Abstract
Organisms are defined by the information encoded in their genomes, and since the evolution of life, this information has been encoded using a two base pair genetic alphabet (A-T and G-C). In vitro, the alphabet has been expanded to include several unnatural base pairs (UBPs)1–3. We have developed a class of UBPs formed between nucleotides bearing hydrophobic nucleobases, exemplified by the pair formed between d5SICS and dNaM (d5SICS-dNaM, Fig. 1a), which is efficiently PCR amplified1 and transcribed4,5 in vitro, and whose unique mechanism of replication has been characterized6,7. However, expansion of a organism’s genetic alphabet presents new and unprecedented challenges: the unnatural nucleoside triphosphates must enter the cell; endogenous polymerases must be able to faithfully incorporate the unnatural triphosphates into DNA within the complex cellular milieu; and finally, the UBP must be stable in the presence of pathways that maintain the integrity of DNA. Here we show that an exogenously expressed algal nucleotide triphosphate transporter efficiently imports the triphosphates of both d5SICS and dNaM (d5SICSTP and dNaMTP) into E. coli, and that the endogenous replication machinery uses them to accurately replicate a plasmid containing d5SICS-dNaM. Neither the presence of the unnatural triphosphates nor the replication of the UBP introduces a significant growth burden. Lastly, we find that the UBP is not efficiently excised by DNA repair pathways. Thus, the resulting bacterium is the first organism to stably propagate an expanded genetic alphabet.
To make the unnatural triphosphates available inside the cell, we previously suggested using passive diffusion of the free nucleosides into the cytoplasm followed by their conversion to the corresponding triphosphate via the nucleoside salvage pathway8. While we have shown that analogs of d5SICS and dNaM are phosphorylated by the nucleoside kinase from D. melanogaster8, monophosphate kinases are more specific9, and in E. coli we found over-expression of the endogenous nucleoside diphosphate kinase results in poor growth. As an alternative, we focused on the nucleotide triphosphate transporters (NTTs) of obligate intracellular bacteria and algal plastids10–14. We expressed eight different NTTs in E. coli C41(DE3)15–17 and measured the uptake of [α-32P]-dATP as a surrogate for the unnatural triphosphates (Extended Data Fig. 1). We confirmed that [α-32P]-dATP is efficiently transported into cells by the NTTs from Phaeodactylum tricornutum (PtNTT2)18 and Thalassiosira pseudonana (TpNTT2)18. While NTTs from Protochlamydia amoebophila (PamNTT2 and PamNTT5)15 also import [α-32P]-dATP, PtNTT2 showed the most activity, and both it and TpNTT2 are known to have broad specificity18, making them the most promising NTTs for further characterization.
Transport via an NTT requires that the unnatural triphosphates are sufficiently stable in culture media; however, preliminary characterization of d5SICSTP and dNaMTP indicated that decomposition occurs in the presence of actively growing E. coli (Extended Data Fig. 2). Similar behavior was observed with [α-32P]-dATP, and the dephosphorylation products detected by thin layer chromatography for [α-32P]-dATP or by HPLC and MALDI for d5SICSTP and dNaMTP, suggest that decomposition is mediated by phosphatases. As no degradation was observed upon incubation in spent media, decomposition appears to occur within the periplasm. No increase in stability was observed in cultures of single-gene deletion mutants of E. coli BW25113 lacking a specific periplasmic phosphatase19 (as identified by the presence of a Sec-type N-terminal leader sequence), including phoA, ushA, appA, aphA, yjjX, surE, yfbR, yjjG, yfaO, mutT, nagD, yggV, yrfG, or ymfB, suggesting that decomposition results from the activity of multiple phosphatases. However, the extracellular stability of [α-32P]-dATP was significantly greater when 50 mM potassium phosphate (KPi) was added to the growth medium (Extended Data Fig. 3). Thus, we measured [α-32P]-dATP uptake from media containing 50 mM KPi after induction of the transporter with IPTG (Extended Data Fig. 4). While induction with 1 mM IPTG resulted in slower growth, consistent with the previously reported toxicity of NTTs17, it also resulted in maximal [α-32P]-dATP uptake. Thus, after addition of 1 mM IPTG, we analyzed the extracellular and intracellular stability of [α-32P]-dATP as a function of time (Extended Data Fig. 5). Cells expressing PtNTT2 were found to have the highest levels of intracellular [α-32P]-dATP, and while both extra- and intercellular dephosphorylation was still observed, the ratio of triphosphate to dephosphorylation products inside the cell remained roughly constant, indicating that the extracellular concentrations and PtNTT2-mediated influx are sufficient to compensate for intracellular decomposition.
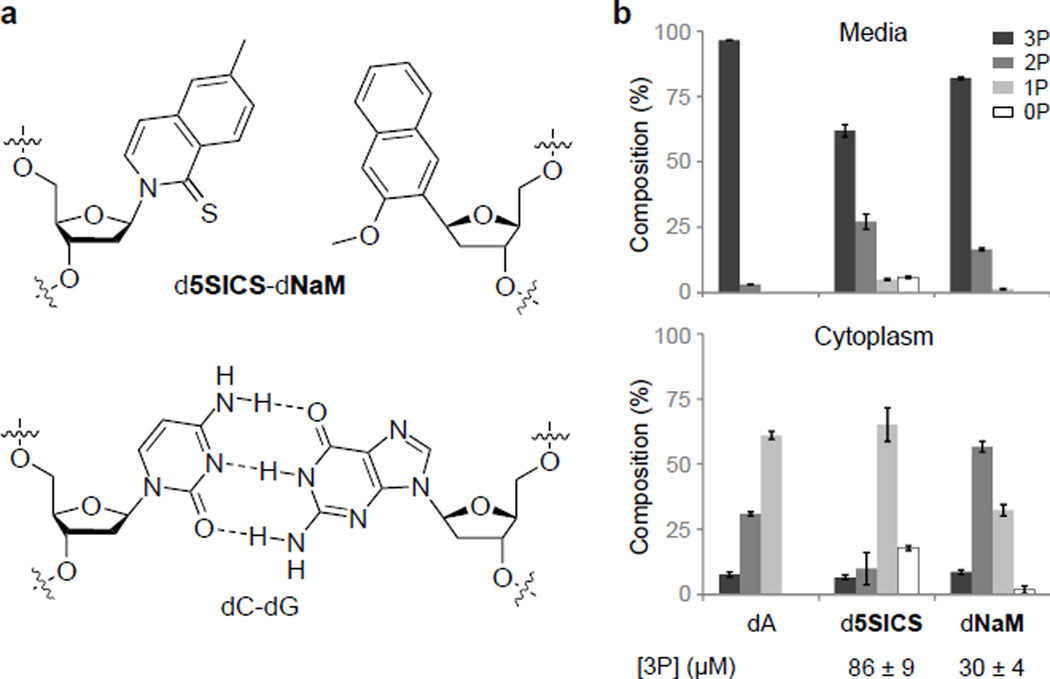
Likewise, we found that the addition of KPi increased the extracellular stability of d5SICSTP and dNaMTP (Extended Data Fig. 2), and when a stationary phase culture was diluted 100-fold into fresh media, the half-lives of both unnatural triphosphates (initial concentrations of 0.25 mM) were found to be ~9 h, which seemed sufficient for our purposes. Thirty minutes after their addition to the media, neither of the unnatural triphosphates was detected in cells expressing TpNTT2; in contrast, 90 µM of d5SICSTP and 30 µM of dNaMTP were found in the cytoplasm of cells expressing PtNTT2 (Fig. 1b). Although intracellular decomposition was still apparent, the intracellular concentrations of intact triphosphate are significantly above the sub-micromolar KM values of the unnatural triphosphates for DNA polymerases20, setting the stage for replication of the UBP in a living bacterial cell.
Figure 1. Nucleoside triphosphate stability and import.
a, Chemical structure of the d5SICS–dNaM UBP compared to the natural dG-dC base pair. b, Composition analysis of d5SICS and dNaM in the media (top) and cytoplasmic (bottom) fractions of cells expressing PtNTT2 after 30 min incubation; dA shown for comparison. 3P, 2P, 1P and 0P correspond to triphosphate, diphosphate, monophosphate and nucleoside, respectively; [3P] is the total intracellular concentration of triphosphate. Errors represent s.d. of the mean, n=3.
The replication of DNA containing d5SICS-dNaM has been validated in vitro with different polymerases, primarily family A polymerases, such as the Klenow fragment of E. coli DNA Polymerase I (Pol I)20,21. As the majority of the E. coli genome is replicated by Pol III, we engineered a plasmid to focus replication of the UBP to Pol I. Plasmid pINF (i.e. the information plasmid) was constructed from pUC19 using DNA solid-phase synthesis and circular-extension PCR to replace the dA-dT pair at position 505 with dNaM paired opposite an analog of d5SICS (dTPT322) (Fig. 2a,b). This positions the UBP downstream of the ColE1 origin of replication where leading-strand replication is mediated by Pol I23, and within the TK-1 Okazaki processing site24, where lagging-strand synthesis is also expected to be mediated by Pol I. Synthetic pINF was constructed using the d5SICS analog because it should be efficiently replaced by d5SICS if replication occurs in vivo, making it possible to differentiate in vivo replicated pINF from synthetic pINF.
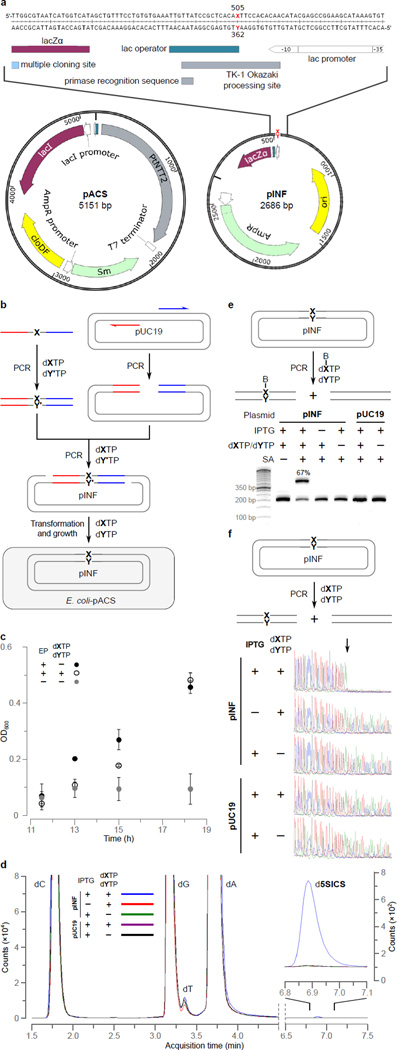
Figure 2. Intracellular UBP replication.
a, Structure of pACS and pINF. dX and dY correspond to dNaM and a d5SICS analog22 that facilitated plasmid construction (see Methods). cloDF = origin of replication; Sm = streptomycin resistance gene; AmpR = ampicillin resistance gene; ori = ColE1 origin of replication; MCS = multiple cloning site; lacZα = β-galactosidase fragment gene. b, Overview of pINF construction. A DNA fragment containing the unnatural nucleotide was synthesized via solid phase DNA synthesis and then used to assemble synthetic pINF via circular-extension PCR29. X = dNaM, Y’ = dTPT3 (an analog of d5SICS22), and Y = d5SICS (see text). Color indicates regions of homology. The doubly-nicked product was used directly to transform E. coli harboring pACS. c, The addition of d5SICSTP and dNaMTP eliminates a growth lag of cells harboring pINF. EP=electroporation. Errors represent s.d. of the mean, n=3. d, LC-MS/MS total ion chromatogram of global nucleoside content in pINF and pUC19 recorded in Dynamic Multiple Reaction Monitoring (DMRM) mode. pINF and pUC19 (control) were propagated in E. coli in the presence or absence of unnatural triphosphates, and with or without PtNTT2 induction. The inset shows a 100× expansion of the mass count axis in the d5SICS region. e, Biotinylation only occurs in the presence of the UBP, the unnatural triphosphates, and transporter induction. After growth, pINF was recovered, and a 194 nt region containing the site of UBP incorporation (nt 437–630) was amplified and biotinylated. B=biotin; SA=streptavidin. The natural pUC19 control plasmid was prepared identically to pINF. 50-bp DNA ladder is shown to the left. f, Sequencing analysis demonstrates retention of the UBP. An abrupt termination in the Sanger sequencing reaction indicates the presence of UBP incorporation (site indicated with arrow).
To determine whether E. coli can use the imported unnatural triphosphates to stably propagate pINF, C41(DE3) cells were first transformed with a pCDF-1b plasmid encoding PtNTT2 (hereafter referred to as pACS, for accessory plasmid, Fig. 2a) and grown in media containing 0.25 mM of both unnatural triphosphates, 50 mM KPi, and 1 mM IPTG to induce transporter production. Cells were then transformed with pINF, and after a 1 h recovery period, cultures were diluted 10-fold with the same media supplemented with ampicillin, and growth was monitored via culture turbidity (Extended Data Table 1). As controls, cells were also transformed with pUC19, or grown without either IPTG or without the unnatural triphosphates. Again, growth was significantly slower in the presence of IPTG, but the addition of d5SICSTP and dNaMTP resulted in only a slight further decrease in growth in the absence of pINF, and interestingly, it eliminated a growth lag in the presence of pINF (Fig. 2c), suggesting that the unnatural triphosphates are not toxic and are required for the efficient replication of pINF.
To demonstrate the replication of pINF, we recovered the plasmid from cells after 15 h of growth. The introduction of the UBP resulted in a small (~2-fold) reduction in the copy number of pINF, as gauged by its ratio to pACS (Extended Data Table 1); we determined that the plasmid was amplified 2 × 107-fold during growth (~24 doublings) based on the amount of recovered plasmid and the transformation efficiency. To determine the level of UBP retention, the recovered plasmid was digested, dephosphorylated to single nucleosides, and analyzed by LC-MS/MS25. While the detection and quantification of dNaM were precluded by its poor fragmentation efficiency and low product ion counts over background, signal for d5SICS was clearly observable (Fig. 2d). External calibration curves were constructed using the unnatural nucleoside and validated by determining its ratio to dA in synthetic oligonucleotides (Extended Data Table 2). Using the resulting calibration curve, we determined the ratio of dA to d5SICS in recovered pINF was 1,106 to 1, which when compared to the expected ratio of 1,325 to 1, suggests the presence of approximately one UBP per plasmid. No d5SICS was detected in control experiments in which the transporter was not induced, nor when the unnatural triphosphates were not added to the media, nor when pUC19 was used instead of pINF (Fig. 2d inset), demonstrating that its presence results from the replication of the UBP and not from misinsertion of the unnatural triphosphates opposite a natural nucleotide. Importantly, as the synthetic pINF contained an analog of d5SICS, and d5SICS was only provided as a triphosphate added to the media, its presence in pINF confirms in vivo replication.
To independently confirm and quantify the retention of the UBP in the recovered plasmid, the relevant region was amplified by PCR in the presence of d5SICSTP and a biotinylated dNaMTP analog4 (Fig. 2e). Analysis by streptavidin gel shift showed that 67% of the amplified DNA contained biotin. No shift was observed in control experiments where the transporter was not induced, nor when unnatural triphosphates were not added, nor when pUC19 was used instead of pINF, demonstrating that the shift results from the presence of the UBP. Based on a calibration curve constructed from the shifts observed with the amplification products of controlled mixtures of DNA containing dNaM or its fully natural counterpart (Methods and Extended Data Fig. 6), the observed gel shift corresponds to a UBP retention of 86%. Similarly, when the amplification product obtained with d5SICSTP and dNaMTP was analyzed by Sanger sequencing in the absence of the unnatural triphoshates1,26,27, the sequencing chromatogram showed complete termination at the position of UBP incorporation, which with an estimated lower limit of read-through detection of 5%, suggests a level of UBP retention in excess of 95% (Fig. 2f). In contrast, amplification products obtained from pINF recovered from cultures grown without PtNTT2 induction, without added unnatural triphosphates, or obtained from pUC19 propagated under identical conditions, showed no termination. Overall, the data unambiguously demonstrate that DNA containing the UBP was replicated in vivo and allow us to estimate that replication occurred with a fidelity (retention per doubling) of at least 99.4% (0.99424 = 0.86). This fidelity corresponds to an error rate of ~10−3, which is comparable to the intrinsic error-rate of some polymerases with natural DNA28.
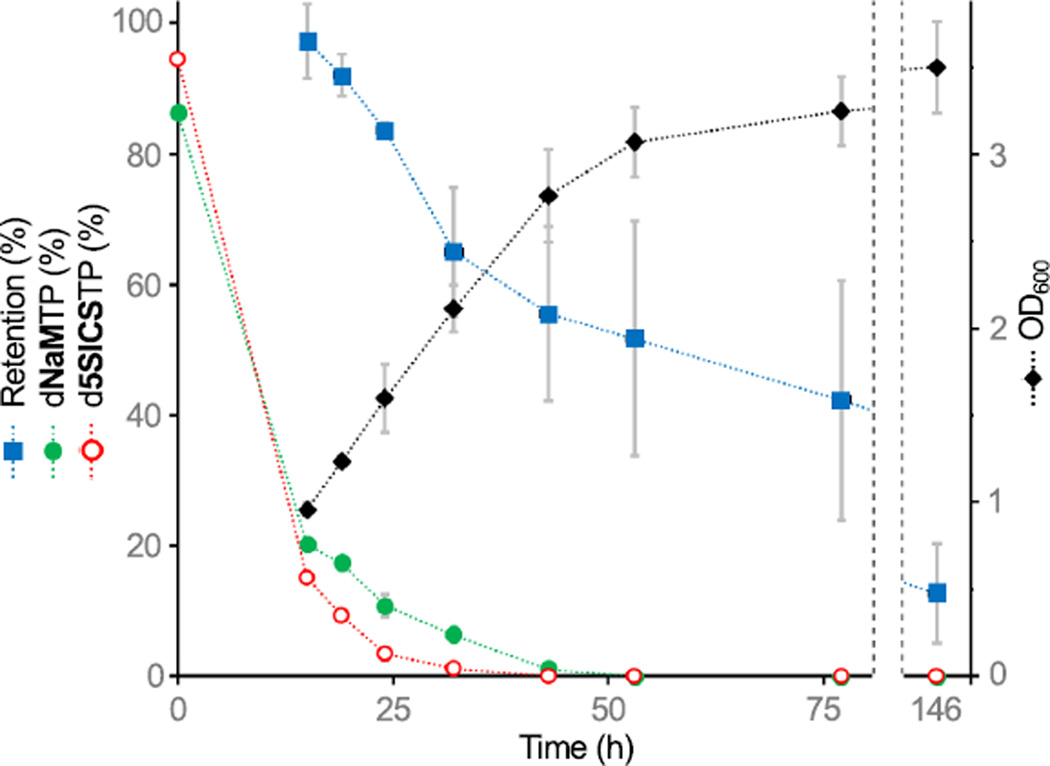
The high retention of the UBP over a 15 h period of growth (~24 doublings) strongly suggests that it is not efficiently excised by DNA repair pathways. To further test this hypothesis and to examine retention during prolonged stationary phase growth, we repeated the experiments, but monitored UBP retention, cell growth, and unnatural triphosphate decomposition for up to 6 days without providing any additional unnatural triphosphates (Fig. 3 and Extended Data Fig 7). At 15 and 19 h of growth, the cultures reached an OD600 of ~0.9 and ~1.2, respectively, and both d5SICSTP and dNaMTP decomposed to 17–20% and 10–16% of their initial 0.25 mM concentrations (Extended Data Fig. 7a). In agreement with the above described experiments, retention of the UBP after 15 h was 97 ± 5% and >95%, as determined by gel shift and sequencing, respectively, and after 19 h it was 91 ± 3% and >95%. As the cultures entered stationary phase and the triphosphates decomposed completely, plasmid loss began to compete with replication (Extended Data Fig. 7b,c,d), but even then, retention of the UBP remained at ~45% and ~15%, at 3 and 6 days respectively. Moreover, when d5SICS-dNaM was lost, it was replaced by dA-dT, which is consistent with the mutational spectrum of DNA Pol I20. Finally, the shape of the retention vs. time curve mirrors that of the growth vs. time curve. Taken together, these data suggest that in the absence of unnatural triphosphates, the UBP is eventually lost by replication-mediated mispairing, and not from the activity of DNA repair pathways.
Figure 3. Intracellular stability of the UBP.
E. coli C41(DE3)-pACS was transformed with pINF and grown after a single dose of d5SICSTP and dNaMTP was provided in the media. UBP retention in recovered pINF (blue squares), OD600 (black diamonds), and relative amount of d5SICSTP and dNaMTP in the media (red and green circles, respectively; 100% = 0.25 mM), were determined as a function of time. Errors represent s.d. of the mean, n=3.
We have demonstrated that PtNTT2 efficiently imports d5SICSTP and dNaMTP into E. coli by PtNTT2 and that an E. coli polymerase, possibly Pol I, is able to efficiently use the unnatural triphosphates to replicate DNA containing the UBP within the cellular environment with reasonable efficiency and fidelity. Moreover, the UBP appears stable during both exponential and stationary phase growth despite the presence of all DNA repair mechanisms. Remarkably, while expression of PtNTT2 results in a somewhat reduced growth rate, neither the unnatural triphosphates nor replication of the UBP results in significant further reduction in growth. The resulting bacterium is the first organism that stably harbors DNA containing three base pairs. In the future, this organism, or a variant with the UBP incorporated at other episomal or chromosomal loci, should provide a synthetic biology platform to orthogonally re-engineer cells, with applications ranging from site-specific labeling of nucleic acids in living cells to the construction of orthogonal transcription networks and eventually the production and evolution of proteins with multiple, different unnatural amino acids.
METHODS
Materials
2×YT, 2×YT-agar, IPTG, ampicillin and streptomycin were obtained from Fisher Scientific. Ampicillin and streptomycin were used at 100 µg/ml and 50 µg/ml, respectively. All pET-16b constructs containing the nucleotide transporters were kindly provided by Dr. Haferkamp (Technische Universität Kaiserslautern, Germany) with the exception of pET16b-RpNTT2, which along with the C41(DE3) E. coli strain, was provided by Dr. Audia (University of South Alabama, USA). Plasmids pUC19 and pCDF-1b were obtained from Thermo Scientific and EMD Millipore, respectively. Plasmids were purified using the PureLink Quick Plasmid DNA Miniprep Kit (Life Technologies). OneTaq, DeepVent, Q5 Hot Start High-Fidelity DNA Polymerases, and all restriction endonucleases were obtained from New England Biolabs. In general, PCR reactions were divided into multiple aliquots with one followed in real time using 0.5× Sybr Green I (Life Technologies); following PCR, the aliquots were recombined, purified by spin column (DNA Clean and Concentrator-5; Zymo Research, Irvine, CA) with elution in 20 µl of water, then separated by agarose gel electrophoresis, followed by band excision and recovery (Zymoclean Gel DNA Recovery Kit), eluting with 20 µl of water unless stated otherwise. Polyacrylamide gels were stained with 1× Sybr Gold (Life Technologies) for 30 min, agarose gels were cast with 1× Sybr Gold. All gels were visualized using a Molecular Imager Gel Doc XR+ equipped with 520DF30 filter (Bio-Rad) and quantified with Quantity One software (Bio-Rad). The sequences of all DNA oligonucleotides used in this study are provided in Supplementary Information. Natural oligonucleotides were purchased from IDT (San Diego, CA). The concentration of dsDNA was measured by fluorescent dye binding (Quant-iT dsDNA HS Assay kit, Life Technologies) unless stated otherwise. The concentration of ssDNA was determined by UV absorption at 260 nm using a NanoDrop 1000 (Thermo Scientific). [α-32P]-dATP (25 µCi) was purchased from PerkinElmer (Shelton, CT). Polyethyleneiminecellulose pre-coated Bakerflex® TLC plates (0.5 mm) were purchased from VWR. dNaM phosphoramidite, dNaM and d5SICS nucleosides were obtained from Berry & Associates Inc (Dexter, MI). Free nucleosides of dNaM and d5SICS (Berry & Associates) were converted to the corresponding triphosphates under Ludwig conditions30. After purification by anion exchange chromatography (DEAE Sephadex A-25) followed by reverse phase (C18) HPLC and elution through a Dowex 50WX2-sodium column, both triphosphates were lyophilized and kept at −20 °C until use. The d5SICSTP analog dTPT3TP22 and the biotinylated dNaMTP analog dMMO2SSBIOTP4 were made as reported previously. MALDI-TOF mass spectrometry (Applied Biosystems Voyager DE-PRO System 6008) was performed at the TSRI Center for Protein and Nucleic Acid Research.
Construction of NTT expression plasmids
The PtNTT2 gene was amplified from plasmid pET-16b-PtNTT2 using primers PtNTT2-fwd and PtNTT2-rev; the TpNTT2 gene was amplified from plasmid pET-16b-TpNTT2 using primers TpNTT2-fwd and TpNTT2-rev. A linear fragment of pCDF-1b was generated using primers pCDF-1b-fwd and pCDF-1b-rev. All fragments were purified as described in Materials. The pCDF-1b fragment (100 ng, 4.4×10−14 mol) and either the PtNTT2 (78 ng, 4.4×10−14 mol) or TpNTT2 (85 ng, 4.4×10−14 mol) fragment were then assembled together using restriction-free circular polymerase extension cloning29 in 1× OneTaq reaction buffer, MgSO4 adjusted to 3.0 mM, 0.2 mM of dNTP, and 0.02 U/µl of OneTaq DNA under the following thermal cycling conditions: Initial denaturation (96 °C, 1 min); 10 cycles of denaturation (96 °C, 30 s), touchdown annealing (54 °C to 49.5 °C for 30 s (−0.5°C per cycle)), extension of 68 °C for 5 min, and final extension (68 °C, 5 min). Upon completion, the samples were purified and used for heat-shock transformation of E. coli XL10. Individual colonies were selected on LB-agar containing streptomycin, and assayed by colony PCR with primers PtNTT2-fwd/rev or TpNTT2-fwd/rev. The presence of the NTT genes was confirmed by sequencing and double digestion with ApaI/EcoO109I restriction endonucleases with the following expected pattern: pCDF-1b-PtNTT2 (2546/2605 bp), pCDF-1b-TpNTT2 (2717/2605 bp), pCDF-1b (1016/2605 bp). The complete nucleotide sequence of the pCDF-1b/PtNTT2 plasmid (i.e. ppACS) is provided in Supplementary Information.
General protocol to quantify nucleoside triphosphate uptake
Growth conditions
E. coli C41(DE3)16 freshly transformed with pCDF-1b-PtNTT2 was grown in 2×YT with streptomycin overnight, then diluted (1:100) into fresh 2×YT medium (1 ml of culture per uptake with [α-32P]-dATP; 2 ml of culture per uptake with d5SICSTP or dNaMTP) supplemented with 50 mM potassium phosphate (KPi) and streptomycin. A negative control with the inactive transporter pET-16b-RpNTT2, was treated identically except ampicillin was used instead of streptomycin. Cells were grown to an OD600 of ~0.6 and the NTT expression was induced by the addition of IPTG (1 mM). The culture was allowed to grow for another hour (final OD600 ~1.2) and then assayed directly for uptake as described below using a method adapted from Haferkamp, et al.15.
Preparation of media fraction
The experiment was initiated by the addition of either dNaMTP or d5SICSTP (10 mM each) directly to the media to a final concentration of 0.25 mM. Cells were incubated with the substrate with shaking at 37 °C for 30 min and then pelleted (8,000 rfu for 5 min, 4 °C). An aliquot of the media fraction (40 µl) was mixed with acetonitrile (80 µl) to precipitate proteins31, and then incubated at RT for 30 min. Samples were either analyzed immediately by HPLC or stored at −80 °C until analysis. Analysis began with centrifugation (12,000 rfu for 10 min, RT), then the pellet was discarded, and the supernatant was reduced to ~20 µl by SpeedVac, resuspended in buffer A (see below) to a final volume of 50 µl, and analyzed by HPLC (see below).
Preparation of cytoplasmic fraction
To analyze the intracellular desphosphorylation of the unnatural nucleoside triphosphate, cell pellets were subjected to 3 ×100 µl washes of ice cold KPi (50 mM). Pellets were then resuspended in 250 µl of ice cold KPi (50 mM) and lysed with 250 µl of Lysis buffer L7 of the PureLink Quick Plasmid DNA Miniprep Kit (200 mM NaOH, 1% w/v SDS), after which the resulting solution was incubated at RT for 5 min. Precipitation buffer N4 (350 µl, 3.1 M potassium acetate, pH 5.5) was added, and the sample was mixed to homogeneity. Following centrifugation (>12,000 rfu for 10 min, RT) the supernatant containing the unnatural nucleotides was applied to a Hypersep C18 solid phase extraction column (Thermo Scientific) prewashed with acetonitrile (1 ml) and buffer A (1 ml, see HPLC Protocol for buffer composition). The column was then washed with buffer A and nucleotides were eluted with 1 ml of 50% acetonitrile:50% triethylammonium bicarbonate (TEAB) 0.1 M (pH 7.5). The eluent was reduced to ~ 50 µl in a SpeedVac and its volume was adjusted to 100 µl with buffer A before HPLC analysis.
HPLC Protocol
Samples were applied to a Phenomenex Jupiter LC column (3 µm C18 300 Å, 250 × 4.6 mm) and subjected to a linear gradient of 0–40% B over 40 min at a flow rate of 1 ml/min. Buffer A: 95% 0.1 M TEAB, pH 7.5; 5% acetonitrile. Buffer B: 20% 0.1 M TEAB, pH 7.5; 80% acetonitrile. Absorption was monitored at 230, 273, 288, 326, 365 nm.
Quantification
Each injection series included two extra control samples containing 5 nmol of dNaMTP or d5SICSTP. The areas under the peaks that corresponded to triphosphate, diphosphate, monophosphate and free nucleoside (confirmed by MALDI-TOF) were integrated for both the control and the unknown samples (described above). After peak integration, the ratio of the unknown peak to the control peak adjusted for the loss from the extraction step (62% and 70% loss for dNaM and d5SICS, respectively, Extended Data Table 3), provided a measure of the amount of each of the moieties in the sample. To determine the relative concentrations of unnatural nucleotide inside the cell, the amount of imported unnatural nucleotide (dXTP, µmol) was then divided by the volume of cells, which was calculated as the product of the volume of a single E. coli cell (1 µm3 based on a reported average value32, i.e. 1×10−9 µl cell−1) and the number of cells in each culture (OD600 of 1.0 equal to 1×109 cells per ml33). The RpNTT2 sample was used as a negative control and its signal was subtracted to account for incomplete washing of nucleotide species from the media.
dATP uptake
To analyze the intracellular desphosphorylation of dATP, after induction of the transporter, the uptake reaction was initiated by the addition of dATP (spiked with [α-32P]-dATP) to a final concentration of 0.25 mM, followed by incubation at 37 °C with shaking for 30 min. The culture was then centrifuged (8,000 rfu for 5 min, RT). Supernatant was analyzed by thin layer chromatography (TLC), while the cell pellets were washed 3 times with ice cold KPi (50 mM, 100 µl) to remove excess radioactive substrate, lysed with NaOH (0.2 M, 100 µl) and centrifuged (10,000 rfu for 5 min, RT) to pellet the cell debris, before the supernatant (1.5 µl) was analyzed by TLC.
TLC analysis
Samples (1 µl) were applied on a 0.5 mm polyethyleneimine cellulose TLC plate and developed with sodium formate pH 3.0 (0.5 M, 30 s; 2.5 M, 2.5 min; 4.0 M, 40 min). Plates were dried using a heat gun and quantified by phosphorimaging (Storm Imager, Molecular Dynamics) and Quantity One software (BioRad).
Optimization of nucleotide extraction from cells for HPLC injection
To minimize the effect of the lysis and triphosphate extraction protocols on the decomposition of nucleoside triphosphate within the cell, the extraction procedure was optimized for the highest recovery with the lowest extent of decomposition (Extended Data Table 3). To test different extraction methods, cells were grown as described above, washed, and then 5 nmol of either dNaMTP or d5SICSTP was added to the pellets, which were then subjected to different extraction protocols including boiling water, hot ethanol, cold methanol, freeze and thaw, lysozyme, glass beads, NaOH, trichloroacetic acid (TCA) with Freon, and perchloric acid (PCA) with KOH34. The recovery and composition of the control was quantified by HPLC as described above to determine the most effective procedure. Method 3, i.e. cell lysis with NaOH (Extended Data Table 3), was found to be most effective and reproducible, thus we further optimized it by resuspension of the pellets in ice cold KPi (50 mM, 250 µl) before addition of NaOH to decrease dephosphorylation after cell lysis (Method 4). Cell pellets were then processed as described above. See above for the final extraction protocol.
Construction of pINF
Preparation of the unnatural insert
The TK-1-dNaM oligonucleotide containing dNaM was prepared using solid phase DNA synthesis with ultra-mild DNA synthesis phosphoramidites on CPG ultramild supports (1 µmol, Glen Research; Sterling, VA) and an ABI Expedite 8905 synthesizer. After the synthesis, the DMT-ON oligonucleotide was deprotected by treatment with concentrated ammonia hydroxide at 50 °C overnight, purified by Glen-Pak cartridge (Glen Research), and then subjected to 8 M urea 8% PAGE. The gel was visualized by UV shadowing, the band corresponding to the 75mer was excised, and the DNA was recovered by crush and soak extraction, filtration (0.45 µm), and final desalting over Sephadex G-25 (NAP-25 Columns, GE Healthcare). The concentration of the single stranded oligonucleotide was determined by UV absorption at 260 nm assuming that the extinction coefficient of dNaM at 260 nm is equal to that of dA. TK-1-dNaM (4 ng) was next amplified by PCR under the following conditions: 1× OneTaq reaction buffer, MgSO4 adjusted to 3.0 mM, 0.2 mM of dNTP, 0.1 mM of dNaMTP, 0.1 mM of the d5SICSTP analog dTPT3TP, 1 µM of each of the primers pUC19-fusion-fwd and pUC19-fusion-rev, and 0.02 U/µl of OneTaq DNA Polymerase (in a total of 4×50 µl reactions) under the following thermal cycling conditions: Initial denaturation (96 °C, 1 min) followed by 12 cycles of denaturation (96 °C, 10 s), annealing (60 °C, 15 s), and extension (68 °C, 2 min). An identical PCR without the unnatural triphosphates was run to obtain fully natural insert under identical conditions for the construction of the natural control plasmid. Reactions were subjected to spin column purification and then the desired PCR product (122 bp) was purified by a 4% agarose gel as described in Materials.
pUC19 linearization
pUC19 (20 ng) was amplified by PCR under the following conditions: 1× Q5 reaction buffer, MgSO4 adjusted to 3.0 mM, 0.2 mM of dNTP, 1 µM of each primers pUC19-lin-fwd and pUC19-lin-rev, and 0.02 U/µl of Q5 Hot Start High-Fidelity DNA Polymerase (in a total of 4×50 µl reactions with one reaction containing 0.5×Sybr Green I) under the following thermal cycling conditions: Initial denaturation (98 °C, 30 s); 20 cycles of denaturation (98°C, 10s), annealing (60 °C, 15 s), and extension (72 °C, 2 min); and final extension (72 °C, 5 min). The desired PCR product (2611 bp) was purified by a 2% agarose gel.
PCR assembly of pINF and the natural control plasmid
A linear fragment was amplified from pUC19 using primers pUC19-lin-fwd and pUC19-lin-rev. The resulting product (800 ng, 4.6×10−13 mol) was combined with either the natural or unnatural insert (see above) (56 ng, 7.0×10−13 mol) and assembled by circular overlap extension PCR under the following conditions: 1× OneTaq reaction buffer, MgSO4 adjusted to 3.0 mM, 0.2 mM of dNTP, 0.1 mM of dNaMTP, 0.1 mM of the d5SICSTP analog dTPT3TP, and 0.02 U/µl of OneTaq DNA Polymerase (in a total of 4×50 µl reactions with one reaction containing 0.5×Sybr Green I) using the following thermal cycling conditions: Initial denaturation (96 °C, 1 min); 12 cycles of denaturation (96 °C, 30 s), annealing 62 °C, 1 min), and extension (68 °C, 5 min); final extension (68 °C, 5 min); and slow cooling (68 °C to 10 °C at a rate of −0.1 °C/s). The PCR product was analyzed by restriction digestion on 1% agarose and used directly for E. coli transformation. The d5SICS analog dTPT322 pairs with dNaM, and dTPT3TP was used in place of d5SICSTP as DNA containing dTPT3-dNaM is better PCR amplified than DNA containing d5SICS-dNaM, and this allowed for differentiation of synthetic and in vivo replicated pINF, as well as facilitated the construction of high-quality pINF (UBP content > 99%).
pINF replication in E. coli
Preparation of electrocompetent cells
C41(DE3) cells were transformed by heat shock33 with 200 ng of pACS plasmid, and the transformants were selected overnight on 2×YT-agar supplemented with streptomycin. A single clone of freshly transformed C41(DE3)-pACS was grown overnight in 2×YT medium (3 ml) supplemented with streptomycin and KPi (50 mM). After 100-fold dilution into the same fresh 2×YT media (300 ml), the cells were grown at 37 °C until they reached an OD600 of 0.20 at which time IPTG was added to a final concentration of 1 mM to induce the expression of PtNTT2. Cells were grown for another 40 min and then growth was stopped by rapid cooling in ice water with intensive shaking. After centrifugation in a prechilled centrifuge (2400 rfu for 10 min, 4 °C), the spent media was removed, and the cells were prepared for electroporation by washing with ice-cold sterile water (3×150 ml). After washing, the cells were resuspended in ice cold 10% glycerol (1.5 ml) and split into 50 µl aliquots. Although we found that dry ice yielded better results than liquid nitrogen for freezing cells to store for later use, freshly prepared cells were used for all reported experiments as they provided higher transformation efficiency of pINF and higher replication fidelity of the UBP.
Electroporation and recovery
The aliquot of cells was mixed with 2 µl of plasmid (400 ng), transferred to 0.2 cm gap electroporation cuvette and electroporated using a Bio-Rad Gene Pulser according to the manufacturer’s recommendations (voltage 25 kV, capacitor 2.5 µF, resistor 200 Ω, time constant 4.8 ms). Pre-warmed 2×YT media (0.95 ml, streptomycin, 1 mM IPTG, 50 mM KPi) was added, and after mixing, 45 µl was removed and combined with 105 µl of the same media (3.33× dilution) supplemented with 0.25 mM of dNaMTP and d5SICSTP. The resulting mixture was allowed to recover for 1 h at 37 °C with shaking (210 rpm). The original transformation media (10 µl) was spread onto 2×YT-agar containing streptomycin with 10× and 50× dilutions for the determination of viable colony forming units after overnight growth at 37 °C to calculate the number of the transformed pINF molecules (see section “Calculation of the plasmid amplification”). Transformation, recovery, and growth was carried out identically for the natural control plasmid. Additionally, a negative control was run and treated identically to pINF transformation except that it was not subjected to electroporation (Extended Data Fig. 7b). No growth in the negative control samples was observed even after 6 days. No PCR amplification of the negative control (refer to “Fidelity measurement” sections) was detected, which confirms that unamplified pINF plasmid is carried through cell growth and later detected erroneously as the propagated plasmid.
Growth and analysis
After recovery, the cells were centrifuged (4000 rfu for 5 min, 4°C), spent media (0.15 ml) was removed and analyzed for nucleotide composition by HPLC (Extended Data Fig. 7a). The cells were resuspended in fresh 2×YT media (1.5 ml, streptomycin, ampicillin, 1 mM IPTG, 50 mM KPi, 0.25 mM dNaMTP, 0.25 mM d5SICSTP) and grown overnight at 37 °C while shaking (250 rpm), resulting in 10× dilution compared to recovery media or 33.3× dilution compared to the originally transformed cells. Aliquots (100 µl) were taken after 15, 19, 24, 32, 43, 53, 77, 146 h, OD600 was determined, and the cells were centrifuged (8000 rfu for 5 min, 4 °C). Spent media was analyzed for nucleotide composition by HPLC (Extended Data Fig. 7a) and the pINF and pACS plasmid mixtures were recovered and linearized with NdeI restriction endonuclease; pINF plasmid was purified by 1% agarose gel electrophoresis (Extended Data Fig. 7b) and analyzed by LC-MS/MS. The retention of the UBP on the pINF plasmid was quantified by biotin gel shift mobility assay and sequencing as described below.
Mass spectrometry
Linearized pINF was digested to nucleosides by treatment with a mixture of nuclease P1 (Sigma-Aldrich), shrimp alkaline phosphatase (NEB), and DNase I (NEB), overnight at 37 °C, following a previously reported protocol25. LC-MS/MS analysis was performed in duplicate by injecting 15 ng of digested DNA on an Agilent 1290 UHPLC equipped with a G4212A diode array detector and a 6490A Triple Quadrupole Mass Detector operating in the positive electrospray ionization mode (+ESI). UHPLC was carried out using a Waters XSelect HSS T3 XP column (2.1 × 100 mm, 2.5 µm) with the gradient mobile phase consisting of methanol and 10 mM aqueous ammonium formate (pH 4.4). MS data acquisition was performed in Dynamic Multiple Reaction Monitoring (DMRM) mode. Each nucleoside was identified in the extracted chromatogram associated with its specific MS/MS transition: dA at m/z 252→136, d5SICS at m/z 292→176, and dNaM at m/z 275→171. External calibration curves with known amounts of the natural and unnatural nucleosides were used to calculate the ratios of individual nucleosides within the samples analyzed. LC-MS/MS quantification was validated using synthetic oligonucleotides1 containing unnatural d5SICS and dNaM (Extended Data Table 2).
Fidelity measurement by gel shift mobility assay (Fig. 2e)
DNA biotinylation by PCR
Purified mixtures of pINF and pACS plasmids (1 ng) from growth experiments were amplified by PCR under the following conditions: 1× OneTaq reaction buffer, MgSO4 adjusted to 3.0 mM, 0.3 mM of dNTP, 0.1 mM of the biotinylated dNaMTP analog dMMO2SSBIOTP, 0.1 mM of d5SICSTP, 1 µM of each of the primers pUC19-seq-fwd and pUC19-seq-rev, 0.02 U/µl of OneTaq DNA Polymerase, and 0.0025 U/µl of DeepVent DNA Polymerase in a total volume of 25 µl in an CFX Connect Real-Time PCR Detection System (Bio-Rad) under the following thermal cycling conditions: Initial denaturation (96 °C, 1 min); 10 cycles of denaturation (96 °C, 30 s), annealing (64 °C, 30 s), and extension (68 °C, 4 min). PCR products were purified as described in Materials, and the resulting biotinylated DNA duplexes (5 µl, 25–50 ng) were mixed with streptavidin (1 µl, 1 µg/µl, Promega) in phosphate buffer (50 mM sodium phosphate, pH 7.5, 150 mM NaCl, 1 mM EDTA), incubated for 30 min at 37 °C, mixed with 5× non-denaturing loading buffer (Qiagen), and loaded onto 6% non-denaturing PAGE. After running at 110 V for 30 min, the gel was visualized and quantified. The resulting fragment (194 bp) with primer regions underlined and the unnatural nucleotide in bold (X = dNaM or its biotinylated analog dMMO2SSBIO) is 5’-GCAGGCATGCAAGCTTGGCGTAATCATGG TCATAGCTGTTTCCTGTGTGAAATTGTTATCCGCTCACAXTTCCACACAACATACGAGCCGGAAGCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCTCACTGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAG
Streptavidin shift calibration
We have already reported a calibration between streptavidin (SA) shift and the fraction of sequences with UBP in the population (UBP), see Fig. S8 in the Supporting Information of Malyshev, et al.1. However, we found that spiking the PCR reaction with DeepVent improves the fidelity with which DNA containing d5SICS-dMMO2SSBIO is amplified, and thus we repeated the calibration with added DeepVent. To quantify the net retention of the UBP, nine mixtures of the TK-1-dNaM template and its fully natural counterpart with a known ratio of unnatural and natural were prepared (Extended Data Fig. 6a), subjected to biotinylation by PCR and analyzed by mobility-shift assay on 6% non-denaturing PAGE as described above. For calibration, the mixtures TK-1-dNaM template and its fully natural counterpart with a known ratio of unnatural and natural templates (0.04 ng) were amplified under the same conditions over nine cycles of PCR with pUC19-fusion primers and analyzed identically to samples from the growth experiment (see section “DNA biotinylation by PCR”). Each experiment was run in triplicate (a representative gel assay is shown in Extended Data Fig. 6b), and the streptavidin shift (SAS, %) was plotted as function of the UBP content (UBP, %). The data was then fit to a linear equation, SAS = 0.77×UBP + 2.0 (R2 = 0.999), where UBP corresponds to the retention of the UBP (%) in the analyzed samples after cellular replication and was calculated from the SAS shift using the equation above.
Calculation of the plasmid amplification
The cells were plated on 2×YT-agar containing ampicillin and streptavidin directly after transformation with pINF, and the colonies were counted after overnight growth at 37 °C. Assuming each cell is only transformed with one molecule of plasmid, colony counts correspond to the original amount of plasmid that was taken up by the cells. After overnight growth, the plasmids were purified from a specific volume of the cell culture and quantified. Since purified plasmid DNA represents a mixture of the pINF and pACS plasmids, digestion restriction analysis with NdeI exonuclease was performed to linearize both plasmids, followed by 1% agarose gel electrophoresis (Extended Data Fig. 7b). An example of calculations for the 19 h time point with one of three triplicates is provided in Supplementary Information.
Fidelity measurement by sequencing (Fig. 2f and Extended Data Figure 7d)
Fragment generation for sequencing
Purified mixtures of pINF and pACS plasmids (1 ng) after the overnight growth were amplified by PCR under the following conditions: 1× OneTaq reaction buffer, MgSO4 adjusted to 3.0 mM, 0.2 mM of dNTP, 0.1 mM of dNaMTP, 0.1 mM of the d5SICSTP analog dTPT3TP, 1 µM of each of the primers pUC19-seq2-fwd and pUC19-seq-rev (see below), and 0.02 U/µl of OneTaq DNA Polymerase in a total volume of 25 µl under the following thermal cycling conditions: denaturation (96 °C, 1 min); and 10 cycles of denaturation (96 °C, 30 s), annealing 64 °C, 30 s), and extension 68 °C, 2 min. Products were purified by spin column, quantified to measure DNA concentration and then sequenced as described below. The sequenced fragment (304 bp) with primer regions underlined and the unnatural nucleotide in bold (X = dNaM) is 5’-GCTGCAAGGCGATTAAGTTGGGTAACGCC AGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTGAATTCGAGCTCGGTACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCAAGCTTGGCGTAATCATGGTCATAGCTGTTTCCTGTGTGAAATTGTTATCCGCTCACAXTTCCACACAACATACGAGCCGGAAGCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCTCACTGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAG
Sanger sequencing
The cycle sequencing reactions (10 µl) were performed on a 9800 Fast Thermal Cycler (Applied Biosystems) with the Cycle Sequencing Mix (0.5 µl) of the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) containing 1 ng template and 6 pmol of sequencing primer pUC19-seq-rev under the following thermal cycling conditions: initial denaturation (98 °C, 1 min); and 25 cycles of denaturation (96 °C, 10 s), annealing (60 °C, 15 s), and extension (68 °C, 2.5 min). Upon completion, the residual dye terminators were removed from the reaction with Agencourt CleanSEQ (Beckman-Coulter, Danvers, MA). Products were eluted off the beads with deionized water and sequenced directly on a 3730 DNA Analyzer (Applied Biosystems). Sequencing traces were collected using Applied Biosystems Data Collection software v3.0 and analyzed with the Applied Biosystems Sequencing Analysis v5.2 software.
Analysis of Sanger Sequencing Traces
Sanger Sequencing traces were analyzed as described previously1,26 to determine the retention of the unnatural base pair. Briefly, the presence of an unnatural nucleotide leads to a sharp termination of the sequencing profile, while mutation to a natural nucleotide results in “read-through”. The extent of this read-through after normalization is inversely correlated with the retention of the unnatural base pair. Raw sequencing traces were analyzed by first adjusting the start and stop points for the Sequencing Analysis software (Applied Biosystems) and then determining the average signal intensity individually for each channel (A, C, G and T) for peaks within the defined points. This was done separately for the parts of the sequencing trace before (section L) and after (section R) the unnatural nucleotide. The R/L ratio after normalization (R/L)norm for sequencing decay and read-through in the control unamplified sample (R/L = 0.55(R/L)norm + 7.2, see Malyshev et al.26 for details) corresponds to the percentage of the natural sequences in the pool. Therefore, an overall retention (F) of the incorporation of the unnatural base pair during PCR is equal to 1–(R/L)norm. Since significant read-through (over 20%) was observed in the direction of the pUC19-seq2-fwd primer even with the control plasmid (synthetic pINF), sequencing of only the opposite direction (pUC19-seq-rev) was used to gauge fidelity. Raw sequencing traces are shown in Fig. 2f and provided as source data for Extended Figure 7d
Supplementary Material
Acknowledgements
We thank I Haferkamp and J Audia for kindly providing the NTT plasmids and helpful discussions, and P Ordoukhanian for providing access to the Center for Protein and Nucleic Acid Research at TSRI. This work was supported by the NIH (GM 060005).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Online Content Full Methods and Extended Data display items are available in the online version of the paper.
Author Contributions. D.A.M., K.D., T.C. and F.E.R. designed the experiments. D.A.M., K.D., T.L. performed the experiments. N.D., J.M.F. and I.R.C.J. performed LC-MS/MS analysis. D.A.M., K.D. and F.E.R. analyzed data and D.A.M. and F.E.R. wrote the manuscript with assistance from the other authors.
Author Information: Reprints and permissions information are available at www.nature.com/reprints. F.E.R. and D.A.M. have filed a patent application based on the use of NTTs for biotechnological applications. F.E.R., and D.A.M. have a financial interest (shares) in Synthorx Inc., a company that has commercial interests in the UBP. D.A.M. is currently employed by Synthorx, Inc. The other authors declare no competing financial interests.
References
- 1.Malyshev DA, et al. Efficient and sequence-independent replication of DNA containing a third base pair establishes a functional six-letter genetic alphabet. Proc. Natl. Acad. Sci. USA. 2012;109:12005–12010. doi: 10.1073/pnas.1205176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Z, Chen F, Alvarado JB, Benner SA. Amplification, mutation, and sequencing of a six-letter synthetic genetic system. J. Am. Chem. Soc. 2011;133:15105–15112. doi: 10.1021/ja204910n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamashige R, et al. Highly specific unnatural base pair systems as a third base pair for PCR amplification. Nucleic Acids Res. 2012;40:2793–2806. doi: 10.1093/nar/gkr1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seo YJ, Malyshev DA, Lavergne T, Ordoukhanian P, Romesberg FE. Site-specific labeling of DNA and RNA using an efficiently replicated and transcribed class of unnatural base pairs. J. Am. Chem. Soc. 2011;133:19878–19888. doi: 10.1021/ja207907d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seo YJ, Matsuda S, Romesberg FE. Transcription of an expanded genetic alphabet. J. Am. Chem. Soc. 2009;131:5046–5047. doi: 10.1021/ja9006996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betz K, et al. Structural insights into DNA replication without hydrogen bonds. J. Am. Chem. Soc. 2013;135:18637–18643. doi: 10.1021/ja409609j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betz K, et al. KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry. Nat. Chem. Biol. 2012;8:612–614. doi: 10.1038/nchembio.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y, Fa M, Tae EL, Schultz PG, Romesberg FE. Enzymatic phosphorylation of unnatural nucleosides. J. Am. Chem. Soc. 2002;124:14626–14630. doi: 10.1021/ja028050m. [DOI] [PubMed] [Google Scholar]
- 9.Yan H, Tsai MD. Nucleoside monophosphate kinases: structure, mechanism, and substrate specificity. Adv. Enzymol. Relat. Areas Mol. Biol. 1999;73:103–134. doi: 10.1002/9780470123195.ch4. [DOI] [PubMed] [Google Scholar]
- 10.Winkler HH, Neuhaus HE. Non-mitochondrial ATP transport. Trends Biochem. Sci. 1999;24:64–68. doi: 10.1016/s0968-0004(98)01334-6. [DOI] [PubMed] [Google Scholar]
- 11.Amiri H, Karlberg O, Andersson SG. Deep origin of plastid/parasite ATP/ADP translocases. J. Mol. Evol. 2003;56:137–150. doi: 10.1007/s00239-002-2387-0. [DOI] [PubMed] [Google Scholar]
- 12.Hatch TP, Al-Hossainy E, Silverman JA. Adenine nucleotide and lysine transport in Chlamydia psittaci. J. Bacteriol. 1982;150:662–670. doi: 10.1128/jb.150.2.662-670.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkler HH. Rickettsial permeability: an ADP-ATP transport system. J. Biol. Chem. 1976;251:389–396. [PubMed] [Google Scholar]
- 14.Horn M, Wagner M. Bacterial endosymbionts of free-living amoebae. J. Eukaryot. Microbiol. 2004;5:509–514. doi: 10.1111/j.1550-7408.2004.tb00278.x. [DOI] [PubMed] [Google Scholar]
- 15.Haferkamp I, et al. Tapping the nucleotide pool of the host: novel nucleotide carrier proteins of Protochlamydia amoebophila . Mol. Microbiol. 2006;60:1534–1545. doi: 10.1111/j.1365-2958.2006.05193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miroux B, Walker JE. Over-production of proteins in Escherichia coli : mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 17.Haferkamp I, Linka N. Functional expression and characterisation of membrane transport proteins. Plant Biol. 2012;14:675–690. doi: 10.1111/j.1438-8677.2012.00591.x. [DOI] [PubMed] [Google Scholar]
- 18.Ast M, et al. Diatom plastids depend on nucleotide import from the cytosol. Proc. Natl. Acad. Sci. USA. 2009;106:3621–3626. doi: 10.1073/pnas.0808862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavergne T, Malyshev DA, Romesberg FE. Major groove substituents and polymerase recognition of a class of predominantly hydrophobic unnatural base pairs. Chem. Eur. J. 2012;18:1231–1239. doi: 10.1002/chem.201102066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo YJ, Hwang GT, Ordoukhanian P, Romesberg FE. Optimization of an unnatural base pair toward natural-like replication. J. Am. Chem. Soc. 2009;131:3246–3252. doi: 10.1021/ja807853m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, et al. Natural-like replication of an unnatural base pair for the expansion of the genetic alphabet and biotechnology applications. J. Am. Chem. Soc. 2014;136:826–829. doi: 10.1021/ja408814g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomizawa J, Selzer G. Initiation of DNA synthesis in Escherichia coli. Annu. Rev. Biochem. 1979;48:999–1034. doi: 10.1146/annurev.bi.48.070179.005031. [DOI] [PubMed] [Google Scholar]
- 24.Allen JM, et al. Roles of DNA polymerase I in leading and lagging-strand replication defined by a high-resolution mutation footprint of ColE1 plasmid replication. Nucleic Acids Res. 2011;39:7020–7033. doi: 10.1093/nar/gkr157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashimoto H, et al. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature. 2014;506:391–395. doi: 10.1038/nature12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malyshev DA, Seo YJ, Ordoukhanian P, Romesberg FE. PCR with an expanded genetic alphabet. J. Am. Chem. Soc. 2009;131:14620–14621. doi: 10.1021/ja906186f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirao I, et al. An unnatural hydrophobic base pair system: site-specific incorporation of nucleotide analogs into DNA and RNA. Nat. Methods. 2006;3:729–735. doi: 10.1038/nmeth915. [DOI] [PubMed] [Google Scholar]
- 28.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 29.Quan J, Tian J. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat. Protoc. 2011;6:242–251. doi: 10.1038/nprot.2010.181. [DOI] [PubMed] [Google Scholar]
- 30.Ludwig J, Eckstein F. Rapid and efficient synthesis of nucleoside 5'-0-(1-thiotriphosphates), 5'-triphosphates and 2',3'-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. J. Org. Chem. 1989;54:631–635. [Google Scholar]
- 31.Alpert A, Shukla A. ABRF 2003: Translating biology using proteomics and functional genomics Poster no. P111-W. 2003 [Google Scholar]
- 32.Kubitschek HE, Friske JA. Determination of bacterial cell volume with the Coulter Counter. J. Bacteriol. 1986;168:1466–1467. doi: 10.1128/jb.168.3.1466-1467.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ausubel FM, et al. Current Protocols in Molecular Biology, Unit 1.8. New York: Wiley; 2013. [Google Scholar]
- 34.Yanes O, Tautenhahn R, Patti GJ, Siuzdak G. Expanding coverage of the metabolome for global metabolite profiling. Anal. Chem. 2011;83:2152–2161. doi: 10.1021/ac102981k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knab S, Mushak TM, Schmitz-Esser S, Horn M, Haferkamp I. Nucleotide parasitism by Simkania negevensis (Chlamydiae) J. Bacteriol. 2011;193:225–235. doi: 10.1128/JB.00919-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Audia JP, Winkler HH. Study of the five Rickettsia prowazekii proteins annotated as ATP/ADP translocases (Tlc): Only Tlc1 transports ATP/ADP, while Tlc4 and Tlc5 transport other ribonucleotides. J. Bacteriol. 2006;188:6261–6268. doi: 10.1128/JB.00371-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofer A, Ekanem JT, Thelander L. Allosteric regulation of Trypanosoma brucei ribonucleotide reductase studied in vitro and in vivo. J. Biol. Chem. 1998;273:34098–34104. doi: 10.1074/jbc.273.51.34098. [DOI] [PubMed] [Google Scholar]
- 38.Reijenga JC, Wes JH, van Dongen CAM. Comparison of methanol and perchloric acid extraction procedures for analysis of nucleotides by isotachophoresis. J. Chromatogr. B: Biomed. Sci. Appl. 1986;374:162–169. [Google Scholar]
- 39.Invitrogen. PureLink Quick Plasmid Miniprep kits, Publication Part Number 25-0789, MAN0003721. 2011 Apr 25; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.