Abstract
Psoriasis is a chronic inflammatory skin disease driven by aberrant signals from the immune system. In this issue Li et al. present the first large RNA-seq analysis of gene expression in normal skin and psoriasis lesions, providing a more comprehensive view of mRNA expression than earlier microarray studies. This study’s size enables gene co-expression analysis, a method illustrating which pathways are altered by the presence of disease.
The skin is a physical barrier, a thermoregulator, a radiation shield, and a barrier against infection. When tissue-resident and circulating immune cells function properly, invading bacteria and viruses are eliminated before they can threaten the host. However, when these powerful protective systems fail, the resulting inflammatory and autoimmune disorders can wreak havoc. Cutaneous psoriasis vulgaris is one of the most common immune-related inflammatory disorders, affecting about 2% of Americans (Nestle et al., 2009). The most visible symptoms of psoriasis are scaly, puritic surface lesions formed by an expansion of the outer cornified layer and a thickened, hyperproliferative epidermal layer that forms long projections down into the dermis. It is now generally agreed that these changes are driven by activated T cells that stimulate antigen-presenting cells in the skin, resulting in hyperplasia and a chronic pro-inflammatory microenvironment (Lowes et al., 2007). Understanding the pathogenesis of psoriasis therefore requires considering the dynamic relationships between innate and adaptive immunity in skin.
A new technique for a familiar question
In vitro and mouse xenotransplantation models have provided important insights into the pathogenesis of psoriasis, but studying human tissue in situ provides the most accurate picture of disease development and progression. Studies of increasing size and statistical power have used microarray technology to survey gene expression by analysis of mRNA in psoriatic lesions, uninvolved tissue from patients with psoriasis, and skin from disease-free individuals (Bowcock et al., 2001; Gudjonsson et al., 2009; Zhou et al., 2003). However, microarrays are not sensitive enough to detect rare transcripts, and it is difficult or impossible to assess the relative abundance of distinct gene isoforms or mRNA transcripts for which an array is not designed. There is evidence that microRNAs and other non-coding transcripts affect keratinocyte proliferation and are also altered in psoriasis (Xu et al., 2011). Thus, there is good reason to believe that a more sensitive and more general technique for measuring mRNA expression would improve our understanding of psoriasis and other skin diseases.
In this issue, Li et al. present the first large study to measure gene expression in psoriasis using RNA-seq on 92 psoriatic patients and 82 normal individuals (Li, 2014). RNA-seq provides greater sensitivity than microarrays, especially for transcripts expressed at very low levels. It can identify the expression of coding and non-coding transcripts absent on microarray platforms. Finally, it has the potential to quantify alternatively spliced RNA isoforms. Li et al. compare the RNA-seq results to their earlier microarray studies in psoriasis (Gudjonsson et al., 2009), and they show that, while differential expression analysis using both methods gives similar results on the most abundant transcripts, RNA-seq identifies a large number of transcripts expressed at low levels that could not be confidently called differentially expressed using a microarray study of equivalent size.
Pathways, not just levels
Consistent with previous findings, Li et al. identify significant up-regulation of genes known to be associated with inflammatory responses, cell proliferation, and keratinization in samples from patients with psoriasis. There is strong evidence that the pathogenesis of psoriasis involves activation of inflammatory signaling by activated TH17 T cells via the IL23 pathway (Di Cesare et al., 2009), and Li et al. report substantial up-regulation of inflammatory signatures and genes related to TH17 and interferon-gamma. They employ both differential expression analysis and Weighted Gene Co-expression Network Analysis, a statistical approach that identifies modules of significantly correlated genes (Figure 1A–B). Genes within co-expression modules are often related functionally or expressed in the same cell type. Network analysis permits the inference that genes in a module whose present function is uncertain may be related functionally to better-studied genes present in the same module. Gene expression network analysis has been applied fruitfully to numerous diseases, including obesity, skin cancer, and Alzheimer’s disease (Emilsson et al., 2008; Quigley et al., 2009; Zhang et al., 2013).
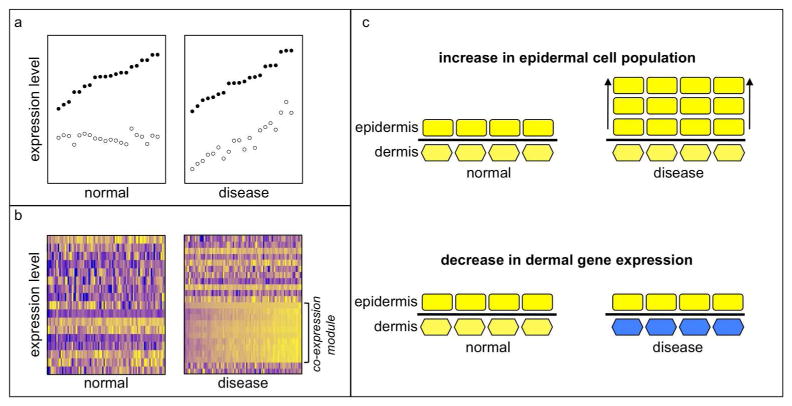
FIGURE 1. Correlation analysis identifies changed relationships between genes.
a) Expression levels of two hypothetical genes (solid and open circles) measured in normal and diseased tissue samples. Although each gene is expressed at the same level on average in both conditions, their expression levels are only correlated in disease. This relationship would not be evident from analysis of gene expression levels. b) Gene expression levels in normal and diseased tissue samples expressed as heat maps, with higher expression brighter yellow and lower expression darker blue. Correlation analysis can identify sets of genes that are significantly correlated with each other in only one state. These genes may be up-regulated, down-regulated, or not significantly different in expression levels. A decrease in gene expression between normal and diseased tissue can be complex to interpret when cell population proportions change. (c) In a schematic illustration, expansion of cells in the epidermis (rectangles) compared to cells in the dermis (hexagons) without a change in expression levels is illustrated at the top. A decrease in expression of dermal cells (colored blue, as in the heat map) without change in the relative cell population proportions is illustrated at the bottom. In both cases, the microarray will report a decrease in dermal gene expression relative to epidermal gene expression; additional analysis is required to understand to what extent each scenario applies.
Li et al. identified motifs in normal skin associated with physical structures such as hair follicles, adipose tissues, and erector pili muscles, as well as with processes such as keratinization. In psoriatic lesions, the most interesting up-regulated motifs were related to myeloid cells and T cells. The authors found significant decreases in expression levels of IL37 and, confirmed by immunostaining, that IL37 protein is not present in psoriatic lesions. IL37 was part of a psoriatic co-expression module enriched for genes expressed in the stratum granulosum, an epithelial layer that is diminished in psoriasis. It may be that disruption in the cell population expressing IL37 is responsible for its reduced expression. However, IL37 is an important anti-inflammatory IL-1 family cytokine (Nold et al., 2010), and reduced IL37 levels are compatible with the pro-inflammatory microenvironment in psoriasis.
A Question of Proportion
A fundamental question in gene expression analysis is the extent to which changes in gene expression are cell-intrinsic, as opposed to changing proportions of cell populations within a tissue (Figure 1C). Histopathological examination of tissue sections gives an indispensible grounding in the proportions of cell populations. Another approach is to sort cells by FACS using markers specific for cell types of interest, but this requires live cells, well-validated markers, and a willingness to discard information from the cells that are excluded by sorting. Given the importance of cross-talk between T cells and keratinocytes in psoriasis, there is value in measuring the expression from all cell types that reside in skin. Li et al. used gene expression measurements taken from dermal and epidermal tissues isolated by laser-capture microscopy to identify genes that are expressed preferentially in each compartment and that are expressed at consistent levels in normal and psoriasis tissues. They identified a decrease in the relative quantity of dermal-specific gene expression in psoriasis, consistent with the known increase in the epidermal compartment compared to the dermis.
For logistical and ethical reasons, until recently there has been a dearth of cohort studies of normal human tissues other than blood, and even fewer studies using RNA-seq. Although many early microarray studies of disease were performed with a small number of normal controls, analysis of a large number of normal tissue samples is crucial to understanding how tissues change during the progression of psoriasis. The ongoing GTEx project to analyze normal tissues in hundreds of healthy individuals will increase considerably the number of normal tissue samples available for analysis (Lonsdale et al., 2013). The value of control tissues can be improved by parallel processing and by using either the patient’s own unaffected tissue or samples appropriately matched to the patient population for important risk factors. Statistical approaches such as gene co-expression analysis require larger sample sizes than can be accommodated by laser capture microscopy, so further development and application of methods that cope with and exploit diverse cell populations within a sample will be quite useful. RNA-seq is now the technology of choice for genomic studies, although microarray will continue to be attractive in large studies where RNA-seq analysis is not economically feasible. Li et al. have illustrated the use of these approaches and the data they present will inform studies of psoriasis and the fields of skin biology and immunology.
Acknowledgments
Thanks to Minh To, Harry Quigley, and Allan Balmain for their comments and support.
References
- Bowcock AM, Shannon W, Du F, et al. Insights into psoriasis and other inflammatory diseases from large-scale gene expression studies. Hum Mol Genet. 2001;10:1793–805. doi: 10.1093/hmg/10.17.1793. [DOI] [PubMed] [Google Scholar]
- Di Cesare A, Di Meglio P, Nestle FO. The IL-23/Th17 axis in the immunopathogenesis of psoriasis. J Invest Dermatol. 2009;129:1339–50. doi: 10.1038/jid.2009.59. [DOI] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–8. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- Gudjonsson JE, Ding J, Li X, et al. Global gene expression analysis reveals evidence for decreased lipid biosynthesis and increased innate immunity in uninvolved psoriatic skin. J Invest Dermatol. 2009;129:2795–804. doi: 10.1038/jid.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides new insights into disease mechanisms. Journal of Investigative Dermatology. 2014 doi: 10.1038/jid.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445:866–73. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361:496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- Nold MF, Nold-Petry CA, Zepp JA, et al. IL-37 is a fundamental inhibitor of innate immunity. Nat Immunol. 2010;11:1014–22. doi: 10.1038/ni.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley D, To M, Pérez-Losada J, et al. Genetic architecture of mouse skin inflammation and tumour susceptibility. Nature. 2009;458:505–8. doi: 10.1038/nature07683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Brodin P, Wei T, et al. MiR-125b, a microRNA downregulated in psoriasis, modulates keratinocyte proliferation by targeting FGFR2. J Invest Dermatol. 2011;131:1521–9. doi: 10.1038/jid.2011.55. [DOI] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–20. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Krueger JG, Kao MC, et al. Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol Genomics. 2003;13:69–78. doi: 10.1152/physiolgenomics.00157.2002. [DOI] [PubMed] [Google Scholar]