A related concept is that genotype may have greater predictive strength when applied to young populations, who collectively might not yet manifest phenotypic T2DM risk factors, such as overweight or dysglycaemia. The prospective cohort studies detailed above applied polygenic T2DM prediction models to populations generally comprising middle-aged adults (aged >30 years).10–12,15,16 Although the use of a polygenic GRS predicted incident T2DM in these studies, the scores only marginally improved the clinical prediction models in these middle-aged populations. This finding is not entirely surprising, as individuals at high risk of T2DM had probably already accumulated obvious, measureable phenotypical T2DM risk factors. For instance, in the Framingham Heart Study,10,15 the 40-locus GRS seemed to enhance the net reclassification improvement index (NRI; Box 1) of clinical prediction models in the subgroup of participants aged <50 years but not in the group aged ≥50 years at baseline. The OR per risk allele was 1.17 and the NRI was 11.9% for individuals aged <50 years (P = 0.009). By contrast, the OR per risk allele was 1.07 and the NRI was 0.47% among individuals aged ≥50 years (P = 0.92).10
The effect of age in a nested case–control study was also tested using data from the prospective EPIC study, a large European population-based cohort.17 A GRS comprising 42 known loci was able to predict T2DM in this population. The C-statistic for the whole cohort was 0.579; after stratification, this value was 0.589 for individuals aged <50 years and 0.577 for those aged ≥50 years.
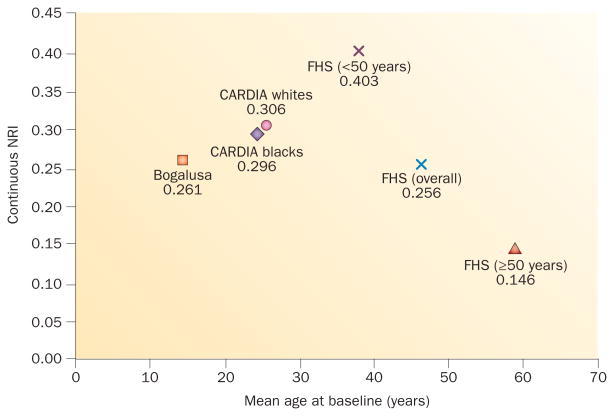
Some studies have specifically examined genetic prediction of T2DM in youth. Polygenic GRS predicted incident T2DM in two US prospective population-based cohorts that were followed up over 25 years: the Bogalusa Heart Study (adolescents aged 12–18 years at baseline) and the CARDIA study (young adults aged 18–30 years at baseline).18,19 The hazard ratio (HR) per risk allele in the demographic model was 1.09 (P = 0.01) among the participants from the Bogalusa Heart Study (adolescents at baseline) and 1.09 (P <0.0001) among young adults from the CARDIA study. The NRI for the GRS in both of these studies (0.261–0.306) seemed to be stronger than in the overall middle-aged Framingham Heart Study prediction model (0.256), but not quite as strong as the NRI in Framingham Heart Study participants aged <50 years (0.403), as hypothesized on the premise of young age (Figure 3). Once again, the GRS significantly predicted T2DM incidence but provided marginal value to improving prediction models based on clinical measurements. Thus, BMI and other phenotypical metabolic risk factors present in childhood and adolescence seem to already be important predictors of T2DM risk later in life, independently of known T2DM genotypes.
Figure 3.
Net NRI values of adding genetic risk scores to clinical prediction models according to the age at baseline. Continuous NRIs for the addition of a 62-locus GRS (FHS and CARDIA studies) and a 38-locus GRS (Bogalusa study) to a clinical prediction model for type 2 diabetes mellitus by mean baseline age. Clinical prediction models comprised age, sex, systolic blood pressure or mean arterial pressure, BMI, fasting plasma glucose, HDL cholesterol and triglyceride levels. NRIs of 0.2, 0.4 and 0.6 indicate weak, moderate, and strong improvement in prediction, respectively. Abbreviations: FHS, Framingham Heart Study; GRS, genetic risk score; NRI, net reclassification improvement index.
Prediction models applied in early childhood or even at birth have appeal for primary prevention. Using only the information that might be available at birth (that is, sex and genotype), the C-statistic in the Framingham Heart Study increased from 0.534 (sex only) to 0.581 (sex plus GRS; P = 0.01), yielding an NRI of 4.1% (P = 0.004).10 Identifying perinatal risk factors for adult T2DM, such as maternal factors and birth weight, has also garnered interest.20 In an analysis of data from 2,003 adults aged 56–70 years in the Helsinki Birth Cohort Study, a study of Finns born between 1934 and 1944, birth weight and genetic variants at nine loci interacted to influence T2DM risk.21 Individuals with low birth weight and high overall genetic risk were most likely to develop T2DM by 70 years of age. How additional maternal and perinatal factors and GRS might compare and interact as important T2DM predictors over the life course is still unknown.
Influence of gestational diabetes mellitus
Some studies have investigated the performance of a GRS to predict T2DM among women with previous gestational diabetes mellitus. This population is generally young, as it comprises women of reproductive age, but recognized to have a very high risk of developing T2DM.22 A GRS based on 13 variants at 11 known T2DM loci was associated with incident T2DM in a cohort of multiethnic women with previous gestational diabetes mellitus who underwent serial oral glucose tolerance tests for up to 5 years (HR = 1.11; P = 1.6 × 10−4).23 Similar findings were reported in a cohort of East Asian women using a 48-locus GRS; adding this GRS increased the C-statistic from 0.741 to 0.775 (P = 0.02), with a continuous NRI of 0.430, indicative of moderate reclassification compared with the clinical model that included age, BMI, family history of T2DM, blood pressure and fasting levels of glucose and insulin.24 Thus, including genetic risk in a prediction model could inform T2DM prediction even among populations already known to be at very high risk of developing this condition.
Influence of ethnicity
Evaluating the role of genetic T2DM prediction in non-European racial and ethnic groups is important, as they are at greater risk of developing T2DM than their European counterparts. Unfortunately, however, study of the genetic architecture of T2DM in these populations has lagged behind that in populations of European ancestry. For example, the 2012 DIAGRAM GWAS meta-analysis comprised almost 150,000 individuals, the vast majority of whom were of European ancestry.1 The largest T2DM GWAS among eastern Asian populations included almost 55,000 individuals and identified eight novel T2DM loci as well as many of the previously described European loci.25 By contrast, the largest GWAS analyses of populations of African and Indian ancestry conducted to date included just over 8,000 and 12,000 people, respectively.26,27
Replication studies have generally found that T2DM-associated loci discovered in European populations are also associated with T2DM in non-European populations,28–30 despite an increased genetic diversity among evolutionarily ancient populations, such as Africans, whose genomes contain large numbers of common variants and shorter length of DNA sequences between recombination locations (also known as linkage disequilibrium blocks). Among adolescents and young adults enrolled in the Bogalusa and CARDIA studies, respectively, the ability of a polygenic GRS to predict incident T2DM did not differ between white and black Americans.18,19 In addition, a case–control study demonstrated that a polygenic GRS comprising loci discovered in Europeans was significantly associated with T2DM among middle-aged African-Americans with a mean age of 56 years.31 Finally, EpiDREAM, a multi-centre prospective study of almost 19,000 adults at risk of dysglycaemia, found no difference in the performance of a polygenic score that included 16 single-nucleotide polymorphisms between individuals of European, South Asian and Latino background, showing consistency of effect of European-based T2DM GRS among diverse ethnicities.32
Prevention of T2DM
Intensive lifestyle intervention
Several randomized controlled trials have demonstrated that T2DM can be prevented in high-risk individuals by intensive lifestyle intervention.4–7 The interventions tested in the Diabetes Prevention Program4 and the Finnish Diabetes Prevention Study5 both included frequent patient contact with a health professional and behavioural support to moderately increase physical activity and decrease caloric intake, aiming for 5–10% weight loss and maintenance. Despite the large benefit for T2DM prevention (relative risk reduction of 58% in both studies), intensive lifestyle interventions are not generally offered in most health-care settings. This discrepancy might reflect a lack of specialist resources such as dieticians, physical activity specialists and behavioural counsellors; in addition, clinicians could have doubts about the ability of patients to change their behaviour. One hope of ‘personalized medicine’ for T2DM is to identify patients who are most likely to benefit from intensive lifestyle intervention either by achieving a large physiological response (for example, substantial weight loss) or else by experiencing an increased psychological effect (for example, increased motivation or adherence) following disclosure of their T2DM genetic risk.
Influence of genetic risk on intervention
Some studies have examined whether genotype—either at a single locus or across several T2DM-associated loci— influences the effectiveness of and biological response to lifestyle modification for T2DM prevention.
TCF7L2
The Diabetes Prevention Program investigated whether the effects of lifestyle intervention differed by TCF7L2 genotype.33 This gene encodes transcription factor 7-like 2 and is the T2DM risk locus identified to date that has the largest effect size per risk allele and that has been the most replicated across studies. Individuals carrying two copies of the T risk allele at rs7903146 had a greater risk of developing T2DM in the control group (HR = 1.81 versus CC homozygotes; P = 0.004) as expected from risk conferred by that allele in observational studies. By contrast, progression to T2DM was not associated with the risk genotype in the intensive lifestyle intervention group (carriers of two copies of the T risk allele had an HR = 1.15; P = 0.60), suggesting that behaviour modification can mitigate the risk conferred by this genetic variant. Similar results were found in the Finnish Diabetes Prevention Study when investigating the same risk variant at TCF7L2.34
Together, the findings of these two studies strongly suggest that intensive lifestyle intervention can efficiently reduce the genetic susceptibility to T2DM attributable to TCF7L2. Conversely, in a single-arm lifestyle intervention study among high-risk individuals, carriers of the T risk allele of TCF7L2 showed smaller decreases in BMI and percentage body fat over 9 months than did those without this risk allele.35 Other intervention studies have not generally observed differential effects by TCF7L2 genotype, perhaps owing to a lack of statistical power or because of differences in the populations studied.36,37
Other known T2DM loci
Many other candidate gene variants for T2DM have been tested for their interaction with lifestyle intervention, with predominantly mixed or contradictory results. For example, the effect of risk alleles in genes encoding peroxisome proliferator-activated receptor γ (PPARG38) and ectonucleotide pyrophosphatase/phosphodiesterase family member 1 (ENPP139) on T2DM incidence seemed to be reduced in the lifestyle intervention arm of the Diabetes Prevention Program but not in other studies.40,41 The Diabetes Prevention Program is currently the largest randomized trial of T2DM prevention, with approximately 1,000 individuals enrolled per arm who have consented to provide DNA samples (for each of the three arms completed: placebo, metformin and intensive lifestyle intervention). Despite moderate statistical power to detect a genetic effect size of similar magnitude to that found in observational studies in the overall cohort, the Diabetes Prevention Program remains limited to the detection of gene–intervention interaction.42 A total of 1,536 single-nucleotide polymorphisms located in 40 candidates genes were tested, but only 23 loci were found to have a nominally significant interaction with lifestyle intervention; none of these loci maintained significance after adjustment for multiple testing.42
Aggregated T2DM genetic burden
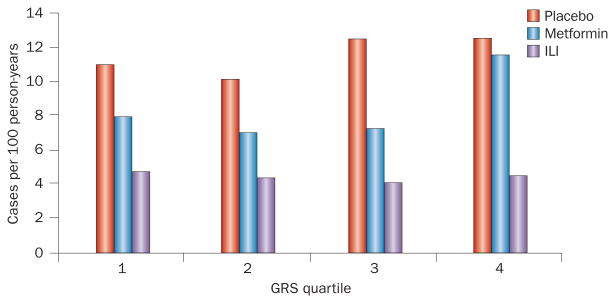
The Diabetes Prevention Program cohort was also examined to determine whether cumulative aggregated T2DM genetic risk, rather than individual loci, influences the effectiveness of lifestyle intervention. A 34-locus GRS predicted the risk of developing T2DM in the overall cohort, with an HR of 1.02 per risk allele (P = 0.03 in the fully adjusted model).43 When participants were classified according to their genetic risk profile within each treatment arm, individuals at the highest genetic risk (top quartile of the GRS) still benefited from the intensive lifestyle intervention. As shown in Figure 4, the T2DM incidence was 12 cases per 100 person-years in the control arm versus five cases per 100 person-years in the lifestyle arm (P <0.001). The results of this analysis also sug gest that the benefit of lifestyle intervention is greatest among individuals with the highest genetic risk; however, the interaction between GRS and the treatment arm was not statistically significant.43
Figure 4.
Outcome of intensive lifestyle intervention in the Diabetes Prevention Program. Intensive lifestyle intervention prevented T2DM in participants, independent of the level of genetic burden. The graph shows the T2DM incidence rate in each arm of the study by quartile of a 34-locus GRS. The results suggest that individuals at high genetic risk of T2DM benefit from intensive lifestyle intervention as much (or even more) than individuals at low genetic risk. Abbreviations: GRS, genetic risk score; ILI, intensive lifestyle intervention; T2DM, type 2 diabetes mellitus.
The Finnish Diabetes Prevention Study investigators demonstrated that lifestyle intervention was beneficial and prevented T2DM independently of the aggregated genetic risk of T2DM estimated by a 19-locus GRS.44 They also reported that the GRS added little to T2DM prediction above known clinical risk factors and the effect of intervention, in line with observational studies.10,11 Overall, these studies reassure clinicians and patients that lifestyle modification and modest weight loss can prevent T2DM, regardless of genetic risk, and possibly benefit individuals at high genetic risk to an even greater degree than those at low genetic risk.
Genetics for motivating behavioural change
Separate from the ability to improve risk stratification, T2DM genetics might also facilitate disease prevention by influencing health psychology. The knowledge of increased genetic susceptibility to T2DM could motivate an individual to implement prevention strategies, such as weight loss or increased physical activity. Indeed, some patients have reported that they would be more motivated to change behaviours to prevent T2DM if they learned that they were at increased genetic risk.9,45–46
In one survey, up to 71% of patients reported that they would be very likely to modify their behaviours if they were tested and told that they were at high genetic risk.9 However, translating such intentions into preventative action requires sustained health behaviour changes in dietary and physical activity habits. In addition, T2DM genetic testing identifies individuals at high, moderate and low genetic risk; the possibility, therefore, exists that individuals may become demotivated to make behavioural changes depending on their risk prediction results. In particular, some sedentary patients with obesity will be classified as low T2DM genetic risk; such a result could adversely influence much needed motivation to lose weight and increase physical activity for improving health among this group.
Observational and intervention trial data do not yet support the use of genetic susceptibility testing to implement behaviour change.47–50 At least three randomized trials have examined the effect of T2DM genetic susceptibility testing on health behaviours and outcomes. The Genetic Counselling and Lifestyle Change for Diabetes Prevention (GC/LC) study allocated patients with features of the metabolic syndrome to T2DM genetic susceptibility testing with a polygenic score comprising 36 single-nucleotide polymorphisms.48 Individuals classified as at either high or low genetic risk for T2DM received a short genetic counselling session to explain their test results, discuss the relative contribution of genetic versus lifestyle factors to T2DM risk, and place their genetic test results in context of overall risk of T2DM (in this study, that related to the metabolic syndrome). The investigators hypothesized that the genetic testing and counselling sessions would enhance patient engagement in riskreduction behaviour changes and increase motivation and confidence to adopt healthier behaviour. A control group did not receive genetic testing or counselling. All study participants were then enrolled in a 12-week group lifestyle modification programme based on the Diabetes Prevention Program.4 Lifestyle modification was associated with a mean weight loss of 3.9 kg, but this amount did not differ between the tested and untested arms, or between individuals who learned they were at high genetic risk versus those who learned they were at low genetic risk. At the end of the 12-week programme, there was no detectable between-groups difference in change in attitude towards T2DM prevention behaviours (perception, motivation, confidence or readiness for behaviour change) or in programme attendance.
Whereas the GC/LC study targeted high-risk individuals and provided substantial support for lifestyle modification, two large trials recruited participants from a general primary care patient population and provided just a single, brief T2DM risk-counselling session highlighting clinical risk factors alone or clinical risk factors plus genetic susceptibility.49,50 The final results of these two trials have not yet been published, but preliminary analyses have demonstrated that none of the changes in behaviours and clinical measurements differed between the two arms.51 Thus, it remains to be seen whether testing to provide knowledge of genetic susceptibility can motivate health behaviour change to prevent T2DM.
Future perspectives
Interest in early life-course determinants of adult chronic diseases, including T2DM, is increasing.52 The epidemiological observations that T2DM is associated with both low and high birth weight have raised numerous hypotheses around what is now called the ‘fetal programming’ of metabolic diseases.53 Both genetic and environmental factors are likely to be involved in these phenomena. Studies of genetic determinants of birth weight by GWAS have revealed that risk variants for T2DM were also associated with low birth weight (ADCY5 [adenylate cyclase type 5] and CDKAL1 [threonylcarbamoyladenosine tRNA methylthiotransferase]) and high birth weight (TCF7L2).54 Conversely, there are clear indications that the in utero environment has a role in fetal programming, as individuals exposed to either famine55,56 or maternal diabetes mellitus (gestational or pre-existing)57 during fetal development are more likely than their unexposed siblings to develop obesity and/or T2DM later in life.
Genetics might help increase understanding of the biological mechanisms underpinning fetal metabolic programming. Of particular interest is the field of epigenetics—the study of changes in gene expression caused by mechanisms other than variation in the underlying DNA sequence. Epigenetic phenomena include DNA methylation and histone modifications, which are malleable but can persist over decades. DNA methylation is particularly sensitive to events during development in utero because DNA is almost fully demethylated during zygote formation and specific methylation is re-established throughout fetal development during tissue differentiation. Patterns of DNA methylation at candidate genes such as LEP (encoding leptin) have been associated with in utero exposure to famine58 and to maternal impaired glucose tolerance during pregnancy,59 suggesting that ‘programmed’ regulation of these genes might lead to certain metabolic outcomes, such as weight regulation in the LEP example, later in life. The field of epigenetics in human fetal metabolic programming is still in its infancy, and more studies are needed to reveal early epigenetic markers that identify individuals affected by adverse in utero or early life events leading to future metabolic diseases, such as T2DM. Once identified, epigenetic markers could help detect children and families for whom intensive lifestyle intervention should be emphasized in the hope of altering the impaired metabolic trajectory for which they are programmed. The dynamic nature of putative epigenetic markers may present some challenges compared to the static nature of genetic variations to predict future disease, but they can also offer interesting avenues to improve understanding of aetiology and the effect of interventions at a molecular level. In this framework, dynamic epigenetic markers could also be used to monitor the effect of lifestyle interventions provided during pregnancy or in early childhood. Interventions aimed at primary prevention are necessary if the current T2DM epidemic is to be slowed.
Conclusions
The use of GRS is associated with increased individual risk of incident T2DM but only marginally improves population discrimination in prediction models that include established clinical risk factors. The optimal number of loci to include for a GRS to be informative will probably reach a threshold given current knowledge of variants frequently associated with T2DM. Clinical trials that incorporated T2DM genetic counselling have not yet shown an enhanced behavioural response to lifestyle intervention aimed at T2DM prevention. Nevertheless, it is encouraging to know that lifestyle interventions are efficacious at preventing T2DM even among individuals at high genetic risk. Future studies should focus on young populations to assess prediction models that include early life determinants of future T2DM and to test interventions in infancy or during pregnancy aiming at primordial prevention of obesity and T2DM.
Key points.
Over the past few years, more than 65 genetic loci associated with type 2 diabetes mellitus (T2DM) have been discovered that reveal novel biologic pathways
Clinical T2DM prediction models are improved by the addition of information about genetic risk variants
Genetic risk scores that aggregate risk variants can predict incident T2DM in populations of diverse ethnic background and age range
Lifestyle interventions targeting moderate weight loss lower the risk of T2DM, independent of the genetic burden, and might have increased benefit among individuals at high genetic risk
Review criteria.
A search for original articles published between 1996 and 2013 was performed in MEDLINE and PubMed. We started with known literature from our group and other colleagues, and completed our search using ‘related articles’ and ‘citing articles’ options from MEDLINE and PubMed. For the ‘diabetes prevention’ section, we also conducted a broader search: the search terms used were “genetics” and “diabetes prevention” or “intensive lifestyle intervention” in addition to a specific search in the literature for the Diabetes Prevention Program and Finnish Diabetes Prevention Study and for any genetics-related study. All articles identified were English-language, full-text papers. We also searched the reference lists of identified articles for further papers.
Footnotes
Competing interests: The authors declare no competing interests.
Author contributions: All authors researched the data for the article and provided a substantial contribution to discussions of the content. M.-F.H. and J.L.V. contributed equally to writing the article. J.B.M. reviewed and edited the manuscript before submission.
Contributor Information
Marie-France Hivert, Department of Population Medicine, Harvard Pilgrim Health Care Institute, Harvard Medical School, 50 Staniford Street, 9th floor, Boston, MA 02114, USA.
Jason L. Vassy, Section of General Internal Medicine, VA Boston Healthcare System, Harvard Medical School, 50 Staniford Street, 9th floor, Boston, MA 02114, USA
James B. Meigs, General Medicine Division, Massachusetts General Hospital, Harvard Medical School, 50 Staniford Street, 9th floor, Boston, MA 02114, USA
References
- 1.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dimas AS, et al. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. doi: 10.2337/db13-0949. http://dx.doi.org/10.2337/db13–0949. [DOI] [PMC free article] [PubMed]
- 3.Ingelsson E, et al. Detailed physiologic characterization reveals diverse mechanisms for novel genetic loci regulating glucose and insulin metabolism in humans. Diabetes. 2010;59:1266–1275. doi: 10.2337/db09-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knowler WC, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuomilehto J, et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med. 2001;344:1343–1350. doi: 10.1056/NEJM200105033441801. [DOI] [PubMed] [Google Scholar]
- 6.Ramachandran A, et al. The Indian Diabetes Prevention Programme shows that lifestyle modification and metformin prevent type 2 diabetes in Asian Indian subjects with impaired glucose tolerance (IDPP-1) Diabetologia. 2006;49:289–297. doi: 10.1007/s00125-005-0097-z. [DOI] [PubMed] [Google Scholar]
- 7.Pan XR, et al. Effects of diet and exercise in preventing NIDDM in people with impaired glucose tolerance. The Da Qing IGT and Diabetes Study. Diabetes Care. 1997;20:537–544. doi: 10.2337/diacare.20.4.537. [DOI] [PubMed] [Google Scholar]
- 8.Herman WH, et al. Effectiveness and cost-effectiveness of diabetes prevention among adherent participants. Am J Manag Care. 2013;19:194–202. [PMC free article] [PubMed] [Google Scholar]
- 9.Grant RW, et al. The clinical application of genetic testing in type 2 diabetes: a patient and physician survey. Diabetologia. 2009;52:2299–2305. doi: 10.1007/s00125-009-1512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meigs JB, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359:2208–2219. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyssenko V, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359:2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 12.Vassy JL, Meigs JB. Is genetic testing useful to predict type 2 diabetes? Best Pract Res Clin Endocrinol Metab. 2012;26:189–201. doi: 10.1016/j.beem.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrison AC, et al. Whole-genome sequence-based analysis of high-density lipoprotein cholesterol. Nat Genet. 2013;45:899–901. doi: 10.1038/ng.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Miguel-Yanes JM, et al. Genetic risk reclassification for type 2 diabetes by age below or above 50 years using 40 type 2 diabetes risk single nucleotide polymorphisms. Diabetes Care. 2011;34:121–125. doi: 10.2337/dc10-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vassy JL, Porneala B, Florez JC, Dupuis J, Meigs JB. Type 2 diabetes prediction with 17-, 40-, and 62-variant genotype risk scores: the Framingham Offspring Study. Circulation. 2013;127:AMP58. abstract. [Google Scholar]
- 17.Mühlenbruch K, Jeppesen C, Joost HG, Boeing H, Schulze MB. The value of genetic information for diabetes risk prediction— differences according to sex, age, family history and obesity. PLoS ONE. 2013;8:e64307. doi: 10.1371/journal.pone.0064307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vassy JL, et al. Genotype prediction of adult type 2 diabetes from adolescence in a multiracial population. Pediatrics. 2012;130:e1235–e1242. doi: 10.1542/peds.2012-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vassy JL, et al. A genotype risk score predicts type 2 diabetes from young adulthood: the CARDIA study. Diabetologia. 2012;55:2604–2612. doi: 10.1007/s00125-012-2637-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berends LM, Ozanne SE. Early determinants of type-2 diabetes. Best Pract Res Clin Endocrinol Metab. 2012;26:569–580. doi: 10.1016/j.beem.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Pulizzi N, et al. Interaction between prenatal growth and high-risk genotypes in the development of type 2 diabetes. Diabetologia. 2009;52:825–829. doi: 10.1007/s00125-009-1291-1. [DOI] [PubMed] [Google Scholar]
- 22.Kim C, Newton KM, Knopp RH. Gestational diabetes and the incidence of type 2 diabetes: a systematic review. Diabetes Care. 2002;25:1862–1868. doi: 10.2337/diacare.25.10.1862. [DOI] [PubMed] [Google Scholar]
- 23.Ekelund M, et al. Genetic prediction of postpartum diabetes in women with gestational diabetes mellitus. Diabetes Res Clin Pract. 2012;97:394–398. doi: 10.1016/j.diabres.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 24.Kwak SH, et al. Prediction of type 2 diabetes in women with a history of gestational diabetes using a genetic risk score. Diabetologia. 2013;56:2556–2563. doi: 10.1007/s00125-013-3059-x. [DOI] [PubMed] [Google Scholar]
- 25.Cho YS, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2011;44:67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lettre G, et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabassum R, et al. Genome-wide association study for type 2 diabetes in Indians identifies a new susceptibility locus at 2q21. Diabetes. 2013;62:977–986. doi: 10.2337/db12-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haiman CA, et al. Consistent directions of effect for established type 2 diabetes risk variants across populations: the Population Architecture using Genomics and Epidemiology (PAGE) Consortium. Diabetes. 2012;61:1642–1647. doi: 10.2337/db11-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waters KM, et al. Consistent association of type 2 diabetes risk variants found in Europeans in diverse racial and ethnic groups. PLoS Genet. 2010;6:e1001078. doi: 10.1371/journal.pgen.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng MC, et al. Transferability and fine mapping of type 2 diabetes loci in African Americans: the Candidate Gene Association Resource Plus Study. Diabetes. 2013;62:965–976. doi: 10.2337/db12-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooke JN, et al. Genetic risk assessment of type 2 diabetes-associated polymorphisms in African Americans. Diabetes Care. 2012;35:287–292. doi: 10.2337/dc11-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anand SS, et al. Genetic information and the prediction of incident type 2 diabetes in a highrisk multiethnic population: the EpiDREAM genetic study. Diabetes Care. 2013;36:2836–2842. doi: 10.2337/dc12-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Florez JC, et al. TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med. 2006;355:241–250. doi: 10.1056/NEJMoa062418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, et al. Variants of transcription factor 7-like 2 (TCF7L2) gene predict conversion to type 2 diabetes in the Finnish Diabetes Prevention Study and are associated with impaired glucose regulation and impaired insulin secretion. Diabetologia. 2007;50:1192–1200. doi: 10.1007/s00125-007-0656-6. [DOI] [PubMed] [Google Scholar]
- 35.Haupt A, et al. Gene variants of TCF7L2 influence weight loss and body composition during lifestyle intervention in a population at risk for type 2 diabetes. Diabetes. 2010;59:747–750. doi: 10.2337/db09-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bo S, et al. Effects of TCF7L2 polymorphisms on glucose values after a lifestyle intervention. Am J Clin Nutr. 2009;90:1502–1508. doi: 10.3945/ajcn.2009.28379. [DOI] [PubMed] [Google Scholar]
- 37.Reinehr T, et al. Evidence for an influence of TCF7L2 polymorphism rs7903146 on insulin resistance and sensitivity indices in overweight children and adolescents during a lifestyle intervention. Int J Obes (Lond) 2008;32:1521–1524. doi: 10.1038/ijo.2008.146. [DOI] [PubMed] [Google Scholar]
- 38.Florez JC, et al. Effects of the type 2 diabetes-associated PPARG P12A polymorphism on progression to diabetes and response to troglitazone. J Clin Endocrinol Metab. 2007;92:1502–1509. doi: 10.1210/jc.2006-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore AF, et al. The association of ENPP1 K121Q with diabetes incidence is abolished by lifestyle modification in the Diabetes Prevention Program. J Clin Endocrinol Metab. 2009;94:449–455. doi: 10.1210/jc.2008-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindi VI, et al. Association of the Pro12Ala polymorphism in the PPAR-γ2 gene with 3-year incidence of type 2 diabetes and body weight change in the Finnish Diabetes Prevention Study. Diabetes. 2002;51:2581–2586. doi: 10.2337/diabetes.51.8.2581. [DOI] [PubMed] [Google Scholar]
- 41.Müssig K, et al. The ENPP1 K121Q polymorphism determines individual susceptibility to the insulin-sensitising effect of lifestyle intervention. Diabetologia. 2010;53:504–509. doi: 10.1007/s00125-009-1612-4. [DOI] [PubMed] [Google Scholar]
- 42.Jablonski KA, et al. Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the Diabetes Prevention Program. Diabetes. 2010;59:2672–2681. doi: 10.2337/db10-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hivert MF, et al. Updated genetic score based on 34 confirmed type 2 diabetes loci is associated with diabetes incidence and regression to normoglycemia in the Diabetes Prevention Program. Diabetes. 2011;60:1340–1348. doi: 10.2337/db10-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uusitupa MI, et al. Impact of positive family history and genetic risk variants on the incidence of diabetes: the Finnish Diabetes Prevention Study. Diabetes Care. 2011;34:418–423. doi: 10.2337/dc10-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vassy JL, Donelan K, Hivert MF, Green RC, Grant RW. Genetic susceptibility testing for chronic disease and intention for behavior change in healthy young adults. J Community Genet. 2013;4:263–271. doi: 10.1007/s12687-013-0140-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vassy JL, et al. Impact of literacy and numeracy on motivation for behavior change after diabetes genetic risk testing. Med Decis Making. 2012;32:606–615. doi: 10.1177/0272989X11431608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bloss CS, Schork NJ, Topol EJ. Effect of direct-to-consumer genomewide profiling to assess disease risk. N Engl J Med. 2011;364:524–534. doi: 10.1056/NEJMoa1011893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grant RW, et al. Personalized genetic risk counseling to motivate diabetes prevention: a randomized trial. Diabetes Care. 2013;36:13–19. doi: 10.2337/dc12-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho AH, et al. Effect of genetic testing for risk of type 2 diabetes mellitus on health behaviors and outcomes: study rationale, development and design. BMC Health Serv Res. 2012;12:16. doi: 10.1186/1472-6963-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voils CI, et al. Examining the impact of genetic testing for type 2 diabetes on health behaviors: study protocol for a randomized controlled trial. Trials. 2012;13:121. doi: 10.1186/1745-6215-13-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho AH, et al. Preliminary outcomes of genetic risk testing in primary care for common DNA variants associated with type 2 diabetes. J Gen Intern Med. 2012;27(Suppl. 2):S278. [Google Scholar]
- 52.Lacroix M, Kina E, Hivert MF. Maternal/fetal determinants of insulin resistance in women during pregnancy and in offspring over life. Curr Diab Rep. 2013;13:238–244. doi: 10.1007/s11892-012-0360-x. [DOI] [PubMed] [Google Scholar]
- 53.Martin-Gronert MS, Ozanne SE. Metabolic programming of insulin action and secretion. Diabetes Obes Metab. 2012;14(Suppl. 3):29–39. doi: 10.1111/j.1463-1326.2012.01653.x. [DOI] [PubMed] [Google Scholar]
- 54.Horikoshi M, et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat Genet. 2013;45:76–82. doi: 10.1038/ng.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lumey LH, Stein AD, Kahn HS, Romijn JA. Lipid profiles in middle-aged men and women after famine exposure during gestation: the Dutch Hunger Winter Families Study. Am J Clin Nutr. 2009;89:1737–1743. doi: 10.3945/ajcn.2008.27038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ravelli AC, van Der Meulen JH, Osmond C, Barker DJ, Bleker OP. Obesity at the age of 50 y in men and women exposed to famine prenatally. Am J Clin Nutr. 1999;70:811–816. doi: 10.1093/ajcn/70.5.811. [DOI] [PubMed] [Google Scholar]
- 57.Dabelea D, et al. Intrauterine exposure to diabetes conveys risks for type 2 diabetes and obesity: a study of discordant sibships. Diabetes. 2000;49:2208–2211. doi: 10.2337/diabetes.49.12.2208. [DOI] [PubMed] [Google Scholar]
- 58.Tobi EW, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bouchard L, et al. Leptin gene epigenetic adaptation to impaired glucose metabolism during pregnancy. Diabetes Care. 2010;33:2436–2441. doi: 10.2337/dc10-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pencina MJ, D'Agostino RB, Sr, Demler OV. Novel metrics for evaluating improvement in discrimination: net reclassification and integrated discrimination improvement for normal variables and nested models. Stat Med. 2012;31:101–113. doi: 10.1002/sim.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]