Abstract
Genetic linkage analyses, genome-wide association studies of single nucleotide polymorphisms, copy number variation surveys, and mutation screenings found the human chromosomal 12q24 locus, with the genes SH2B3 and ATXN2 in its core, to be associated with an exceptionally wide spectrum of disease susceptibilities. Hematopoietic traits of red and white blood cells (like erythrocytosis and myeloproliferative disease), autoimmune disorders (like type 1 diabetes, coeliac disease, juvenile idiopathic arthritis, rheumatoid arthritis, thrombotic antiphospholipid syndrome, lupus erythematosus, multiple sclerosis, hypothyroidism and vitiligo), also vascular pathology (like kidney glomerular filtration rate deficits, serum urate levels, plasma beta-2-microglobulin levels, retinal microcirculation problems, diastolic and systolic blood pressure and hypertension, cardiovascular infarction), furthermore obesity, neurodegenerative conditions (like the polyglutamine-expansion disorder spinocerebellar ataxia type 2, Parkinson’s disease, the motor-neuron disease amyotrophic lateral sclerosis, and progressive supranuclear palsy), and finally longevity were reported. Now it is important to clarify, in which ways the loss or gain of function of the locally encoded proteins SH2B3/LNK and ataxin-2, respectively, contribute to these polygenic health problems. SH2B3/LNK is known to repress the JAK2/ABL1 dependent proliferation of white blood cells. Its null mutations in human and mouse are triggers of autoimmune traits and leukemia (acute lymphoblastic leukemia or chronic myeloid leukemia-like), while missense mutations were found in erythrocytosis-1 patients. Ataxin-2 is known to act on RNA-processing and trophic receptor internalization. While its polyglutamine-expansion mediated gain-of-function causes neuronal atrophy in human and mouse, its deletion leads to obesity and insulin resistance in mice. Thus, it is conceivable that the polygenic pathogenesis of type 1 diabetes is enhanced by an SH2B3-dysregulation-mediated predisposition to autoimmune diseases that conspires with an ATXN2-deficiency-mediated predisposition to lipid and glucose metabolism pathology.
Keywords: Diabetes mellitus type 1, 12q24, ATXN2, Obesity, SH2B3, Autoimmune
Core tip: Within the multifactorial pathogenesis of type 1 diabetes mellitus (T1D), a genetic risk mediated by the chromosome 12q24 locus was consistently observed. Mutations in the ATXN2 gene there trigger the pathogenesis of obesity, while mutations in the SH2B3 gene there trigger the pathogenesis of autoimmune processes. Given that both genes show co-regulated expression, their combined effects may drive these two core aspects of T1D. Tissue and phenotype studies of mouse mutants will identify molecular targets for causal therapies.
INTRODUCTION
The pathogenesis of many common multifactorial diseases was successfully elucidated over the past years, principally through genome-wide association studies (GWAS) in many thousands of sporadic patients vs control individuals. For diabetes mellitus type 1 (T1D), more than 40 chromosomal loci were uncovered to modulate disease risk[1,2]. However, now the challenge consists in establishing causality between one of the multiple genes contained in any locus and one of the disease features. One promising approach is the careful consideration of phenotypes and pathology caused by disruption or overexpression of any candidate gene, e.g., in mouse, and the subsequent comparison with relevant traits that occur within the first years of the disease course. Thus, clinical information may help to guide the characterization of mutant animals, while conversely the tissue analysis of mutant animals may help to elucidate presymptomatic stages of disease. A particularly complex example is the subject of this review-the association of T1D and many other medical conditions with mostly two single nucleotide polymorphisms (SNPs) on chromosome 12q24-rs3184504 and rs653178.
THE EXCEPTIONALLY PLEIOTROPIC DISEASE SUSCEPTIBILITY LOCUS ON CHROMOSOME 12Q24 EXTENDS FROM THE SH2B3 GENE ACROSS THE ATXN2 GENE, BUT MAY STRETCH BEYOND THESE BORDERS
Chromosome 12q contains one of the largest blocks of linkage disequilibrium (LD) in the human genome[3]. It was observed early on in European/Asian/African populations and found to span > 1 Megabase pairs (Mbp) across several genes including the growth repressor SH2B3, the RNA processing factor ATXN2, the nuclear localization inhibitor BRAP, the mitochondrial fatty acid beta-oxidation enzyme ACAD10, the alcohol metabolism enzyme ALDH2, and the stress kinase MAPKAPK5[4]. The core LD block was localized to exon 1 of the ATXN2 gene in a population of European ancestry, and was explained by positive selection of the (CAG)-repeat size in this exon[4]. Indeed, the most frequently observed disease associations at this 12q24 locus are within a 200000 basepairs (bp) fragment, which comprises the ATXN2 gene and the immediately adjacent SH2B3 gene (Figure 1). According to the United States National Center for Biotechnology Information reference sequences, human SH2B3 is transcribed in orientation from the centromere, covering about 46000 bp, and spans 9 predicted exons to constitute an mRNA of 5425 nucleotides, which encodes a protein of 575 amino acids. ATXN2 is transcribed in orientation from the telomere, covering about 147000 bp, and spans 24 predicted exons with several splice-isoforms, of which the longest constitutes an mRNA of 4712 nucleotides and encodes a protein of 1313 amino acids. The missense SNP rs3184504 in SH2B3 open reading frame (resulting in the substitution W262R) was observed in perfect cosegregation (r2 = 1) with the SNP rs653178 deep within intron 2 of the ATXN2 gene[5], in spite of a physical distance of 123148 bp. Since rs653178 is far away from ATXN2 splice sites and since the W262 codon in SH2B3 is not conserved between human and mouse[6], both of these polymorphisms are probably innocent bystanders and are noticed only through their frequency, depending on their random distribution within population stratifications. They are presumably coinherited with other rare sequence variants, e.g., within the promoters or within the mRNA 3’-untranslated regions, which alter the transcript expression levels slightly upwards or downwards. Indeed, both of these cosegregating SH2B3 and ATXN2 variants correlated with significant changes in the expression of both ATXN2 and SH2B3 mRNAs[7]. This coinheritance together with correlated expression changes makes it inherently difficult to establish causality between any of the individual traits within a complex disease and any of the neighbouring genes. This is exemplified by the allocation of six hematologic and three blood pressure traits to the region from SH2B3 to ATXN2 by genome-wide studies, reflecting the exceptional pleiotropy of this locus[8]. The 12q24 linkage disequilibrium block in some studies of restricted populations included further genes, namely CUTL2, FAM109A, SH2B3, ATXN2, BRAP, ACAD10, ALDH2, MAPKAPK5, TMEM116, ERP29[9], NAA25/C12orf30, TRAFD1, HECTD4/C12orf51, RPL6, PTPN11[10-12], thus extending across 1.5 Mbp. For these reasons it is crucial to consider monogenic mutants for each gene and their phenotypic effects, so as to decide which of them might contribute to each of the diseases. However, for most of these genes the relevant mouse mutants are not yet characterized.
Figure 1.
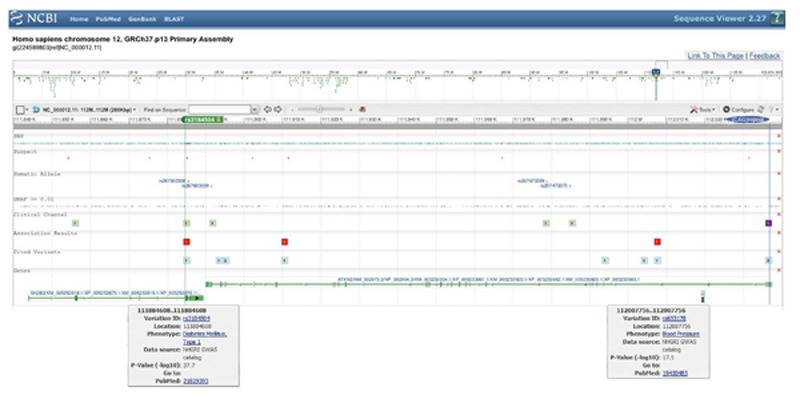
The core 200000 bp region of the chromosome 12q24 locus covering the immediately adjacent SH2B3 and ATXN2 genes, with an illustration of the single nucleotide polymorphism rs3184504 encoding the W272R missense variant of the SH2B3/LNK protein (as shown in the United States National Center for Biotechnology Information database) as well as the (CAG)-repeat structure encoding the unstable polyglutamine domain of the ataxin-2 protein.
NULL MUTATIONS IN MOUSE AND HUMAN DEMONSTRATE SH2B3 TO REPRESS THE PROLIFERATION OF WHITE BLOOD CELLS, IN PARTICULAR B-LYMPHOCYTES
The generation of mice with deletion of SH2B3 (also called Lnk) demonstrated primary splenomegaly and extramedullary hematopoiesis with progenitor hypersensitivity to various cytokines[13]. It caused the accumulation of pre-B and immature B-lymphocytes in enlarged spleens as well as an increase in B-lineage cells in the bone marrow, in parallel to unimpaired T-cell development in thymus[14]. It accelerated and exacerbated oncogenic JAK2-induced myeloproliferative diseases through an expansion of myeloid progenitors, accelerated myelofibrosis and finally features of chronic myeloid leukemia (CML). These murine data supported notions that SH2B3 directly inhibits oncogenic JAK2 and cooperates with the BCR/ABL oncogene in the development of CML[15]. Deletion of SH2B3 was also observed in a genomic and transcriptomic study of patients with BCR-ABL1-positive acute lymphoblastic leukemia with poor outcome (Ph-like ALL), together with promising therapeutic benefits from tyrosine kinase inhibitors[16]. Human germline homozygous SH2B3 mutations including a frameshift with translation stop resulted in growth retardation, high white cell counts in parallel to anemia and thrombocytopenia, splenomegaly and liver cirrhosis, autoimmune Hashimoto thyroiditis, speech delay and ALL. In addition, this study identified homozygous somatic SH2B3 frameshift mutations in ALL cases[17]. A 5 bp deletion of SH2B3, which was predicted to affect both the PH domain and the SH2 domain, manifested clinically as primary myelofibrosis. In contrast, a somatic E208Q missense mutation in the PH domain was observed in a patient with essential thrombocythemia[18]. SH2B3 was also shown to interact with platelet-derived growth factor receptor and repress its downstream signaling[19]. Interestingly, a selective increase in red blood cells (isolated erythrocytosis) was observed in two individuals with the SH2B3 missense mutations E208X and A215V[20]. However, SH2B3 sequencing in 23 erythrocytosis patients uncovered only one non-synonymous polymorphism of unclear relevance[6]. Systematic SH2B3 sequencing analysis in 42 patients with chronic phase myeloproliferative neoplasms detected a missense mutation in 7% of cases, either in the SH2 domain or in the C-terminal domain, which were always accompanied by a JAK2 mutation[21]. Myeloproliferative SH2B3 mutations within the PH domain were also shown to reduce SH2B3 function without altering its binding properties to JAK2, CBL and 14-3-3[22]. An analysis of peripheral mononuclear blood cells stimulated with anti-CD28 and anti-CD3 antibodies detected an increased proliferation of T-lymphocytes in carriers of the W262R missense SH2B3 variant, independent of the presence of juvenile type 1 diabetes[23]. In vitro studies had previously shown SH2B3 to attenuate the ability of SH2B1 to promote JAK2 activation and subsequent tyrosine phosphorylation of insulin receptor substrate-1 by JAK2[24]. SH2B3-deficient hematopoietic stem cells displayed an increased postnatal expansion and enhanced thrombopoietin responsiveness[25]. In subsequent studies they showed increased resistance to apoptosis due to enhanced expression of Bcl-xL upon thrombopoietin stimulation[26]. A limitation of growth by SH2B3 was also observed in the rat neuronal PC12 cell line and in primary cortical neurons, where neurotrophin-induced neurite outgrowth was downregulated by the binding of SH2B3 to the phosphorylated neurotrophin receptor TrkA and the repression of downstream signaling[27].
AUTOIMMUNE DISEASES (EOSINOPHIL NUMBERS, COELIAC DISEASE, JUVENILE IDIOPATHIC ARTHRITIS, RHEUMATOID ARTHRITIS, THROMBOTIC ANTIPHOSPHOLIPID SYNDROME, LUPUS ERYTHEMATOSUS, MULTIPLE SCLEROSIS, HYPOTHYROIDISM, VITILIGO) MAY BE MODULATED BY SH2B3
Possibly as an effect of SH2B3 on B-lymphocyte proliferation, the 12q24 locus modulates the risk for various autoimmune diseases. A GWAS in the Icelandic population studying eosinophil counts observed association with the SH2B3 SNP rs3184504[28]. A GWAS into coeliac disease found the SH2B3 SNP rs3184504 and the ATXN2 intronic SNP rs653178 to be associated[29]. Follow up studies of coeliac disease focusing on 9 and 11 candidate SNPs confirmed the association with SH2B3[30,31], and reported upregulation of SH2B3 mRNA expression levels in intestinal mucosa to be triggered by coeliac disease and by the risk allele T of the SH2B3 SNP rs3184504[31]. Further haplotype studies were confirmatory, and functional experiments indicated that carriers of the rs3184504 risk allele show stronger activation of the NOD2 recognition pathway in response to lipopolysaccharides and muramyl dipeptide[32]. A candidate study of sixteen SNPs known from coeliac disease and from T1D found an association of the ATXN2 SNP rs653178 with juvenile idiopathic arthritis[33]. GWAS studies into rheumatoid arthritis indicated association with SH2B3 particularly among rheumatoid-factor-positive patients[34]. A GWAS meta-analysis confirmed that the ATXN2 intronic SNP rs653178 is associated not only with coeliac disease, but also with rheumatoid arthritis[35]. A study of thrombophilia in antiphospholipid antibody positive individuals by array-comparative genomic hybridization analysis of copy number variations with subsequent fine mapping identified a risk haplotype comprising one SH2B3 SNP and two ATXN2 SNPs[36]. A GWAS of systemic lupus erythematosus observed association with the SNP rs17696736 within the ERP29 gene downstream from SH2B3[9]. A candidate study of 12 SNPs in almost 3000 Spanish multiple sclerosis patients detected association with the SH2B3 SNP rs3184504[37]. A GWAS into hypothyroidism reported the SH2B3 SNP rs3184504 to be associated, with autoimmune Hashimoto thyroiditis as a likely explanation for this observation[38]. A GWAS into the autoimmune skin disease vitiligo reported an association with the 12q24 locus extending from the SH2B3 across the ATXN2 gene[39].
T1D MELLITUS
The first GWAS into T1D encountered a maximal association with the 12q24 SNP rs17696736 in an intron of the C12ORF30/NAA25 gene, while the effect was consistently observed also in its neighbourhood across a 1.5 Mbp LD block[10]. An extended GWAS confirmed this observation and pointed out that the association with the W272R missense variant encoded in exon 3 of SH2B3 was sufficient to model the regional effect[40]. GWAS of additional cases corroborated the association with SH2B3[41], a further GWAS with meta-analysis and combined comparisons supported the association with rs3184504[42], and also a GWAS of affected sib-pair families showed association with the region from the SH2B3 SNP rs739496 across the ERP29 SNP rs17696736 until the SNP rs10850061 beyond PTPN11[11,43]. GWAS of autoantibody positive T1D patients again detected the association with SH2B3[44,45]. GWAS of soluble intercellular adhesion molecule-1 levels as an endothelium-derived inflammatory biomarker in diabetes and infarction also showed the association with the SH2B3 SNP rs3184504[46]. Candidate studies of 2 and 21 SNPs in T1D cases from Russia and United States, respectively, replicated the SH2B3 association[47,48]. Since the effect is so consistent, SH2B3 SNP genotyping was integrated into a signature of 8 polymorphisms that provide optimal prediction of T1D risk[49]. However, it is likely that the SH2B3 sequence variant rs3184504 is not biologically responsible by itself, since sequencing studies failed to find similar SH2B3 variants in NOD mice that model many T1D features[50].
EVIDENCE FROM MOUSE MUTANTS IMPLICATES ATXN2 IN METABOLIC SYNDROME
While the autoimmune component of T1D might be explained by the SH2B3 effect on lymphocyte proliferation, some metabolic features of T1D might be exacerbated by the ataxin-2 effect on glucose and lipid metabolism. Mice with targeted deletion of Atxn2 exon 1 and frameshift in homozygous state displayed marked obesity and infertility in two independently generated mutant lines[51,52]. Hepatic lipid and glycogen accumulation was evident already at age 6 mo. As in other insulin resistance syndromes, pancreatic and blood serum insulin levels were increased, in parallel to a reduction of insulin receptor (IR) protein levels in the liver, in spite of increased IR mRNA levels. Serum cholesterol was significantly increased[52]. Although ataxin-2 is mostly localized at the rough endoplasmic reticulum and has strong effects on mRNA processing[53-59], its effect on the IR is possibly explained through interactions with the endocytic internalization machinery of receptor tyrosine kinases[60,61]. TDP-43 is an interactor protein of ataxin-2 via joint RNA-binding[57], was also demonstrated to regulate glucose homeostasis and fat deposition, with its levels showing direct correlation with the expression levels of the obesity gene Tbc1d1, while its deletion affects the splicing of apolipoprotein A-II[62-64].
EVIDENCE FROM HUMAN MUTATIONS IMPLICATES ATXN2 IN OBESITY
The investigation of obesity in 92 children by systematic sequencing of the ATXN2 coding regions demonstrated a greatly increased frequency of the SNP rs695872 allele C and an overrepresentation of (CAG)-repeat sizes > 22[65]. Indeed, obesity and polyphagia were marked features of infants in middle stages of the neurodegenerative process caused by (CAG)-repeat expansions in ATXN2[66]. Thus, monogenic evidence links obesity to ATXN2 both in mice and in human. This is possibly reflected by a genome-wide SNP genotyping analysis, where SH2B3 variants were associated with low-density lipoprotein (LDL) cholesterol[67]. Interestingly, an association with obesity was also observed for the ataxin-2 binding protein 1 (A2BP1 or RBFOX1) both in a GWAS among Pima Indians and in a candidate approach among French Caucasian adults[68].
ATXN2 IS IMPORTANT FOR NEURODEGENERATIVE DISEASES
The polyglutamine (polyQ) domain at the N-terminal end of ataxin-2 normally has a size of Q22-23, usually encoded by a (CAG)8CAA(CAG)4CAA(CAG)8 sequence in exon 1 of the ATXN2 gene on chromosome 12q24. Its unstable expansion to large sizes beyond (CAG)31 is the monogenic cause of an autosomal dominant multi-system atrophy of the nervous system, which was named spinocerebellar ataxia type 2[69-86]. CAG-repeat expansions with cytosine adenosine adenosine (CAA) interruptions may also manifest as Parkinson’s disease[87,88]. Intermediate CAG-repeat sizes of 26-31 units, sometimes with CAA interruptions, act as polygenic risk factor for the motor-neuron disease amyotrophic lateral sclerosis[57,89]. Intermediate CAG-repeat expansions enhance also the risk for progressive supranuclear palsy[90]. Published evidence suggests that the polyglutamine expansions increase the half-life of ataxin-2 and that a gain-of-toxic-function through accumulation of ataxin-2 aggregates with sequestration of interactor proteins such as the poly(A)-binding-protein PABPC1 underlies the neurodegenerative process[57,91]. In spite of the vast evidence that excess ataxin-2 is the biological cause for neuronal death, SNP genotyping and association studies curiously found an SH2B3 allele haplotype to be more informative and to better predict amyotrophic lateral sclerosis risk than the ATXN2 alleles[92]. This observation underscores old experiences that maximal linkage logarithm of odds scores and maximal haplotype association scores within any chromosomal region depend on random population stratification effects and on the frequency/informativity of alleles. Thus, they are not suitable for the fine mapping of disease genes.
LONGEVITY
Interestingly, the discovery set of a GWAS of exceptional longevity in centenarians detected a significant association with the ATXN2 SNP rs653178, in parallel to several other disease associated SNPs, while the strongest effect correlated with the SNP rs2075650 at the TOMM40/apolipoprotein E (APOEO locus. TOMM40 encodes the channel forming subunit of the translocase across the mitochondrial outer membrane, while APOE encodes the apolipoprotein E, which mediates the binding and clearance of lipoprotein particles such as chylomicrons and very LDLs. Apolipoprotein E polymorphisms are the main known genetic factors associated with the risk of Alzheimer’s disease[93,94]. While it remained unclear in this longevity GWAS, whether an LD effect was consistently observed also for SNPs that surround ATXN2, and whether blood cell traits, autoimmune disorders, obesity, neurodegenerative processes or vascular pathology were underlying this observation, the authors reported their observation of a reduced frequency of the ATXN2 SNP rs653178 allele T among centenarians [with a log10(BayesFactor) of 1.2] in the light of previous ATXN2 GWAS association data with hypertension[93,94].
KIDNEY DISEASE, MICROCIRCULATION, HYPERTENSION AND CARDIOVASCULAR INFARCTION
Indeed, several independent GWAS found renal function (estimated glomerular filtration rate on the basis of cystatin c) and chronic kidney disease to be modulated by the rs653178 variant within an intron of the ATXN2 gene in populations of European and African ancestry[5,95-97]. Also a GWAS into plasma levels of beta-2-microglobulin as a biomarker of kidney function, cardiovascular diseases and mortality reported an association with the ATXN2 SNP rs653178[98]. Furthermore, a recent GWAS into serum urate concentrations uncovered an association with the ATXN2 SNP rs653178[99]. The analysis of 83 candidate SNPs showed kidney disease variants to be associated with vascular phenotypes only in the case of rs653178 within the ATXN2 gene and two SNPs at the SH2B3 locus[100]. A GWAS studying microcirculation as measured by retinal venular caliber reported 4 loci, with only the rs10774625 SNP within an ATXN2 intron showing also significant association with hypertension and coronary heart disease[12]. The ATXN2 SNP rs653178 and the SH2B3 SNP rs3184504 association with diastolic as well as systolic blood pressure, mean arterial pressure and pulse pressure was reported in three independent GWAS of populations with European and African ancestry[7,101-103]. Similarly, an association of the SH2B3 SNP rs3184504 with diastolic and systolic blood pressure and hypertension was detected in a GWAS of 200000 individuals of European descent[104]. A GWAS association of the ATXN2 SNP rs653178 with myocardial infarction was shown in Icelandic individuals[28]. A recent candidate SNP study replicated the association between the SH2B3 SNP rs3184504 and coronary heart disease also in South Asian patients[105]. Thus, it appears that the 12q24 locus has a marked effect on vascular pathology.
RED BLOOD CELL TRAITS
It is unclear whether the above vascular disorders are consequences of vessel wall pathology or of blood cell pathology. It may therefore be relevant that a GWAS into the genetic basis of six traits of erythrocytes (including hemoglobin concentration, hematocrit, mean corpuscular volume, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration and red blood cell count) also showed associations with the 12q24 locus from SH2B3 across the ATXN2 gene[106].
CONCLUSION
For further mechanistic insights it will be important to generate and characterize rodent mutants for each of the genes in the pleiotropic 12q24 disease susceptibility locus.
With the limited knowledge available so far, it is credible that SH2B3 modulates B-lymphocyte proliferation and autoimmune traits. Ataxin-2 gain-of-function is a well-established modulator of several neurodegenerative diseases, while its deficiency appears to predispose to insulin resistance, blood cholesterol elevation, hepatic glycogen and lipid accumulation with overall obesity. Thus, downstream effects of both genes might cooperate to enhance the risk for type 1 diabetes.
Since T1D is an age-associated disease, it will be important to age Atxn2-null mice beyond 6 mo to the end of their natural lifespan around 2 years. This will allow us to assess whether their obesity leads to hypertension and vascular pathology, e.g., in kidneys, whether red blood cell traits are altered, and whether their longevity is abnormal. In particular, the insulin resistance/obesity/dyslipidemia/hepatosteatosis induced by Atxn2-null mutations should be studied regarding their long-term consequences. Mechanistically, it will be intriguing to elucidate how the RNA processing effects of ataxin-2 lead to this pathology.
In view of the polyQ expansion effects extending the protein half-life and causing a gain-of-function of ataxin-2, it is conceivable that the polyQ shrinkage sizes (Q13-21) could mediate a decreased half-life of the protein and a partial loss-of-function. Thus, these rare variants might be associated with phenotypes that were observed in the Atxn2-null mouse, such as obesity, insulin-resistance and diabetes mellitus.
ACKNOWLEDGMENTS
We are grateful to Ulrich Müller and Jan Rozman for critical reading of the manuscript.
Footnotes
P- Reviewer: Joseph P S- Editor: Song XX L- Editor: A E- Editor: Liu SQ
References
- 1.Polychronakos C, Li Q. Understanding type 1 diabetes through genetics: advances and prospects. Nat Rev Genet. 2011;12:781–792. doi: 10.1038/nrg3069. [DOI] [PubMed] [Google Scholar]
- 2.Noble JA, Erlich HA. Genetics of type 1 diabetes. Cold Spring Harb Perspect Med. 2012;2:a007732. doi: 10.1101/cshperspect.a007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scherer SE, Muzny DM, Buhay CJ, Chen R, Cree A, Ding Y, Dugan-Rocha S, Gill R, Gunaratne P, Harris RA, Hawes AC, Hernandez J, Hodgson AV, Hume J, Jackson A, Khan ZM, Kovar-Smith C, Lewis LR, Lozado RJ, Metzker ML, Milosavljevic A, Miner GR, Montgomery KT, Morgan MB, Nazareth LV, Scott G, Sodergren E, Song XZ, Steffen D, Lovering RC, Wheeler DA, Worley KC, Yuan Y, Zhang Z, Adams CQ, Ansari-Lari MA, Ayele M, Brown MJ, Chen G, Chen Z, Clerc-Blankenburg KP, Davis C, Delgado O, Dinh HH, Draper H, Gonzalez-Garay ML, Havlak P, Jackson LR, Jacob LS, Kelly SH, Li L, Li Z, Liu J, Liu W, Lu J, Maheshwari M, Nguyen BV, Okwuonu GO, Pasternak S, Perez LM, Plopper FJ, Santibanez J, Shen H, Tabor PE, Verduzco D, Waldron L, Wang Q, Williams GA, Zhang J, Zhou J, Allen CC, Amin AG, Anyalebechi V, Bailey M, Barbaria JA, Bimage KE, Bryant NP, Burch PE, Burkett CE, Burrell KL, Calderon E, Cardenas V, Carter K, Casias K, Cavazos I, Cavazos SR, Ceasar H, Chacko J, Chan SN, Chavez D, Christopoulos C, Chu J, Cockrell R, Cox CD, Dang M, Dathorne SR, David R, Davis CM, Davy-Carroll L, Deshazo DR, Donlin JE, D’Souza L, Eaves KA, Egan A, Emery-Cohen AJ, Escotto M, Flagg N, Forbes LD, Gabisi AM, Garza M, Hamilton C, Henderson N, Hernandez O, Hines S, Hogues ME, Huang M, Idlebird DG, Johnson R, Jolivet A, Jones S, Kagan R, King LM, Leal B, Lebow H, Lee S, LeVan JM, Lewis LC, London P, Lorensuhewa LM, Loulseged H, Lovett DA, Lucier A, Lucier RL, Ma J, Madu RC, Mapua P, Martindale AD, Martinez E, Massey E, Mawhiney S, Meador MG, Mendez S, Mercado C, Mercado IC, Merritt CE, Miner ZL, Minja E, Mitchell T, Mohabbat F, Mohabbat K, Montgomery B, Moore N, Morris S, Munidasa M, Ngo RN, Nguyen NB, Nickerson E, Nwaokelemeh OO, Nwokenkwo S, Obregon M, Oguh M, Oragunye N, Oviedo RJ, Parish BJ, Parker DN, Parrish J, Parks KL, Paul HA, Payton BA, Perez A, Perrin W, Pickens A, Primus EL, Pu LL, Puazo M, Quiles MM, Quiroz JB, Rabata D, Reeves K, Ruiz SJ, Shao H, Sisson I, Sonaike T, Sorelle RP, Sutton AE, Svatek AF, Svetz LA, Tamerisa KS, Taylor TR, Teague B, Thomas N, Thorn RD, Trejos ZY, Trevino BK, Ukegbu ON, Urban JB, Vasquez LI, Vera VA, Villasana DM, Wang L, Ward-Moore S, Warren JT, Wei X, White F, Williamson AL, Wleczyk R, Wooden HS, Wooden SH, Yen J, Yoon L, Yoon V, Zorrilla SE, Nelson D, Kucherlapati R, Weinstock G, Gibbs RA; Baylor College of Medicine Human Genome Sequencing Center Sequence Production Team. The finished DNA sequence of human chromosome 12. Nature. 2006;440:346–351. doi: 10.1038/nature04569. [DOI] [PubMed] [Google Scholar]
- 4.Yu F, Sabeti PC, Hardenbol P, Fu Q, Fry B, Lu X, Ghose S, Vega R, Perez A, Pasternak S, et al. Positive selection of a pre-expansion CAG repeat of the human SCA2 gene. PLoS Genet. 2005;1:e41. doi: 10.1371/journal.pgen.0010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Köttgen A, Pattaro C, Boger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMullin MF, Wu C, Percy MJ, Tong W. A nonsynonymous LNK polymorphism associated with idiopathic erythrocytosis. Am J Hematol. 2011;86:962–964. doi: 10.1002/ajh.22154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganesh SK, Tragante V, Guo W, Guo Y, Lanktree MB, Smith EN, Johnson T, Castillo BA, Barnard J, Baumert J, et al. Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum Mol Genet. 2013;22:1663–1678. doi: 10.1093/hmg/dds555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang J, Johnson AD, O’Donnell CJ. PRIMe: a method for characterization and evaluation of pleiotropic regions from multiple genome-wide association studies. Bioinformatics. 2011;27:1201–1206. doi: 10.1093/bioinformatics/btr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, Ortmann W, Kosoy R, Ferreira RC, Nordmark G, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper JD, Walker NM, Smyth DJ, Downes K, Healy BC, Todd JA. Follow-up of 1715 SNPs from the Wellcome Trust Case Control Consortium genome-wide association study in type I diabetes families. Genes Immun. 2009;10 Suppl 1:S85–S94. doi: 10.1038/gene.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikram MK, Sim X, Jensen RA, Cotch MF, Hewitt AW, Ikram MA, Wang JJ, Klein R, Klein BE, Breteler MM, et al. Four novel Loci (19q13, 6q24, 12q24, and 5q14) influence the microcirculation in vivo. PLoS Genet. 2010;6:e1001184. doi: 10.1371/journal.pgen.1001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velazquez L, Cheng AM, Fleming HE, Furlonger C, Vesely S, Bernstein A, Paige CJ, Pawson T. Cytokine signaling and hematopoietic homeostasis are disrupted in Lnk-deficient mice. J Exp Med. 2002;195:1599–1611. doi: 10.1084/jem.20011883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takaki S, Sauer K, Iritani BM, Chien S, Ebihara Y, Tsuji K, Takatsu K, Perlmutter RM. Control of B cell production by the adaptor protein lnk. Definition Of a conserved family of signal-modulating proteins. Immunity. 2000;13:599–609. doi: 10.1016/s1074-7613(00)00060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bersenev A, Wu C, Balcerek J, Jing J, Kundu M, Blobel GA, Chikwava KR, Tong W. Lnk constrains myeloproliferative diseases in mice. J Clin Invest. 2010;120:2058–2069. doi: 10.1172/JCI42032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, Chen SC, Payne-Turner D, Churchman ML, Harvey RC, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Garcia A, Ambesi-Impiombato A, Hadler M, Rigo I, LeDuc CA, Kelly K, Jalas C, Paietta E, Racevskis J, Rowe JM, et al. Genetic loss of SH2B3 in acute lymphoblastic leukemia. Blood. 2013;122:2425–2432. doi: 10.1182/blood-2013-05-500850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh ST, Simonds EF, Jones C, Hale MB, Goltsev Y, Gibbs KD, Merker JD, Zehnder JL, Nolan GP, Gotlib J. Novel mutations in the inhibitory adaptor protein LNK drive JAK-STAT signaling in patients with myeloproliferative neoplasms. Blood. 2010;116:988–992. doi: 10.1182/blood-2010-02-270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gueller S, Hehn S, Nowak V, Gery S, Serve H, Brandts CH, Koeffler HP. Adaptor protein Lnk binds to PDGF receptor and inhibits PDGF-dependent signaling. Exp Hematol. 2011;39:591–600. doi: 10.1016/j.exphem.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Lasho TL, Pardanani A, Tefferi A. LNK mutations in JAK2 mutation-negative erythrocytosis. N Engl J Med. 2010;363:1189–1190. doi: 10.1056/NEJMc1006966. [DOI] [PubMed] [Google Scholar]
- 21.Ha JS, Jeon DS. Possible new LNK mutations in myeloproliferative neoplasms. Am J Hematol. 2011;86:866–868. doi: 10.1002/ajh.22107. [DOI] [PubMed] [Google Scholar]
- 22.Koren-Michowitz M, Gery S, Tabayashi T, Lin D, Alvarez R, Nagler A, Koeffler HP. SH2B3 (LNK) mutations from myeloproliferative neoplasms patients have mild loss of function against wild type JAK2 and JAK2 V617F. Br J Haematol. 2013;161:811–820. doi: 10.1111/bjh.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavrikova EY, Nikitin AG, Kuraeva TL, Peterkova VA, Tsitlidze NM, Chistiakov DA, Nosikov VV. The carriage of the type 1 diabetes-associated R262W variant of human LNK correlates with increased proliferation of peripheral blood monocytes in diabetic patients. Pediatr Diabetes. 2011;12:127–132. doi: 10.1111/j.1399-5448.2010.00656.x. [DOI] [PubMed] [Google Scholar]
- 24.Li M, Li Z, Morris DL, Rui L. Identification of SH2B2beta as an inhibitor for SH2B1- and SH2B2alpha-promoted Janus kinase-2 activation and insulin signaling. Endocrinology. 2007;148:1615–1621. doi: 10.1210/en.2006-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buza-Vidas N, Antonchuk J, Qian H, Månsson R, Luc S, Zandi S, Anderson K, Takaki S, Nygren JM, Jensen CT, et al. Cytokines regulate postnatal hematopoietic stem cell expansion: opposing roles of thrombopoietin and LNK. Genes Dev. 2006;20:2018–2023. doi: 10.1101/gad.385606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki N, Yamazaki S, Ema H, Yamaguchi T, Nakauchi H, Takaki S. Homeostasis of hematopoietic stem cells regulated by the myeloproliferative disease associated-gene product Lnk/Sh2b3 via Bcl-xL. Exp Hematol. 2012;40:166–174.e3. doi: 10.1016/j.exphem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Wang TC, Chiu H, Chang YJ, Hsu TY, Chiu IM, Chen L. The adaptor protein SH2B3 (Lnk) negatively regulates neurite outgrowth of PC12 cells and cortical neurons. PLoS One. 2011;6:e26433. doi: 10.1371/journal.pone.0026433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, Thorleifsson G, Helgadottir H, Steinthorsdottir V, Stefansson H, Williams C, Hui J, Beilby J, Warrington NM, James A, Palmer LJ, Koppelman GH, Heinzmann A, Krueger M, Boezen HM, Wheatley A, Altmuller J, Shin HD, Uh ST, Cheong HS, Jonsdottir B, Gislason D, Park CS, Rasmussen LM, Porsbjerg C, Hansen JW, Backer V, Werge T, Janson C, Jönsson UB, Ng MC, Chan J, So WY, Ma R, Shah SH, Granger CB, Quyyumi AA, Levey AI, Vaccarino V, Reilly MP, Rader DJ, Williams MJ, van Rij AM, Jones GT, Trabetti E, Malerba G, Pignatti PF, Boner A, Pescollderungg L, Girelli D, Olivieri O, Martinelli N, Ludviksson BR, Ludviksdottir D, Eyjolfsson GI, Arnar D, Thorgeirsson G, Deichmann K, Thompson PJ, Wjst M, Hall IP, Postma DS, Gislason T, Gulcher J, Kong A, Jonsdottir I, Thorsteinsdottir U, Stefansson K. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 29.Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, Romanos J, Dinesen LC, Ryan AW, Panesar D, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romanos J, Barisani D, Trynka G, Zhernakova A, Bardella MT, Wijmenga C. Six new coeliac disease loci replicated in an Italian population confirm association with coeliac disease. J Med Genet. 2009;46:60–63. doi: 10.1136/jmg.2008.061457. [DOI] [PubMed] [Google Scholar]
- 31.Plaza-Izurieta L, Castellanos-Rubio A, Irastorza I, Fernández-Jimenez N, Gutierrez G, Bilbao JR. Revisiting genome wide association studies (GWAS) in coeliac disease: replication study in Spanish population and expression analysis of candidate genes. J Med Genet. 2011;48:493–496. doi: 10.1136/jmg.2011.089714. [DOI] [PubMed] [Google Scholar]
- 32.Zhernakova A, Elbers CC, Ferwerda B, Romanos J, Trynka G, Dubois PC, de Kovel CG, Franke L, Oosting M, Barisani D, et al. Evolutionary and functional analysis of celiac risk loci reveals SH2B3 as a protective factor against bacterial infection. Am J Hum Genet. 2010;86:970–977. doi: 10.1016/j.ajhg.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinks A, Martin P, Flynn E, Eyre S, Packham J, Barton A, Worthington J, Thomson W. Investigation of type 1 diabetes and coeliac disease susceptibility loci for association with juvenile idiopathic arthritis. Ann Rheum Dis. 2010;69:2169–2172. doi: 10.1136/ard.2009.126938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coenen MJ, Trynka G, Heskamp S, Franke B, van Diemen CC, Smolonska J, van Leeuwen M, Brouwer E, Boezen MH, Postma DS, et al. Common and different genetic background for rheumatoid arthritis and coeliac disease. Hum Mol Genet. 2009;18:4195–4203. doi: 10.1093/hmg/ddp365. [DOI] [PubMed] [Google Scholar]
- 35.Zhernakova A, Stahl EA, Trynka G, Raychaudhuri S, Festen EA, Franke L, Westra HJ, Fehrmann RS, Kurreeman FA, Thomson B, et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ochoa E, Iriondo M, Bielsa A, Ruiz-Irastorza G, Estonba A, Zubiaga AM. Thrombotic antiphospholipid syndrome shows strong haplotypic association with SH2B3-ATXN2 locus. PLoS One. 2013;8:e67897. doi: 10.1371/journal.pone.0067897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alcina A, Vandenbroeck K, Otaegui D, Saiz A, Gonzalez JR, Fernandez O, Cavanillas ML, Cénit MC, Arroyo R, Alloza I, et al. The autoimmune disease-associated KIF5A, CD226 and SH2B3 gene variants confer susceptibility for multiple sclerosis. Genes Immun. 2010;11:439–445. doi: 10.1038/gene.2010.30. [DOI] [PubMed] [Google Scholar]
- 38.Eriksson N, Tung JY, Kiefer AK, Hinds DA, Francke U, Mountain JL, Do CB. Novel associations for hypothyroidism include known autoimmune risk loci. PLoS One. 2012;7:e34442. doi: 10.1371/journal.pone.0034442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin Y, Birlea SA, Fain PR, Ferrara TM, Ben S, Riccardi SL, Cole JB, Gowan K, Holland PJ, Bennett DC, et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet. 2012;44:676–680. doi: 10.1038/ng.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JH, Howson JM, Stevens H, McManus R, Wijmenga C, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooper JD, Walker NM, Healy BC, Smyth DJ, Downes K, Todd JA. Analysis of 55 autoimmune disease and type II diabetes loci: further confirmation of chromosomes 4q27, 12q13.2 and 12q24.13 as type I diabetes loci, and support for a new locus, 12q13.3-q14.1. Genes Immun. 2009;10 Suppl 1:S95–120. doi: 10.1038/gene.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howson JM, Rosinger S, Smyth DJ, Boehm BO, Todd JA. Genetic analysis of adult-onset autoimmune diabetes. Diabetes. 2011;60:2645–2653. doi: 10.2337/db11-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plagnol V, Howson JM, Smyth DJ, Walker N, Hafler JP, Wallace C, Stevens H, Jackson L, Simmonds MJ, Bingley PJ, et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 2011;7:e1002216. doi: 10.1371/journal.pgen.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paré G, Ridker PM, Rose L, Barbalic M, Dupuis J, Dehghan A, Bis JC, Benjamin EJ, Shiffman D, Parker AN, et al. Genome-wide association analysis of soluble ICAM-1 concentration reveals novel associations at the NFKBIK, PNPLA3, RELA, and SH2B3 loci. PLoS Genet. 2011;7:e1001374. doi: 10.1371/journal.pgen.1001374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nikitin AG, Lavrikova EIu, Seregin IuA, Zil’berman LI, Tsitlidze NM, Kuraeva TL, Peterkova VA, Dedov II, Nosikov VV. [Association of the polymorphisms of the ERBB3 and SH2B3 genes with type 1 diabetes] Mol Biol (Mosk) 2010;44:257–262. [PubMed] [Google Scholar]
- 48.Reddy MV, Wang H, Liu S, Bode B, Reed JC, Steed RD, Anderson SW, Steed L, Hopkins D, She JX. Association between type 1 diabetes and GWAS SNPs in the southeast US Caucasian population. Genes Immun. 2011;12:208–212. doi: 10.1038/gene.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winkler C, Krumsiek J, Lempainen J, Achenbach P, Grallert H, Giannopoulou E, Bunk M, Theis FJ, Bonifacio E, Ziegler AG. A strategy for combining minor genetic susceptibility genes to improve prediction of disease in type 1 diabetes. Genes Immun. 2012;13:549–555. doi: 10.1038/gene.2012.36. [DOI] [PubMed] [Google Scholar]
- 50.Li YJ, Li XY, Guo XR, Li Y, Shen BF, Shi YC, Zhang JY. Absence of SH2B3 mutation in nonobese diabetic mice. Genet Mol Res. 2012;11:1266–1271. doi: 10.4238/2012.May.9.6. [DOI] [PubMed] [Google Scholar]
- 51.Kiehl TR, Nechiporuk A, Figueroa KP, Keating MT, Huynh DP, Pulst SM. Generation and characterization of Sca2 (ataxin-2) knockout mice. Biochem Biophys Res Commun. 2006;339:17–24. doi: 10.1016/j.bbrc.2005.10.186. [DOI] [PubMed] [Google Scholar]
- 52.Lastres-Becker I, Brodesser S, Lütjohann D, Azizov M, Buchmann J, Hintermann E, Sandhoff K, Schürmann A, Nowock J, Auburger G. Insulin receptor and lipid metabolism pathology in ataxin-2 knock-out mice. Hum Mol Genet. 2008;17:1465–1481. doi: 10.1093/hmg/ddn035. [DOI] [PubMed] [Google Scholar]
- 53.Satterfield TF, Pallanck LJ. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum Mol Genet. 2006;15:2523–2532. doi: 10.1093/hmg/ddl173. [DOI] [PubMed] [Google Scholar]
- 54.Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, Lehrach H, Krobitsch S. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tharun S. Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int Rev Cell Mol Biol. 2009;272:149–189. doi: 10.1016/S1937-6448(08)01604-3. [DOI] [PubMed] [Google Scholar]
- 56.van de Loo S, Eich F, Nonis D, Auburger G, Nowock J. Ataxin-2 associates with rough endoplasmic reticulum. Exp Neurol. 2009;215:110–118. doi: 10.1016/j.expneurol.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 57.Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCann C, Holohan EE, Das S, Dervan A, Larkin A, Lee JA, Rodrigues V, Parker R, Ramaswami M. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc Natl Acad Sci USA. 2011;108:E655–E662. doi: 10.1073/pnas.1107198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lim C, Allada R. ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science. 2013;340:875–879. doi: 10.1126/science.1234785. [DOI] [PubMed] [Google Scholar]
- 60.Nonis D, Schmidt MH, van de Loo S, Eich F, Dikic I, Nowock J, Auburger G. Ataxin-2 associates with the endocytosis complex and affects EGF receptor trafficking. Cell Signal. 2008;20:1725–1739. doi: 10.1016/j.cellsig.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 61.Drost J, Nonis D, Eich F, Leske O, Damrath E, Brunt ER, Lastres-Becker I, Heumann R, Nowock J, Auburger G. Ataxin-2 modulates the levels of Grb2 and SRC but not ras signaling. J Mol Neurosci. 2013;51:68–81. doi: 10.1007/s12031-012-9949-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiang PM, Ling J, Jeong YH, Price DL, Aja SM, Wong PC. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc Natl Acad Sci USA. 2010;107:16320–16324. doi: 10.1073/pnas.1002176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stallings NR, Puttaparthi K, Dowling KJ, Luther CM, Burns DK, Davis K, Elliott JL. TDP-43, an ALS linked protein, regulates fat deposition and glucose homeostasis. PLoS One. 2013;8:e71793. doi: 10.1371/journal.pone.0071793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mercado PA, Ayala YM, Romano M, Buratti E, Baralle FE. Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 2005;33:6000–6010. doi: 10.1093/nar/gki897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Figueroa KP, Farooqi S, Harrup K, Frank J, O’Rahilly S, Pulst SM. Genetic variance in the spinocerebellar ataxia type 2 (ATXN2) gene in children with severe early onset obesity. PLoS One. 2009;4:e8280. doi: 10.1371/journal.pone.0008280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abdel-Aleem A, Zaki MS. Spinocerebellar ataxia type 2 (SCA2) in an Egyptian family presenting with polyphagia and marked CAG expansion in infancy. J Neurol. 2008;255:413–419. doi: 10.1007/s00415-008-0690-4. [DOI] [PubMed] [Google Scholar]
- 67.Talmud PJ, Drenos F, Shah S, Shah T, Palmen J, Verzilli C, Gaunt TR, Pallas J, Lovering R, Li K, et al. Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip. Am J Hum Genet. 2009;85:628–642. doi: 10.1016/j.ajhg.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma L, Hanson RL, Traurig MT, Muller YL, Kaur BP, Perez JM, Meyre D, Fu M, Körner A, Franks PW, et al. Evaluation of A2BP1 as an obesity gene. Diabetes. 2010;59:2837–2845. doi: 10.2337/db09-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Imbert G, Saudou F, Yvert G, Devys D, Trottier Y, Garnier JM, Weber C, Mandel JL, Cancel G, Abbas N, et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat Genet. 1996;14:285–291. doi: 10.1038/ng1196-285. [DOI] [PubMed] [Google Scholar]
- 70.Pulst SM, Nechiporuk A, Nechiporuk T, Gispert S, Chen XN, Lopes-Cendes I, Pearlman S, Starkman S, Orozco-Diaz G, Lunkes A, et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet. 1996;14:269–276. doi: 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]
- 71.Sanpei K, Takano H, Igarashi S, Sato T, Oyake M, Sasaki H, Wakisaka A, Tashiro K, Ishida Y, Ikeuchi T, et al. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat Genet. 1996;14:277–284. doi: 10.1038/ng1196-277. [DOI] [PubMed] [Google Scholar]
- 72.Auburger G, Diaz GO, Capote RF, Sanchez SG, Perez MP, del Cueto ME, Meneses MG, Farrall M, Williamson R, Chamberlain S. Autosomal dominant ataxia: genetic evidence for locus heterogeneity from a Cuban founder-effect population. Am J Hum Genet. 1990;46:1163–1177. [PMC free article] [PubMed] [Google Scholar]
- 73.Orozco Diaz G, Nodarse Fleites A, Cordovés Sagaz R, Auburger G. Autosomal dominant cerebellar ataxia: clinical analysis of 263 patients from a homogeneous population in Holguín, Cuba. Neurology. 1990;40:1369–1375. doi: 10.1212/wnl.40.9.1369. [DOI] [PubMed] [Google Scholar]
- 74.Gispert S, Twells R, Orozco G, Brice A, Weber J, Heredero L, Scheufler K, Riley B, Allotey R, Nothers C. Chromosomal assignment of the second locus for autosomal dominant cerebellar ataxia (SCA2) to chromosome 12q23-24.1. Nat Genet. 1993;4:295–299. doi: 10.1038/ng0793-295. [DOI] [PubMed] [Google Scholar]
- 75.Estrada R, Galarraga J, Orozco G, Nodarse A, Auburger G. Spinocerebellar ataxia 2 (SCA2): morphometric analyses in 11 autopsies. Acta Neuropathol. 1999;97:306–310. doi: 10.1007/s004010050989. [DOI] [PubMed] [Google Scholar]
- 76.Lastres-Becker I, Rüb U, Auburger G. Spinocerebellar ataxia 2 (SCA2) Cerebellum. 2008;7:115–124. doi: 10.1007/s12311-008-0019-y. [DOI] [PubMed] [Google Scholar]
- 77.Rüb U, Schöls L, Paulson H, Auburger G, Kermer P, Jen JC, Seidel K, Korf HW, Deller T. Clinical features, neurogenetics and neuropathology of the polyglutamine spinocerebellar ataxias type 1, 2, 3, 6 and 7. Prog Neurobiol. 2013;104:38–66. doi: 10.1016/j.pneurobio.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 78.Gierga K, Bürk K, Bauer M, Orozco Diaz G, Auburger G, Schultz C, Vuksic M, Schöls L, de Vos RA, Braak H, et al. Involvement of the cranial nerves and their nuclei in spinocerebellar ataxia type 2 (SCA2) Acta Neuropathol. 2005;109:617–631. doi: 10.1007/s00401-005-1014-8. [DOI] [PubMed] [Google Scholar]
- 79.Velázquez-Pérez L, Seifried C, Santos-Falcón N, Abele M, Ziemann U, Almaguer LE, Martínez-Góngora E, Sánchez-Cruz G, Canales N, Pérez-González R, et al. Saccade velocity is controlled by polyglutamine size in spinocerebellar ataxia 2. Ann Neurol. 2004;56:444–447. doi: 10.1002/ana.20220. [DOI] [PubMed] [Google Scholar]
- 80.Rüb U, Del Turco D, Del Tredici K, de Vos RA, Brunt ER, Reifenberger G, Seifried C, Schultz C, Auburger G, Braak H. Thalamic involvement in a spinocerebellar ataxia type 2 (SCA2) and a spinocerebellar ataxia type 3 (SCA3) patient, and its clinical relevance. Brain. 2003;126:2257–2272. doi: 10.1093/brain/awg234. [DOI] [PubMed] [Google Scholar]
- 81.Hernández A, Magariño C, Gispert S, Santos N, Lunkes A, Orozco G, Heredero L, Beckmann J, Auburger G. Genetic mapping of the spinocerebellar ataxia 2 (SCA2) locus on chromosome 12q23-q24.1. Genomics. 1995;25:433–435. doi: 10.1016/0888-7543(95)80043-l. [DOI] [PubMed] [Google Scholar]
- 82.Gispert S, Lunkes A, Santos N, Orozco G, Ha-Hao D, Ratzlaff T, Aguiar J, Torrens I, Heredero L, Brice A. Localization of the candidate gene D-amino acid oxidase outside the refined I-cM region of spinocerebellar ataxia 2. Am J Hum Genet. 1995;57:972–975. [PMC free article] [PubMed] [Google Scholar]
- 83.Tuin I, Voss U, Kang JS, Kessler K, Rüb U, Nolte D, Lochmüller H, Tinschert S, Claus D, Krakow K, et al. Stages of sleep pathology in spinocerebellar ataxia type 2 (SCA2) Neurology. 2006;67:1966–1972. doi: 10.1212/01.wnl.0000247054.90322.14. [DOI] [PubMed] [Google Scholar]
- 84.Riess O, Laccone FA, Gispert S, Schöls L, Zühlke C, Vieira-Saecker AM, Herlt S, Wessel K, Epplen JT, Weber BH, et al. SCA2 trinucleotide expansion in German SCA patients. Neurogenetics. 1997;1:59–64. doi: 10.1007/s100480050009. [DOI] [PubMed] [Google Scholar]
- 85.Belal S, Cancel G, Stevanin G, Hentati F, Khati C, Ben Hamida C, Auburger G, Agid Y, Ben Hamida M, Brice A. Clinical and genetic analysis of a Tunisian family with autosomal dominant cerebellar ataxia type 1 linked to the SCA2 locus. Neurology. 1994;44:1423–1426. doi: 10.1212/wnl.44.8.1423. [DOI] [PubMed] [Google Scholar]
- 86.Schöls L, Gispert S, Vorgerd M, Menezes Vieira-Saecker AM, Blanke P, Auburger G, Amoiridis G, Meves S, Epplen JT, Przuntek H, et al. Spinocerebellar ataxia type 2. Genotype and phenotype in German kindreds. Arch Neurol. 1997;54:1073–1080. doi: 10.1001/archneur.1997.00550210011007. [DOI] [PubMed] [Google Scholar]
- 87.Charles P, Camuzat A, Benammar N, Sellal F, Destée A, Bonnet AM, Lesage S, Le Ber I, Stevanin G, Dürr A, et al. Are interrupted SCA2 CAG repeat expansions responsible for parkinsonism? Neurology. 2007;69:1970–1975. doi: 10.1212/01.wnl.0000269323.21969.db. [DOI] [PubMed] [Google Scholar]
- 88.Gispert S, Kurz A, Waibel S, Bauer P, Liepelt I, Geisen C, Gitler AD, Becker T, Weber M, Berg D, et al. The modulation of Amyotrophic Lateral Sclerosis risk by ataxin-2 intermediate polyglutamine expansions is a specific effect. Neurobiol Dis. 2012;45:356–361. doi: 10.1016/j.nbd.2011.08.021. [DOI] [PubMed] [Google Scholar]
- 89.Lee T, Li YR, Ingre C, Weber M, Grehl T, Gredal O, de Carvalho M, Meyer T, Tysnes OB, Auburger G, et al. Ataxin-2 intermediate-length polyglutamine expansions in European ALS patients. Hum Mol Genet. 2011;20:1697–1700. doi: 10.1093/hmg/ddr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ross OA, Rutherford NJ, Baker M, Soto-Ortolaza AI, Carrasquillo MM, DeJesus-Hernandez M, Adamson J, Li M, Volkening K, Finger E, et al. Ataxin-2 repeat-length variation and neurodegeneration. Hum Mol Genet. 2011;20:3207–3212. doi: 10.1093/hmg/ddr227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Damrath E, Heck MV, Gispert S, Azizov M, Nowock J, Seifried C, Rüb U, Walter M, Auburger G. ATXN2-CAG42 sequesters PABPC1 into insolubility and induces FBXW8 in cerebellum of old ataxic knock-in mice. PLoS Genet. 2012;8:e1002920. doi: 10.1371/journal.pgen.1002920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lahut S, Ömür Ö, Uyan Ö, Ağım ZS, Özoğuz A, Parman Y, Deymeer F, Oflazer P, Koç F, Özçelik H, et al. ATXN2 and its neighbouring gene SH2B3 are associated with increased ALS risk in the Turkish population. PLoS One. 2012;7:e42956. doi: 10.1371/journal.pone.0042956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sebastiani P, Solovieff N, Puca A, Hartley SW, Melista E, Andersen S, Dworkis DA, Wilk JB, Myers RH, Steinberg MH, et al. Genetic signatures of exceptional longevity in humans. Science. 2010;2010 doi: 10.1126/science.1190532. [DOI] [PubMed] [Google Scholar]
- 94.Sebastiani P, Solovieff N, Dewan AT, Walsh KM, Puca A, Hartley SW, Melista E, Andersen S, Dworkis DA, Wilk JB, et al. Genetic signatures of exceptional longevity in humans. PLoS One. 2012;7:e29848. doi: 10.1371/journal.pone.0029848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Köttgen A. Genome-wide association studies in nephrology research. Am J Kidney Dis. 2010;56:743–758. doi: 10.1053/j.ajkd.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 96.Böger CA, Heid IM. Chronic kidney disease: novel insights from genome-wide association studies. Kidney Blood Press Res. 2011;34:225–234. doi: 10.1159/000326901. [DOI] [PubMed] [Google Scholar]
- 97.Liu CT, Garnaas MK, Tin A, Kottgen A, Franceschini N, Peralta CA, de Boer IH, Lu X, Atkinson E, Ding J, et al. Genetic association for renal traits among participants of African ancestry reveals new loci for renal function. PLoS Genet. 2011;7:e1002264. doi: 10.1371/journal.pgen.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tin A, Astor BC, Boerwinkle E, Hoogeveen RC, Coresh J, Kao WH. Genome-wide association study identified the human leukocyte antigen region as a novel locus for plasma beta-2 microglobulin. Hum Genet. 2013;132:619–627. doi: 10.1007/s00439-013-1274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Köttgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, Pistis G, Ruggiero D, O’Seaghdha CM, Haller T, et al. Liu K, D’Adamo P, Ulivi S, Rotter JI, Psaty BM, Vollenweider P, Waeber G, Campbell S, Devuyst O, Navarro P, Kolcic I, Hastie N, Balkau B, Froguel P, Esko T, Salumets A, Khaw KT, Langenberg C, Wareham NJ, Isaacs A, Kraja A, Zhang Q, Wild PS, Scott RJ, Holliday EG, Org E, Viigimaa M, Bandinelli S, Metter JE, Lupo A, Trabetti E, Sorice R, Doring A, Lattka E, Strauch K, Theis F, Waldenberger M, Wichmann HE, Davies G, Gow AJ, Bruinenberg M, Stolk RP, Kooner JS, Zhang W, Winkelmann BR, Boehm BO, Lucae S, Penninx BW, Smit JH, Curhan G, Mudgal P, Plenge RM, Portas L, Persico I, Kirin M, Wilson JF, Mateo Leach I, van Gilst WH, Goel A, Ongen H, Hofman A, Rivadeneira F, Uitterlinden AG, Imboden M, von Eckardstein A, Cucca F, Nagaraja R, Piras MG, Nauck M, Schurmann C, Budde K, Ernst F, Farrington SM, Theodoratou E, Prokopenko I, Stumvoll M, Jula A, Perola M, Salomaa V, Shin SY, Spector TD, Sala C, Ridker PM, Kahonen M, Viikari J, Hengstenberg C, Nelson CP, Meschia JF, Nalls MA, Sharma P, Singleton AB, Kamatani N, Zeller T, Burnier M, Attia J, Laan M, Klopp N, Hillege HL, Kloiber S, Choi H, Pirastu M, Tore S, Probst-Hensch NM, Volzke H, Gudnason V, Parsa A, Schmidt R, Whitfield JB, Fornage M, Gasparini P, Siscovick DS, Polasek O, Campbell H, Rudan I, Bouatia-Naji N, Metspalu A, Loos RJ, van Duijn CM, Borecki IB, Ferrucci L, Gambaro G, Deary IJ, Wolffenbuttel BH, Chambers JC, Marz W, Pramstaller PP, Snieder H, Gyllensten U, Wright AF, Navis G, Watkins H, Witteman JC, Sanna S, Schipf S, Dunlop MG, Tonjes A, Ripatti S, Soranzo N, Toniolo D, Chasman DI, Raitakari O, Kao WH, Ciullo M, Fox CS, Caulfield M, Bochud M, Gieger C. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013;45:145–154. doi: 10.1038/ng.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Olden M, Teumer A, Bochud M, Pattaro C, Köttgen A, Turner ST, Rettig R, Chen MH, Dehghan A, Bastardot F, et al. Overlap between common genetic polymorphisms underpinning kidney traits and cardiovascular disease phenotypes: the CKDGen consortium. Am J Kidney Dis. 2013;61:889–898. doi: 10.1053/j.ajkd.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fox ER, Young JH, Li Y, Dreisbach AW, Keating BJ, Musani SK, Liu K, Morrison AC, Ganesh S, Kutlar A, et al. Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Hum Mol Genet. 2011;20:2273–2284. doi: 10.1093/hmg/ddr092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.International Consortium for Blood Pressure Genome-Wide Association Studies; Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O’Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sober S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjogren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimaki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND, Aspelund T, Garcia M, Chang YP, O’Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Hopewell JC, Ongen H, Dreisbach AW, Li Y, Young JH, Bis JC, Kahonen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Bolton JA, Kottgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grassler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Sun YV, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Soler Artigas M, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, McKenzie CA, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stancakova A, Raffel LJ, Yao J, Kathiresan S, O’Donnell CJ, Schwartz SM, Ikram MA, Longstreth WT, Jr. Mosley TH, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli CS, Mani KR, Yajnik CS, Hofman A, Mattace-Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, Ala-Korpela M, Kangas AJ, Lyytikainen LP, Soininen P, Tukiainen T, Wurtz P, Ong RT, Dorr M, Kroemer HK, Volker U, Volzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G, Meschia JF, Nalls MA, Sharma P, Terzic J, Kumar MV, Denniff M, Zukowska-Szczechowska E, Wagenknecht LE, Fowkes FG, Charchar FJ, Schwarz PE, Hayward C, Guo X, Rotimi C, Bots ML, Brand E, Samani NJ, Polasek O, Talmud PJ, Nyberg F, Kuh D, Laan M, Hveem K, Palmer LJ, van der Schouw YT, Casas JP, Mohlke KL, Vineis P, Raitakari O, Ganesh SK, Wong TY, Tai ES, Cooper RS, Laakso M, Rao DC, Harris TB, Morris RW, Dominiczak AF, Kivimaki M, Marmot MG, Miki T, Saleheen D, Chandak GR, Coresh J, Navis G, Salomaa V, Han BG, Zhu X, Kooner JS, Melander O, Ridker PM, Bandinelli S, Gyllensten UB, Wright AF, Wilson JF, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller PP, Elosua R, Soranzo N, Sijbrands EJ, Altshuler D, Loos RJ, Shuldiner AR, Gieger C, Meneton P, Uitterlinden AG, Wareham NJ, Gudnason V, Rotter JI, Rettig R, Uda M, Strachan DP, Witteman JC, Hartikainen AL, Beckmann JS, Boerwinkle E, Vasan RS, Boehnke M, Larson MG, Jarvelin MR, Psaty BM, Abecasis GR, Chakravarti A, Elliott P, van Duijn CM, Newton-Cheh C, Levy D, Caulfield MJ, Johnson T. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lian J, Huang Y, Huang RS, Xu L, Le Y, Yang X, Xu W, Huang X, Ye M, Zhou J, et al. Meta-analyses of four eosinophil related gene variants in coronary heart disease. J Thromb Thrombolysis. 2013;36:394–401. doi: 10.1007/s11239-012-0862-z. [DOI] [PubMed] [Google Scholar]
- 106.Ganesh SK, Zakai NA, van Rooij FJ, Soranzo N, Smith AV, Nalls MA, Chen MH, Kottgen A, Glazer NL, Dehghan A, et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet. 2009;41:1191–1198. doi: 10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]


