Abstract
The ribosomal stalk complex binds and recruits translation factors to the ribosome during protein biosynthesis. In Escherichia coli the stalk is composed of protein L10 and four copies of L7/L12. Despite the crucial role of the stalk, mechanistic details of L7/L12 subunit exchange are not established. By incubating isotopically labeled intact ribosomes with their unlabeled counterparts we monitored the exchange of the labile stalk proteins by recording mass spectra as a function of time. Based on kinetic analysis we proposed a mechanism whereby exchange proceeds via L7/L12 monomers and dimers. We also compared exchange of L7/L12 from free ribosomes with exchange from ribosomes in complex with elongation factor G (EF–G), trapped in the posttranslocational state by fusidic acid. Results showed that binding of EF–G reduces the L7/L12 exchange reaction of monomers by ~27% and of dimers by ~47% compared with exchange from free ribosomes. This is consistent with a model in which binding of EF–G does not modify interactions between the L7/L12 monomers but rather one of the four monomers, and as a result one of the two dimers, become anchored to the ribosome-EF–G complex preventing their free exchange. Overall therefore our results not only provide mechanistic insight into the exchange of L7/L12 monomers and dimers and the effects of EF–G binding but also have implications for modulating stability in response to environmental and functional stimuli within the cell.
Protein biosynthesis is carried out by the ribosomal machinery and requires interaction of several translation factors with the ribosomal stalk complex. In bacteria the base of the stalk region, interacting with the rRNA of the 50S large ribosomal subunit, is composed of adjacent proteins L11 and L10 (1). The C-terminal domain (CTD) of L10 is a long and mobile helix which interacts with two dimers of protein L12 (2). Three pairs of L12 are observed in thermophilic bacteria and archaea (3-6) and L12 is the only protein present as multiple copies on the ribosome. The N-terminal domain (NTD) of L12 is responsible for dimerization and for anchoring of the protein to the ribosome (7). In Escherichia coli (E. coli), L12 is partially N-acetylated to form protein L7 (8). The ratio of L7 to L12 is known to change throughout the growth phase (8-10) and has been linked to variation in the stability of the stalk complex via hydrogen exchange mass spectrometry (MS) experiments (11). L12 CTD and NTD are connected via a flexible hinge region providing mobility to the highly structured CTD (12, 13). These highly mobile proteins have eluded high resolution X-ray analysis for many years. Only recently the atomic structure of a truncated stalk complex of a thermophilic bacterium revealed the binding mode of the NTD of L12 proteins to L10 (4). The tight antiparallel binding of the L12 NTDs is similar to that observed by NMR for the E. coli L7/L12 dimer in solution (14). The mobility and dynamics of the stalk proteins have been linked to their ability to bind to several translation factors, trapping them in distinct conformational states for catalysis of essential steps of the translation process (15).
Intrinsic flexibility is coupled with rapid subunit exchange of E. coli L7/L12 in the free dimeric L7/L12 complex, occurring in vitro in a matter of seconds (16). By contrast the L10-L7/L12 interactions in the stalk complex have been shown to be remarkably stable once formed (17). There is evidence from a fluorescence assay that in vitro ribosome-bound E. coli L7/L12 proteins are exchanged with a pool of free L7/L12 proteins, albeit on a relatively slow timescale (4). This is of interest since the process of assembly and disassembly of the stalk proteins during translation has been proposed to modulate ribosomal activity in yeast (18, 19). The mechanistic details of the process of exchange however, are not yet established in any species. Since L7/L12 dimer formation is suggested to be the first step in the stalk assembly process (20, 21) it is possible that these proteins could exchange solely via dimers. Moreover as L7/L12 proteins are involved in translation factor recruitment, their exchange is likely affected by binding of translation factors to the stalk (22).
Considerable insight into the interaction of stalk proteins with translation factors has arisen from recent atomic resolution images of the ribosome where labile binding is trapped using antibiotics (23, 24). Among others, fusidic acid can be used to trap the ribosome in the posttranslocational state. Fusidic acid binds to elongation factor G (EF–G) on the ribosome, allows translocation and GTP cleavage but prevents the release of EF–G from the ribosome (25, 26). A recent X-ray structure of EF–G bound to the ribosome in the posttranslocational state demonstrates large scale movements in response to EF–G binding and reveals that a copy of an L12 CTD makes contact with EF–G (24), as suggested by previous cryo-electron microscopy data (27). The effects of this L12 CTD-EF–G interaction on the process of subunit exchange are currently unknown.
We have shown previously that intact ribosomes and their subunits can be observed by MS (3, 6, 28, 29), and that the intact stalk complex dissociates from the large subunit while retaining its non-covalent interactions (3, 6, 11). This allows us to study the composition of the stalk, without prior separation in solution, within the context of the intact ribosome. Using ribosomes with natural abundance isotopes and ribosomes uniformly labeled with 13C and 15N isotopes we can follow in real-time formation of heterogeneous assemblies, containing both unlabeled and isotopically labeled proteins, due to the exchange of labile proteins between ribosomes. This strategy enables us to determine the composition of the complexes quantitatively and to deduce the kinetics of the exchange process. Here we first establish exchange kinetics in free ribosomes and use modeling to distinguish between the various exchange scenarios. We extend this to study ribosomes in complex with EF–G, inhibited in the posttranslocational state with fusidic acid. The results show that the E. coli L7/L12 exchange approaches equilibrium after a period of ~6h. We also find that the stalk complex can exchange both monomeric and dimeric L7/L12 proteins. Translation factor binding slows the exchange, through deactivating the dissociation of EF–G bound L7/L12 monomer as well as trapping the dimer that includes the EF–G bound monomer. We discuss the implications of this study on the structure of the stalk complex and its role in modulating ribosomal stability and interactions.
RESULTS AND DISCUSSION
Isotopically labeled ribosomes maintain interactions
The dynamics of the L7/L12 proteins were investigated using an MS strategy that has proven successful to elucidate the subunit exchange dynamics of other protein complexes (30-33). Specifically, ribosomes are uniformly labeled with stable isotopes (13C and 15N) to provide sufficient mass differences to resolve heterocomplexes formed with wild-type ribosomes. After mixing [12C,14N] wild-type ribosomes (l-) with [13C,15N]-labeled ribosomes (h-) hetero stalk complexes will form of the composition [l/hL10(lL7/L12)x(hL7/L12)y] where x+y = 4. To confirm that isotopic labeling method does not affect the subunit interactions within the ribosome we first recorded mass spectra of h-ribosomes. We found that the spectra are closely similar to those of unlabeled E. coli ribosomes with all proteins shifted to higher m/z values, consistent with >99 % incorporation of isotopic label (Supplementary Figure S1 and Table S1). We conclude that protein interactions within the stalk pentamer [hL10(L7/L12)4] and hexamer [hL11·L10(L7/L12)4] are unaffected by the labeling process.
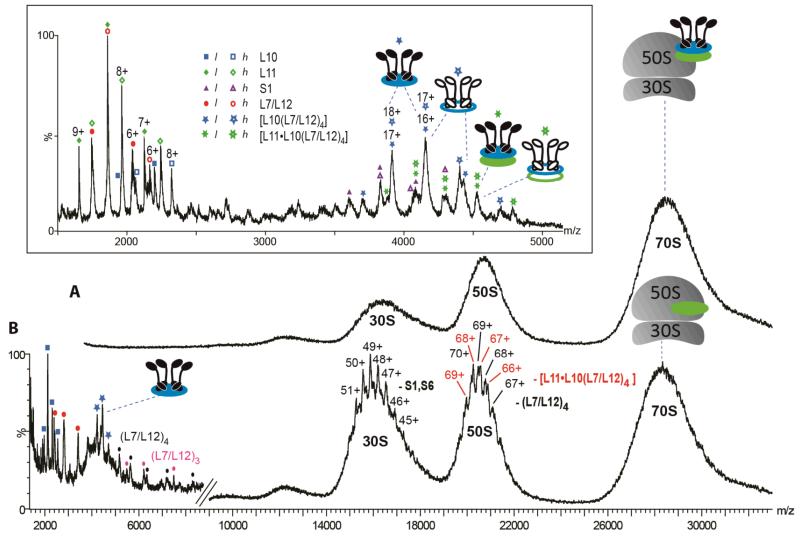
To maintain the integrity of the 70S particles, a high [Mg2+] is required together with a low ionic strength (28, 34). From solutions containing 5 mM Mg2+, mass spectra confirm the presence of the intact 70S assembly (Figure 1, panel A) (28). At higher collision energy, from the same Mg2+ solution, charge states from the 30S and 50S subunits are partially resolved, enabling mass measurement (Figure 1, panel B, Supplementary Table S1). The molecular mass of the 30S subunit corresponds to loss of S1 and S6, as observed previously (35), while the two 50S subunit complexes are consistent with collision-induced dissociation (CID) of the hexameric stalk [L11·L10(L7/L12)4], and of four copies of L7/L12. The low m/z region is dominated by the stalk proteins (L10, L7/L12) and complexes thereof. This spectrum therefore confirms the presence of the stalk on the intact 70S and highlights a means of studying the intact stalk complex within the context of the intact ribosome.
Figure 1.
Mass spectra of ribosomes in the presence of 5 mM Mg2+. A) Upper spectrum was acquired under ‘soft’ MS conditions (collision energy 5 V without collision gas) and reveals a large unresolved signal at 28000 m/z assigned to the intact 70S assembly. B) Lower spectrum was acquired under dissociating MS conditions (collision energy 100 V and 10 psi argon collision gas). Charge states of both the 30S and 50S particles are partially resolved. The mass of the 30S subunit is consistent with loss of S1 and S6 as observed previously (35). For the 50S subunit, two charge state series are observed which are consistent with loss of hexameric stalk complex and four copies of L7/L12. Inset, expansion of the spectrum obtained with an equimolar solution of l- and h-ribosomes in 10 mM ammonium acetate and 6 mM Mg2+. Peaks are assigned to dissociated proteins as well as two populations of intact l- and h-stalk complexes, [L10(L7/L12)4] and [L11·L10(L7/L12)4]. [12C,14N] and [14C,15N] species are indicated by filled and empty symbols respectively.
To determine the dynamics of the L7/L12 proteins, an equimolar solution of l- and h- ribosomes was monitored at 37°C and pH 7.0. Aliquots of the solution were withdrawn and reduction of the [Mg2+] was carried out immediately prior to analysis. This procedure allows the dissociation of proteins via CID, and yet preserves solution-phase interactions during the exchange reaction. The spectrum recorded at t=0 for an equimolar mixture of l and h-ribosomes reveals well-resolved charge state assigned to individual l- and h-stalk proteins (Figure 1 inset, Supplementary Table S1). At ~4000 m/z the most intense series is assigned to the l- and h [L10(L7/L12)4] pentameric stalk complexes. We applied tandem MS to investigate the composition of these intact pentameric stalk complexes (Supplementary Figure S2). Observation of both labeled and unlabeled proteins during CID led us to conclude that the peaks from the pentamers overlap. Although their masses differ, their m/z values are coincident. In the light of this result we reasoned that if the homopentamers overlapped, incorporation of labeled and unlabeled proteins into the ribosome would yield populations of heteropentamers between the homopentamers. We examined the 17+ charge state assigned to the pentamer [L10(L7/L12)4] as a function of incubation time at 37°C (Supplementary Figure S3A). After 1h new peaks are observed corresponding to l- and h-heterostalk complexes. Their intensity increases until equilibrium is approached after ~6h. This exchange reaction results in twelve possible compositions of the pentameric stalk with unique masses. The dominant peaks arise from the overlap of several products (Supplementary Table S2). Because of this overlap it is not possible to quantify the various populations.
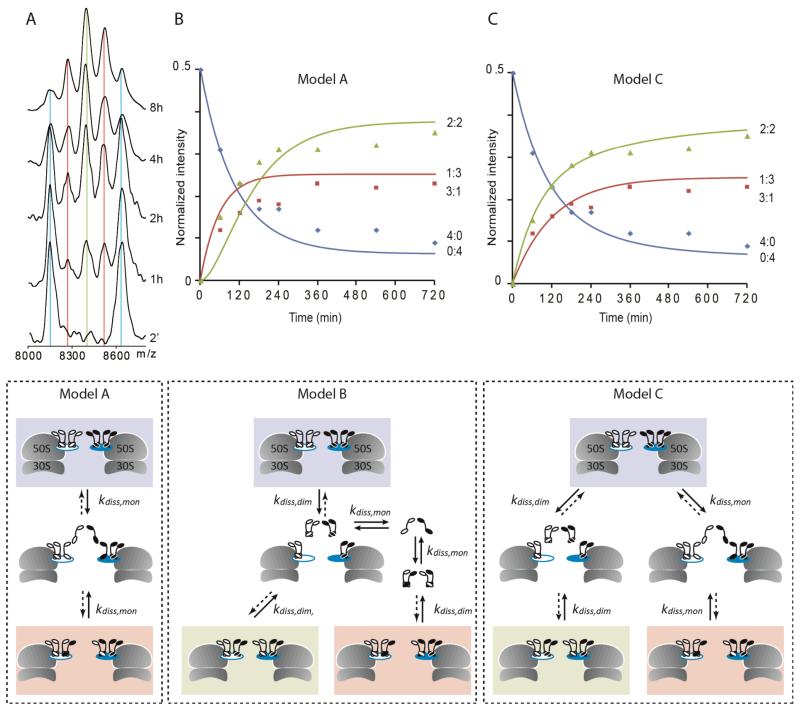
Gas-phase dissociation of the [L10(L7/L12)4] stalk complex is effected under activating MS conditions (36) to reduce spectral complexity (Supplementary Figure S4). This procedure allows resolution of different populations of stripped complexes at higher m/z values than the stalk complex. The most prominent stripped complex [L10(L7/L12)3], formed in the gas phase, arises from loss of an individual L7/L12 protein (Supplementary Figure S3B). Peaks corresponding to these stripped complexes are more readily assigned than those recorded without activation (Supplementary Figure S3A). All the peaks between the two homocomplexes, however, can be assigned to at least two different species containing l- or hL10 with different numbers of l- and hL7/L12 subunits (Supplementary Table S2). We therefore considered a further series of charge states assigned to the loss of protein L10 in the gas phase. This CID process results in the formation of a gas phase complex consisting of the four L7/L12 subunits (Figure 2, panel A). While this assembly has no significance from a structural point of view, since it does not exist in solution, our ability to remove L10 from the stalk complex enables us to assign all the (L7/L12)4 peaks to unique compositions (Supplementary Table S2). As such we can follow the exchange process by assigning the new peaks that are observed between the homo-(L7/L12)4 peaks (Figure 2, panel A). The spectrum acquired after 8h shows the statistical distribution of the 5 tetramers with the central peak corresponding to ribosomes with 2:2 (l:h) L7/L12 stoichiometry predominating. The observation of 3:1 and 1:3 (l:h) compositions however implies that exchange cannot proceed solely via dimers.
Figure 2.
L7/L12 exchange from free ribosomes. A) CID spectra of an equimolar solution of l- and h-ribosomes at various time intervals after mixing showing the 6+ charge state of the (L7/L12)4 stripped complex. Homotetramers are shown in blue, heterotetramers with a 3:1 or 1:3 (l:h) L7/L12 composition in red and a 2:2 (l:h) L7/L12 stoichiometry in green. B–C) Relative intensities of the tetramers with 4:0/0:4, 3:1/1:3 and 2:2 (l:h) L7/L12 ratio are indicated by blue diamond, red square and green triangle, respectively. Data are compared with different models considered (blue, red and green lines represent the modeled relative intensities): B) model A, in which only monomer dissociation is allowed and C) model C in which both monomers and dimers dissociate independently. Model B in which only dimers dissociate but are subsequently allowed to exchange before re-association has a relatively large RMSD between the data and the model (Supplementary Figure S5).
Kinetic analysis reveals mechanisms of L7/L12 subunit exchange
To investigate possible mechanisms of subunit exchange occurring on the intact 70S ribosome we plotted the intensity of the various homo- and hetero-(L7/L12)4 complexes as a function of time. We then computed average values for the 4:0 (l:h) and 0:4 (l:h) homocomplexes and 3:1 (l:h) and 1:3 (l:h) heterocomplexes. We considered three different models for our simulation (Figure 2, full details of the modeling are given in the Supplementary Information). First, the stalk complex was allowed to exchange via monomers only (Model A). The second model considers generation of a stable dimeric intermediate which is competent to either reassociate into the stalk complex, or to exchange monomeric L7/L12 subunits followed by subsequent reassembly into the stalk (Model B). The final model considers subunit exchange via dimer and monomer intermediates without interconversion between dimers (Model C). For each model, 10,000 sets of values of kdiss,mon and kdiss,dim were considered. The values that minimize the root mean square deviation (RMSD) between the simulation and the experimental data were used to determine the model which had the best fit (Supplementary Figure S5 and Table S3). From this best fit we then extracted rate constants for dissociative events experienced by the stalk complex.
For model A, which considered only the dissociation of monomeric L7/L12, we varied the rate constant kdiss,mon, setting the dimer dissociation rate constant kdiss,dim to zero. This model agrees well with the ratio of L7/L12 tetramers at equilibrium but is unable to account for the experimental finding that the rate of formation of the 2:2 complex is faster than that of the 3:1 and 1:3 at the early stage of the reaction (Figure 2, panel B). For model B, exchange of the two independent dimers, followed by monomeric exchange of the dissociated dimer, can be modeled to fit the data. However the RMSD is relatively large in this case (Supplementary Figure S5 and Table S3). For model C, which allows exchange of both monomers and dimers between tetramers, much closer agreement with the experimental data is achieved (Figure 2, panel C, Supplementary Figure S5 and Table S3). From this fit, we extract two rate constants for kdiss,mon and kdiss,dim which are 0.0081 min−1 and 0.0070 min−1 respectively. They indicate that the exchange via monomer and dimer occurs at similar rates. The two dissociation events can be summed to provide the overall rate of disappearance of the homostalk complexes of 0.015 min−1.
Binding of EF–G reduces L7/L12 subunit exchange
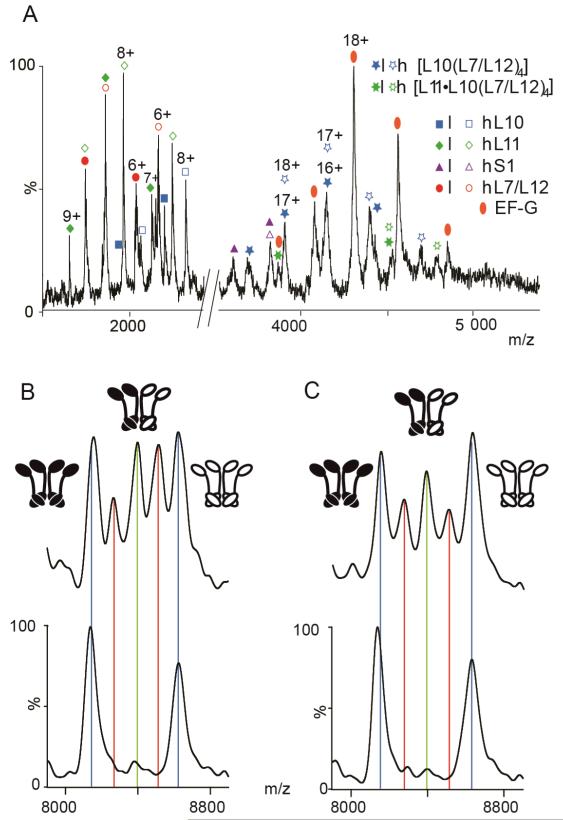
The influence of EF–G binding on L7/L12 subunit exchange was determined by incubating ribosomes with 6 equivalents of the translation factor and an excess of fusidic acid and GTP. These conditions trap the ribosome-EF–G complex in the posttranslocational state (25, 26). We added EF–G, GTP and fusidic acid to a solution containing equimolar quantities of l- and h-ribosomes. As demonstrated previously (37), the MS spectrum of ribosome-EF–G complex is largely indistinguishable from spectra recorded for native ribosome save for the additional peaks assigned to EF–G (Figure 3, panel A). We also prepared a control experiment, containing ribosomes, GTP and fusidic acid, but in the absence of EF–G. This was examined in parallel using identical MS and solution conditions. Comparing the MS of the control and EF–G complex peaks assigned to heterocomplexes confirm exchange of L7/L12 subunits has taken place (Figure 3, panels B and C, Supplementary Figure S6). The control experiment established that the presence of excess fusidic acid and GTP did not affect subunit exchange, which was found to be closely similar to that observed for free ribosomes. By contrast, in the presence of EF–G after 3 h of exchange, heterospecies are less intense than in the control. This result clearly indicates that for the ribosome-EF–G complex, exchange of L7/L12 is significantly slower.
Figure 3.
L7/L12 exchange from ribosomes trapped in the posttranslocational state. A) Low m/z region of a mass spectrum recorded for an equimolar solution of l- and h-ribosome-EF–G complex trapped in the posttranslocational state by fusidic acid, prior to subunit exchange. B–C) The 6+ charge state of (L7/L12)4 stripped complex at time t=0 (lower spectra) and after 3h incubation at 37°C (upper spectra) for ribosomes in: B) the absence of EF–G and C) for the EF–G complex. Peaks corresponding to tetramers with different ratios of (l:h) L7/L12 are highlighted in blue (4:0/0:4), red (3:1/1:3) and green (2:2).
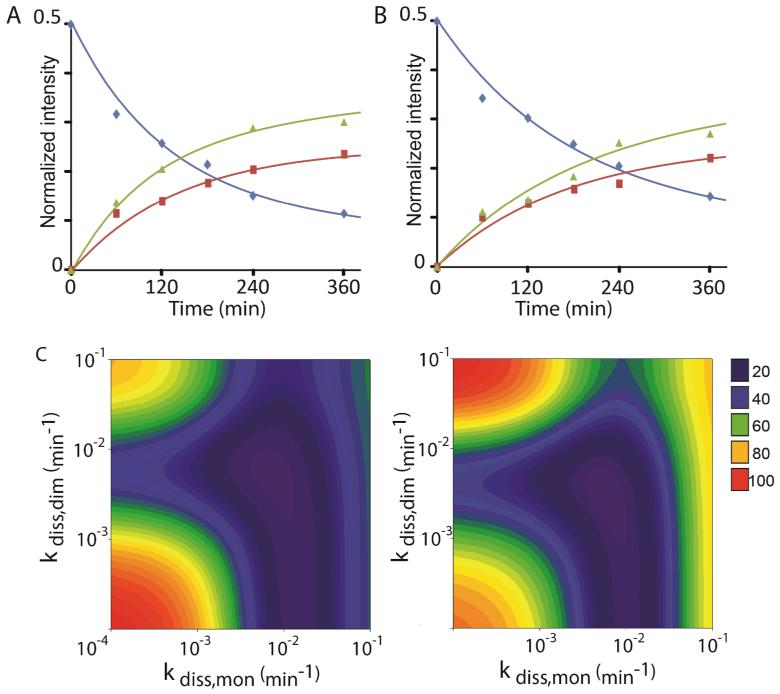
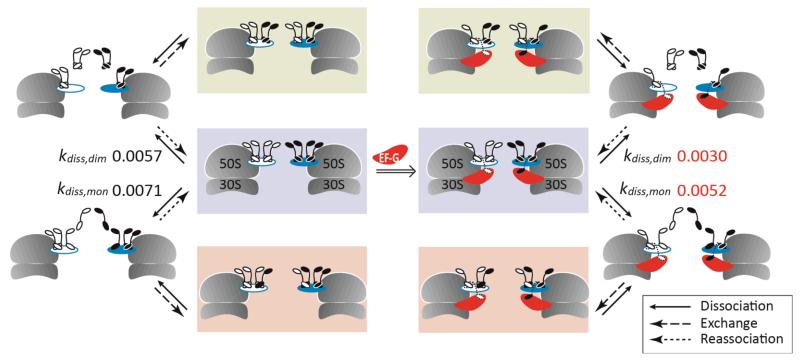
We measured the normalized intensity of each (L7/L12)4 species for both reactions and applied the hybrid model C (Figure 2). For the control reaction (Figure 4, panel A), we extract rate constants via monomers (kdiss,mon = 0.0071 min−1) and via two independent dimers (kdiss,dim = 0.0057 min−1) with an overall rate of disappearance of the homostalk of 0.013 min−1. These rate constants display a slight decrease compared to the solution examined above, without GTP and fusidic acid, assigned to differences in the solution conditions employed. For the ribosome-EF–G complex (Figure 4, panel B), results show that both rates of exchange are significantly lower than in the control, kdiss,mon = 0.0052 min−1 and kdiss,dim = 0.0030 min−1 giving an overall rate of disappearance of the homostalk complex of 0.0082 min−1. Interestingly the rate constants for dissociation of a monomer and a dimer are lowered by ~27% and ~47% respectively, in the EF–G complex compared to the control experiment, demonstrating that both monomeric and dimeric exchange pathways are affected (Figure 5).
Figure 4.
Binding of EF–G slows down L7/L12 exchange. Kinetic modeling of L7/L12 subunit exchange in: A) the control solution in the absence of factor and B) the ribosome-EF–G complex. Relative intensities of the (l:h) L7/L12 tetramers with 4:0/0:4, 3:1/1:3 and 2:2 are indicated by blue diamond, red square and green triangle, respectively. Data are compared with the intensities predicted from the hybrid monomeric and dimeric exchange Model C (corresponding lines). C) The optimization procedure is illustrated by showing the RMSDs between both sets of experimental data and Model C for 10,000 kdiss,mon and kdiss,dim values.
Figure 5.
Schematic representation of the L7/L12 exchange mechanism which takes place between ribosomes in the absence of factors (left) and in the ribosome-EF–G complex (right). The schematic shows all events that were considered in our model. For the ribosome-EF–G complex slower rate constants (min−1) are observed for dissociation of monomers and dimers.
Discussion
We have shown that ribosomes remain intact in the gas phase, enabling us to monitor exchange of stalk proteins in solution, in situ on the ribosome. Well-resolved mass spectra allow us to quantify the incorporation of l- and h-proteins as a function of time. Detailed kinetic modeling enabled us to delineate different pathways of disassembly within the context of the intact ribosome and to determine that exchange occurs via both monomers and dimers (Figure 5). Our data are consistent with two independent L7/L12 dimers, in line with available X-ray data (4). It is interesting to note that in free ribosomes, the rate constants for monomer and dimer exchange are similar. This implies that the dimers exist as independent units, competent to dissociate and re-associate almost as rapidly as the monomers. In E. coli, multiple L7/L12 dimers are essential for efficient protein synthesis (38-40). Despite evidence of the structural and functional significance of the dimeric unit, and given that all the bacterial stalks studied so far have either a 4:1 and/or 6:1 (L7/L12:L10) ratio, direct evidence of the role of dimers in subunit exchange, from intact ribosomes, has not been demonstrated previously.
In the ribosome-EF–G complex, structural studies show that binding of EF–G affects the long α-helix at the C-terminus of L10 (24). This leads to bending of L10 towards L11 relative to its structure in the isolated [L10(L12NTD)6] structure (4). As a result of this bending, one copy of L7/L12 CTD interacts directly with both EF–G and the NTD of L11 (24). Given these interactions, it is anticipated that exchange of monomeric L7/L12 would be affected. Kinetic modeling of our experimental data indicates that disassembly via monomers is reduced by 27% when EF–G is trapped on the ribosome in the posttranslocational state. This is consistent with one of the four copies of L7/L12 becoming trapped on the ribosome, unable to participate in the exchange reaction due to its interaction with EF–G. The binding sites of the two L7/L12 dimers are separated by a kink in the helix of L10 (4). Additional bending of the L10 helix, as a result of EF–G binding as seen in the X-ray structure (24) would likely increase the independence of the two binding sites. In our model we considered the two dimers to act independently. If interactions in the L7/L12 dimers were unchanged as a result of EF–G binding, we would anticipate a reduction in the amplitude of exchange proceeding via dimers by ~50%. That exchange via dimers is reduced by ~47% implies that interactions with EF–G prevent one of the dimers from undergoing exchange and that the interactions between independent dimers are unchanged. Since several factors are known to be recruited by L7/L12 during translation, and given the high concentrations of these factors in cells (41), L7/L12 will likely be involved in repeated interactions, preventing their free exchange. From X-ray data (24), however we know that only two L7/L12 subunits are affected by translation factor binding, the remaining stalk proteins are thus available for exchange as observed here.
Bacterial ribosomes are known to be stable for at least 24h, throughout cellular growth, and while engaged in translation (42, 43). In our experiments after ~2h of exchange >50% of ribosomes have undergone exchange of the L7/L12 proteins. The L12:L7 ratio is modified as a function of the growth phase (9, 10) but it has been established that once incorporated L12 and L7 do not undergo interconversion, via acetylation or deacetylation (44). Since L7 has increased interactions with L10, compared with those of L12 (11), it is entirely feasible for exchange of L12 for L7 to take place in cells. While such fine-tuning is likely to have minor effects on the overall composition of the stalk, subtle changes could form part of a strategy for survival, conferring enhanced stability to the stalk under conditions of stress (11) in-line with proposals from other studies (45). Comparing the exchange of bacterial ribosomes with their eukaryotic counterpart, reveals that yeast P1 and P2 proteins are in more rapid exchange leading to greater heterogeneity in stalk complexes (46). This would allow for better regulation of protein synthesis in the more complex eukaryotic translation system than its bacterial equivalent. However it has been demonstrated that the number of L7/L12 dimers does affect ribosomal function (40). This implies therefore that fine-tuning the protein composition in the bacterial stalk could also provide a mechanism for adapting to a particular cellular environment.
METHODS
Materials
Labeled ribosomes were purified from MRE600 E. coli strain grown in M9 minimal media employing 13C-D-glucose (Sigma) for carbon labeling and 15N-ammonium chloride (Cambridge Isotope Laboratories, Inc) for nitrogen labeling according to the same protocol (47). Labeling of the ribosomal proteins was higher than 99% as identified by MS (Supplementary Table S1). Aliquots were stored at −80°C in 10 mM Tris-HCl (pH 7.5), 10 mM magnesium chloride, 50 mM ammonium chloride, 6 mM β-mercaptoethanol. EF–G was expressed and purified following a previously described procedure (15). Chemicals were purchased from Sigma unless otherwise stated.
Subunit exchange reaction
For subunit exchange without EF–G, an equimolar solution (1 μM total ribosome concentration) of [12C,14N]-70S and [13C,15N]-70S ribosomes was incubated at 37°C in 6 mM magnesium acetate, 10 mM ammonium acetate (pH 7.0). For comparison of L7/L12 subunit exchange with and without translation factor, 25 μL of a mixture of EF–G (25 μM), fusidic acid (FA) (130 μM) and GTP (130 μM) in 10 mM ammonium acetate were incubated at room temperature for 20 mins. To this solution was added 95 μL of an equimolar solution (1.1 μM total ribosome concentration) of [12C,14N]-70S and [13C,15N]-70S ribosomes in 10 mM ammonium acetate, 7.7 mM magnesium acetate, pH 7.0, to give a final volume of 120 μl. The molar ratio of 70S/EF–G/GTP/FA was 1:6:31:31, closely similar to that employed in the determination of the X-ray structure of the ribosome-EF–G complex (24). The control experiment was prepared identically except that EF–G was omitted from the FA/GTP solution. Both reactions were incubated at 37°C. Aliquots (10 μL) were buffer-exchanged immediately prior to analysis using micro BioSpin 6 columns into 10 mM ammonium acetate solution pH 7.0 to remove excess Mg2+, GTP and FA. Aliquots were withdrawn from t=2 minutes to 12h and stored on ice prior to analysis.
Mass spectrometry
MS and tandem MS analysis was carried out on a Synapt HDMS quadrupole IM–MS instrument (48) and spectra in Figure 1, panels A and B, were acquired on the QTOF2 (Waters). MS parameters were optimised for the transmission of large non-covalent complexes. Typical values are: capillary voltage 1.7 kV, cone voltage 80 V, cone gas 40 litres h−1, extractor 1 V, ion transfer stage pressure 5.70 mbar, transfer voltage 12 V, bias voltage 35 V, trap voltage 15 V, trap and transfer pressure 5.3×10−2 mbar, IMS pressure 5.0×10−1 mbar, ToF analyser pressure 1.2×10−6 mbar. For spectra recorded under dissociating conditions the trap collision voltage was increased to 100 V. In tandem MS mode ions of a narrow range of m/z were selected in the first quadrupole mass analyser, prior to accumulation in the ion trap. These ions were then subjected to increasing collision voltage in the ion trap just prior to the mobility cell (15–70 V) while keeping other settings constant. Nanoflow electrospray capillaries were prepared in-house as previously described (49). Mass spectra were calibrated externally using a solution of caesium iodide (100 mg ml−1) and analysed using Masslynx 4.1 software (Waters). Spectra are shown with minimal smoothing. The protein molecular masses were obtained from the UniProt database (www.uniprot.org). The relative abundance of the different types of (L7/L12)4 stripped complexes was calculated by fitting Gaussians to each peak of the 6+ charge state. The intensity is expressed as a ratio of the total intensity of the peaks assigned to these tetramers.
Kinetic modeling
The kinetic model describing the L7/L12 exchange process was modified from an approach developed previously (31, 33). The stripped complex (L7/L12)4 represented the distribution of l- and hL7/L12 subunits on the intact ribosome. Models were produced in-house using Mathematica 6.0. Further details are provided in the Supplementary Information.
Supplementary Material
ACKNOWLEDGEMENTS
S.D. acknowledges the Biotechnology and Biological Sciences Research Council. C.V.R. is a Professor of the Royal Society. Y.G. and J.M. acknowledge support from the Wellcome Trust and European Union 7th Framework Program PROSPECTS (Proteomics Specification in Space and Time Grant HEALTH-F4-2008-201648). S.S. acknowledges research supports from Swedish Research Council, Carl-Tryggers Foundation, Wenner-Gren Foundation and Knut and Alice Wallenberg Foundation.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet.
References
- 1.Beauclerk AAD, Cundliffe E, Dijk J. The binding site for ribosomal protein complex L8 within 23 S ribosomal RNA of Escherichia coli. J. Biol. Chem. 1984;259:6559–6563. [PubMed] [Google Scholar]
- 2.Österberg R, Sjöberg B. Small-angle x-ray scattering study of the protein complex of L7/L12 and L10 from Escherichia coli ribosomes. FEBS Lett. 1977;73:22–24. doi: 10.1016/0014-5793(77)80006-9. [DOI] [PubMed] [Google Scholar]
- 3.Ilag LL, Videler H, McKay AR, Sobott F, Fucini P, Nierhaus KH, Robinson CV. Heptameric (L12)6/L10 rather than canonical pentameric complexes are found by tandem MS of intact ribosomes from thermophilic bacteria. Proc. Natl. Acad. Sci. USA. 2005;102:8192–8197. doi: 10.1073/pnas.0502193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaconu M, Kothe U, Schlünzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/L12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 5.Maki Y, Hashimoto T, Zhou M, Naganuma T, Ohta J, Nomura T, Robinson CV, Uchiumi T. Three binding sites for stalk protein dimers are generally present in ribosomes from archaeal organism. J. Biol. Chem. 2007;282:32827–32833. doi: 10.1074/jbc.M705412200. [DOI] [PubMed] [Google Scholar]
- 6.Gordiyenko Y, Videler H, Zhou M, McKay AR, Fucini P, Biegel E, Muller V, Robinson CV. Mass spectrometry defines the stoichiometry of ribosomal stalk complexes across the phylogenetic tree. Mol. Cell Proteomics. 2010;9:1774–1783. doi: 10.1074/mcp.M000072-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudkov AT, Budovskaya EV, Sherstobaeva NM. The first 37 residues are sufficient for dimerization of ribosomal L7/L12 protein. FEBS Lett. 1995;367:280–282. doi: 10.1016/0014-5793(95)00564-p. [DOI] [PubMed] [Google Scholar]
- 8.Terhorst C, Wittmann-Liebold B, Möller W. 50-S ribosomal proteins - Peptide studies on two acidic proteins, A1 and A2, isolated from 50-S ribosomes of Escherichia coli. Eur. J. Biochem. 1972;25:13–19. doi: 10.1111/j.1432-1033.1972.tb01661.x. [DOI] [PubMed] [Google Scholar]
- 9.Deusser E, Wittmann HG. Ribosomal proteins: variation of the protein composition in Escherichia coli ribosomes as function of growth rate. Nature. 1972;238:269–270. doi: 10.1038/238269a0. [DOI] [PubMed] [Google Scholar]
- 10.Ramagopal S, Subramanian AR. Alteration in the acetylation level of ribosomal protein L12 during growth cycle of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1974;71:2136–2140. doi: 10.1073/pnas.71.5.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordiyenko Y, Deroo S, Zhou M, Videler H, Robinson CV. Acetylation of L12 increases interactions in the Escherichia coli ribosomal stalk complex. J. Mol. Biol. 2008;380:404–414. doi: 10.1016/j.jmb.2008.04.067. [DOI] [PubMed] [Google Scholar]
- 12.Gudkov AT, Bubunenko MG, Gryaznova OI. Overexpression of L7/L12 protein with mutations in its flexible region. Biochimie. 1991;73:1387–1389. doi: 10.1016/0300-9084(91)90168-z. [DOI] [PubMed] [Google Scholar]
- 13.Bubunenko MG, Chuikov SV, Gudkov AT. The length of the interdomain region of the L7/L12 protein is important for its function. FEBS Lett. 1992;313:232–234. doi: 10.1016/0014-5793(92)81198-u. [DOI] [PubMed] [Google Scholar]
- 14.Bocharov EV, Sobol AG, Pavlov KV, Korzhnev DM, Jaravine VA, Gudkov AT, Arseniev AS. From Structure and dynamics of protein L7/L12 to molecular switching in ribosome. J. Biol. Chem. 2004;279:17697–17706. doi: 10.1074/jbc.M313384200. [DOI] [PubMed] [Google Scholar]
- 15.Helgstrand M, Mandava CS, Mulder FAA, Liljas A, Sanyal S, Akke M. The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. J. Mol. Biol. 2007;365:468–479. doi: 10.1016/j.jmb.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 16.Hamman BD, Oleinikov AW, Jokhadze GG, Traut RR, Jameson DM. Dimer/monomer equilibrium and domain separations of Escherichia coli ribosomal protein L7/L12. Biochemistry. 1996;35:16680–16686. doi: 10.1021/bi9624189. [DOI] [PubMed] [Google Scholar]
- 17.Pettersson I, Hardy SJS, Liljas A. The ribosomal protein L8 is a complex of L7/L12 and L10. FEBS Lett. 1976;64:135–138. doi: 10.1016/0014-5793(76)80267-0. [DOI] [PubMed] [Google Scholar]
- 18.Briceño V, Camargo H, Remacha M, Santos C, Ballesta JPG. Structural and functional characterization of the amino terminal domain of the yeast ribosomal stalk P1 and P2 proteins. Int. J. Biochem. Cell Biol. 2009;41:1315–1322. doi: 10.1016/j.biocel.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Cardenas D, Revuelta-Cervantes J, Jimenez-Diaz A, Camargo H, Remacha M, Ballesta JP. P1 and P2 protein heterodimer binding to the P0 protein of Saccharomyces cerevisiae is relatively non-specific and a source of ribosomal heterogeneity. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks036. Epub Jan 24, 2012. DOI: 2010.1093/nar/gks2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caldwell P, Luk DC, Weissbach H, Brot N. Oxidation of the methionine residues of Escherichia coli ribosomal protein L12 decreases the protein’s biological activity. Proc. Natl. Acad. Sci. USA. 1978;75:5349–5352. doi: 10.1073/pnas.75.11.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nomura T, Mochizuki R, Dabbs ER, Shimizu Y, Ueda T, Hachimori A, Uchiumi T. A point mutation in ribosomal protein L7/L12 reduces its ability to form a compact dimer structure and to assemble into the GTPase center. Biochemistry. 2003;42:4691–4698. doi: 10.1021/bi027087g. [DOI] [PubMed] [Google Scholar]
- 22.Sanyal S, Liljas A. The end of the beginning: structural studies of ribosomal proteins. Cur. Op. Struct. Biol. 2000;10:633–636. doi: 10.1016/s0959-440x(00)00143-3. [DOI] [PubMed] [Google Scholar]
- 23.Schmeing TM, Voorhees RM, Kelley AC, Gao Y-G, Murphy FV, IV, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and Aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y-G, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bodley JW, Zieve FJ, Lin L, Zieve ST. Studies on translocation. III. Conditions necessary for the formation and detection of a stable ribosome-G factor-guanosine diphosphate complex in the presence of fusidic acid. J. Biol. Chem. 1970;245:5656–5661. [PubMed] [Google Scholar]
- 26.Bodley JW, Zieve FJ, Lin L. Studies on translocation. IV. The hydrolysis of a single round of guanosine triphosphate in the presence of fusidic acid. J. Biol. Chem. 1970;245:5662–5667. [PubMed] [Google Scholar]
- 27.Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK. Interaction of the G’ domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translation as revealed by cryo-EM. Mol. Cell. 2005;20:723–731. doi: 10.1016/j.molcel.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 28.Rostom AA, Fucini P, Benjamin DR, Juenemann R, Nierhaus KH, Hartl FU, Dobson CM, Robinson CV. Detection and selective dissociation of intact ribosomes in a mass spectrometer. Proc. Natl. Acad. Sci. USA. 2000;97:5185–5190. doi: 10.1073/pnas.97.10.5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Videler H, Ilag LL, McKay AR, Hanson CL, Robinson CV. Mass spectrometry of intact ribosomes. FEBS Lett. 2005;579:943–947. doi: 10.1016/j.febslet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Sobott F, Benesch JLP, Vierling E, Robinson CV. Subunit exchange of multimeric protein complexes. J. Biol. Chem. 2002;277:38921–38929. doi: 10.1074/jbc.M206060200. [DOI] [PubMed] [Google Scholar]
- 31.Keetch CA, Bromley EHC, McCammon MG, Wang N, Christodoulou J, Robinson CV. L55P transthyretin accelerates subunit exchange and leads to rapid formation of hybrid tetramers. J. Biol. Chem. 2005;280:41667–41674. doi: 10.1074/jbc.M508753200. [DOI] [PubMed] [Google Scholar]
- 32.Natan E, Hirschberg D, Morgner N, Robinson CV, Fersht AR. Ultraslow oligomerization equilibria of p53 and its implications. Proc. Natl. Acad. Sci. USA. 2009;106:14327–14332. doi: 10.1073/pnas.0907840106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyung S-J, Deroo S, Robinson CV. Retinol and retinol-binding protein stabilize transthyretin via formation of retinol transport complex. ACS Chem. Biol. 2010;5:1137–1146. doi: 10.1021/cb100144v. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto T, Shimizu Y, Ueda T, Shiro Y. Mg2+ dependence of 70 S ribosomal protein flexibility revealed by hydrogen/deuterium exchange and mass spectrometry. J. Biol. Chem. 2010;285:5646–5652. doi: 10.1074/jbc.M109.081836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKay AR, Ruotolo BT, Ilag LL, Robinson CV. Mass measurements of increased accuracy resolve heterogeneous populations of intact ribosomes. J. Am. Chem. Soc. 2006;128:11433–11442. doi: 10.1021/ja061468q. [DOI] [PubMed] [Google Scholar]
- 36.Benesch JLP. Collisional activation of protein complexes: Picking up the pieces. J. Am. Soc. Mass Spectrom. 2009;20:341–348. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 37.Hanson CL, Fucini P, Ilag LL, Nierhaus KH, Robinson CV. Dissociation of intact Escherichia coli ribosomes in a mass spectrometer. J. Biol. Chem. 2003;278:1259–1267. doi: 10.1074/jbc.M208966200. [DOI] [PubMed] [Google Scholar]
- 38.Hamel E, Koka M, Nakamoto T. Requirement of an Escherichia coli 50 S ribosomal protein component for effective interaction of the ribosome with T and G factors and with guanosine triphosphate. J. Biol. Chem. 1972;247:805–814. [PubMed] [Google Scholar]
- 39.Griaznova O, Traut RR. Deletion of C-terminal residues of Escherichia coli ribosomal protein L10 causes the loss of binding of one L7/L12 dimer: ribosomes with one L7/L12 dimer are active. Biochemistry. 2000;39:4075–4081. doi: 10.1021/bi992621e. [DOI] [PubMed] [Google Scholar]
- 40.Mandava CS, Peisker K, Ederth J, Kumar R, Ge X, Szaflarski W, Sanyal S. Bacterial ribosome requires multiple L12 dimers for efficient initiation and elongation of protein synthesis involving IF2 and EF-G. Nucleic Acids Res. 2012;40:2054–64. doi: 10.1093/nar/gkr1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit Rev Biochem Mol Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 42.Deutscher MP. Degradation of stable RNA in bacteria. J. Biol. Chem. 2003;278:45041–45044. doi: 10.1074/jbc.R300031200. [DOI] [PubMed] [Google Scholar]
- 43.Zundel MA, Basturea GN, Deutscher MP. Initiation of ribosome degradation during starvation in Escherichia coli. RNA. 2009;15:977–983. doi: 10.1261/rna.1381309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramagopal S, Subramanian AR. Growth-dependent regulation in production and utilization of acetylated ribosomal protein L7. J. Mol. Biol. 1975;94:633–641. doi: 10.1016/0022-2836(75)90327-7. [DOI] [PubMed] [Google Scholar]
- 45.Wada A. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells. 1998;3:203–208. doi: 10.1046/j.1365-2443.1998.00187.x. [DOI] [PubMed] [Google Scholar]
- 46.Cardenas D, Revuelta-Cervantes J, Jimenez-Diaz A, Camargo H, Remacha M, Ballesta JP. P1 and P2 protein heterodimer binding to the P0 protein of accharomyces cerevisiae is relatively non-specific and a source of ribosomal heterogeneity. Nucleic Acids Res. 2012 Jan 24; doi: 10.1093/nar/gks036. 2012. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christodoulou J, Larsson G, Fucini P, Connell S, Pertinhez TA, Hanson CL, Redfield C, Nierhaus KH, Robinson CV, Schleucher J, Dobson CM. Heteronuclear NMR investigations of dynamic regions of intact Escherichia coli ribosomes. Proc. Natl. Acad. Sci. USA. 2004;101:10949–10954. doi: 10.1073/pnas.0400928101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-TOF instrument. Int. J. Mass Spectrom. 2007;261:1–12. [Google Scholar]
- 49.Hernández H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat. Protocol. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.