Abstract
Cell and islet microencapsulation in synthetic hydrogels provide an immunoprotective and cell-supportive microenvironment. A microfluidic strategy for the genaration of biofunctionalized, synthetic microgel particles with precise control over particle size and molecular permeability for cell and protein delivery is presented. These engineered capsules support high cell viability and function of encapsulated human stem cells and islets.
Keywords: microfluidics, cell microencapsulation, synthetic microgel, pancreatic islets, hydrogels
Hydrogel microencapsulation of cells is a promising strategy for immunoprotection after transplantation. Since the development of alginate-poly-L-lysine encapsulation by Lim and Sun in 1980,[1] their approach has remained the standard for cell encapsulation, although major efforts have led to significant improvements.[2] The ease of alginate microencapsulation, along with alginate’s inherent biotolerance in vivo, have led to its prevalence,[3] even though the ability to control local cellular environment via incorporation of bioactive molecules (e.g., adhesive peptides) is limited. Highly tunable, synthetic hydrogel encapsulation is attractive for various regenerative medicine applications,[4, 5] not only for immunoisolation, but also for directing cell behavior and fate.[4] Several groups have developed more complex encapsulation configurations, such as cell encapsulation in natural hydrogel fibers,[6] but the benefits of added geometric complexity remain to be established. Minimization of encapsulation volume is also important in many regenerative medicine scenarios, including pancreatic islet transplantation. In an effort to reduce the high polydispersity present in electrostatically generated alginate droplets with diameters <200 µm,[7] microfluidic droplet generation has been explored.[8] Microfluidic devices have also been used to generate synthetic hydrogel particles.[9–11] Even for synthetic polymer encapsulation, control of cellular microenvironment by functionalization of polymers with bioactive molecules remains a significant challenge. Current synthetic polymer encapsulation strategies typically rely on cytotoxic crosslinking mechanisms such as UV- or thermal-based free radical polymerization. Here, we present a facile, modular microfluidics-based technology for the generation of microgels of controlled size and microencapsulation of clinically relevant cells, such as human pancreatic islets and human mesenchymal stem cells (hMSCs). Peptides and proteins are also easily incorporated into 4-arm PEG maleimide (PEG-4MAL) microgels, which allows for the presentation of a highly controlled cellular microenvironment. The PEG-4MAL hydrogel system has significant advantages over other hydrogel chemistries, such as well-defined hydrogel structure, facile and stoichiometric incorporation of bioligands, increased cytocompatibility, improved crosslinking efficiency, and tunable reaction rates.[12] Additionally, the PEG-4MAL macromer exhibits minimal toxicity and inflammation in vivo and is rapidly excreted via the urine,[13] which are important considerations in translating this material to in vivo applications. The crosslinking scheme for this system utilizes a Michael-type addition reaction, is cytocompatible, and does not rely on free radical polymerization. By varying device geometry and fluid flow rates, monodisperse microgels with a wide range of diameters can be produced. Kinetic studies of the release of microgel-encapsulated biomolecules demonstrate not only the immunoisolation potential of the microgels, but also the capability of tuning critical network parameters that cannot be easily tuned in natural polymers, such as macromer molecular weight, for rate-controlled release of peptide therapeutics. Importantly, the microencapsulation process does not affect the viability or function of human pancreatic islets and mesenchymal stem cells (hMSCs).
Pioneering studies by Thorsen demonstrated controlled generation of emulsions using T-junctions in microfluidic devices,[14] and the work of Stone established the ability to generate droplets using flow focusing microfluidic geometries.[15] Weitz established encapsulation of cells inside emulsions for high throughput cell-based assays. [16] Translating this work into covalently crosslinking of microgels within microfluidic devices adds significant complexity because polymer precursors must be liquid while flowing through the focusing nozzle, but droplets must crosslink rapidly after being generated to prevent them from merging. Recently, synthetic polymer microgels have been generated, including cell-laden microgels.[9, 10, 11, 17, 18] However, most of these schemes require crosslinking using UV-based free radical polymerization, resulting in potentially cytotoxic effects on encapsulated cells. Although cell encapsulation in synthetic microgels crosslinked without free radicals has been reported, the polymer cannot easily be functionalized with bioactive molecules.[9, 17] This major limitation makes the maintenance of cells requiring adhesive ligands for viability and function difficult. Recently, Lutolf devised an elegant microfluidic scheme to generate surface-modifiable synthetic microgels that does not utilize free radical polymerization, but neither bulk modification with bioactive molecules nor cell encapsulation was shown.[11] Microfluidic encapsulation of large clusters of cells, such as human islets, is more challenging than single cell encapsulation, because the larger particles tend to clog microfluidic channels. For these reasons, synthetic polymer microencapsulation of islets using microfluidics has not been shown. To minimize encapsulation volume while avoiding microfluidics altogether, investigators have explored conformal coating of islets.[19] Whereas conformal coating minimizes transplant volume, the immunoisolation potential of such thin polymer membranes remains unknown.
To address these limitations, we generated a strategy (Figure 1) to produce cell- and cell aggregate-laden synthetic PEG-4MAL-based microgels, functionalized with cell adhesive peptides, by producing droplets using a flow focusing microfluidic device and subsequently covalently crosslinking the droplets with the small molecule dithiothreitol (DTT). Three independent flows of (1) mineral oil containing SPAN80 (a surfactant), (2) a crosslinker phase containing mineral oil and SPAN80 with an emulsion of aqueous DTT solution, and (3) PEG-4MAL macromer in aqueous physiological buffer were pumped into the microfluidic chip using syringe pumps. As the macromer phase approached the flow-focusing nozzle, a co-flowing continuous phase of oil shielded the macromer from contact with the crosslinker-laden oil phase. Because crosslinker could not reach the macromer before flow instability occurred, monodisperse, spherical droplets were formed. The crosslinker then rapidly diffused into droplets, covalently crosslinking the PEG-4MAL macromer into the microgel network via Michael-type addition reaction of the maleimide groups on the macromer and thiols on the crosslinker. The PEG-4MAL hydrogel platform used for this system is easily modified with thiol-containing molecules, including cysteine-containing adhesive ligands and growth factors, due to the high specificity of the maleimide groups for thiols at physiological pH. This Michael-type addition reaction requires no free radicals and is cytocompatible.[12] Furthermore, fast reaction kinetics render this hydrogel ideal for microfluidic encapsulation, allowing for short residence time on chip, and minimizing cell stress.
Figure 1.
PEG-4MAL for microencapsulation of cells and proteins in a flow-focusing microfluidic chip using a cytocompatible crosslinking reaction. (a) PEG-4MAL macromer consists of a 4-arm branched PEG backbone modified with a maleimide group terminating each arm. At physiological pH, free thiol-containing molecules undergo a Michael-type addition reaction with maleimides, forming a covalent bond to macromer. This reaction is facilitated by nucleophilic buffers such as triethanolamine (TEA), and can be used to either functionalize the macromer or crosslink macromer into a hydrogel network. (b) A microfluidic device with flow focusing geometry is utilized to produce microgels. A co-flowing oil phase shields an aqueous macromer solution, containing cells and/or proteins, from the crosslinker-containing oil phase as the macromer solution approaches the flow focusing nozzle. After droplet formation, the DTT emulsion rapidly crosslinks macromer solution into cell- or protein-laden microgels.
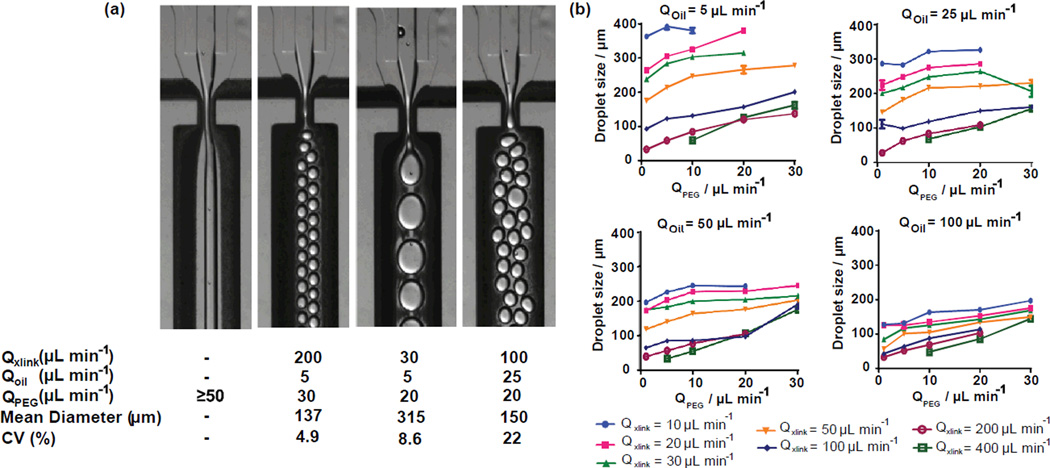
Precise control of particle size and monodispersity are critical for many applications of microgels, and the microfluidic platform affords this control over a wide range of particle sizes. We varied macromer solution and continuous phase flow rates for a device with a 300 µm nozzle, and measured the corresponding droplet size for each flow rate (Figure 2). No cells were encapsulated in this application. Microgels with a wide range of sizes, ranging from 20 to 400 µm, could be produced on the same device; however, several flow regimes produced microgels with undesirable polydisperse distributions (coefficient of variation, CV>10%). Importantly, flow rate combinations were identified that produced a range of microgel sizes from 135 – 325 µm with monodisperse populations (CV< 5%). An example of one of these flow rates is shown in figure 2, along with several other representative flow regimes, including one regime that does not produce droplets and one that produces a very polydisperse (CV = 22%) microgel population. Although a device with fixed geometry is capable of producing a wide range of particle sizes, droplets should be generated with diameters that are 50–100% of the nozzle width to obtain a monodisperse population. Even if polydisperse populations are acceptable, device throughout is limited, because no droplets were formed for any PEG-4MAL macromer flow rates exceeding 50 µL min−1. If monodisperse populations of microgels are required that are outside the 135 – 325 µm range, the microfluidic device nozzle can be scaled up or down so that nozzle is roughly equal to the desired microgel size.
Figure 2.
Microgel size and polydispersity can be controlled by altering macromer solution and continuous phase flow rates. (a) Representative images and quantification of microgel diameters are shown for varied flow rates. (l-r) No droplets are produced for PEG-4MAL flow rates of 50 µL min−1 or greater. Monodisperse (CV < 5%) populations can be generated for a range of sizes, one of which is shown here. Polydispersity is driven by a complex combination of factors as seen in the final 2 images. (b) Microgel diameter was measured for fluid flow rates that were varied for all combinations of: QPEG (µL min−1) = 1, 5, 10, 20, 30; Qoil (µL min−1) = 5, 25, 50, 100; Qxlink (µL min−1) = 10, 20, 30, 50, 100, 200, 400. Mean and standard error were plotted as calculated from a minimum of 30 measurements for each condition.
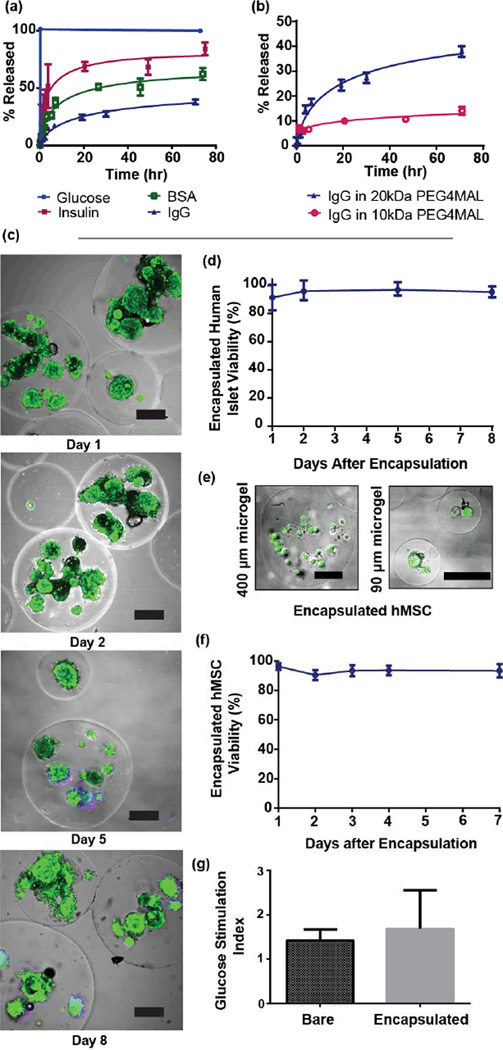
A tunable hydrogel network with selective permeability to biomolecules is essential for cell microencapsulation, because antibodies and immune cells (relatively large objects) must be prevented from reaching encapsulated cells, while nutrients, signaling molecules, and waste (relatively small molecules) must be easily transported across the microgel capsule. Therefore, we investigated the suitability of our microgels for biomolecule release and cell encapsulation by measuring their permeability to relevant molecules of various sizes that were labeled with fluorescent tags. These molecules were encapsulated within microgels generated from a 20 kDa PEG-4MAL macromer, and the rate of their release into buffer was used as a metric of permeability (Figure 3a). 2-NBD-glucose (342 Da) was rapidly released from the gels, fully equilibrating concentration with the buffer by the first fluorescence measurement, 5 minutes after swelling. Similarly, insulin-AlexaFluor488 (5.8 kDa) was rapidly released from the microgels upon swelling, indicating that relevant functional molecules diffuse quickly through the microgel. In contrast, encapsulated IgG-AF488 (~160 kDa) was released from the microgels at a slow rate, suggesting that the microgel capsules are capable of preventing transport and binding of antibodies to encapsulated cells. Release kinetics for BSA (66.5 kDa) fell between IgG and insulin, suggesting that physical molecular entanglement due to the network structure is the determining factor for permeability in our hydrogel network. These results show nearly 100% release for the smaller molecules glucose and insulin. For the larger proteins BSA and IgG, the release did not reach 50% of the incorporated amount because these larger molecules remain trapped within the tight network structure of the PEG hydrogel. These results for reduced transport and entrapment of IgG support the use of these materials for immuno-encapsulation applications.
Figure 3.
PEG-4MAL microgels exhibit selective permeability to biomolecules and retain viability and function of encapsulated cells. (a) Release kinetics for biomolecules of varying size from microgels (made with 20 kDa macromers) demonstrate selective permeability. IgG was released from microgels slowly and incompletely. Conversely, glucose and insulin were rapidly released, indicating that mass transport of these smaller, function-preserving molecules is not grossly limited. (b) IgG was encapsulated in PEG-4MAL prepared from macromers of 10 kDa or 20 kDa, and the tighter network structure generated with smaller macromer decreased permeability of microgel to IgG. (c) Human islets maintain high viability in culture after encapsulation. On days 1, 2, 5, and 8 after microencapsulation, viability of human pancreatic islets was imaged (c) (scale bars = 200µm) and quantified (d) using fluorescent area ratios between TOTO-3 iodide (dead, purple) and calcein AM (live, green). (e) Human MSCs were encapsulated in microgels of various sizes (scale bars = 100 µm), and viability of hMSCs encapsulated in 400 µm microgels was quantified for 7 days post encapsulation. (f) No significant loss in viability was noted for hMSCs. (g) A glucose-stimulated insulin secretion assay, performed one day after human islet encapsulation, shows no significant difference between bare and encapsulated islets, demonstrating no functional losses in microencapsulated cells.
As a demonstration of the ability to tune network structure for protein release applications, we encapsulated IgG in microgels made with PEG macromers of different (10 kDa vs. 20 kDa) molecular weights (Figure 3b). As expected, the tighter network mesh of the microgels based on 10 kDa macromers slowed the release of IgG compared to microgels made with 20 kDa macromer. Because altering macromer size results in drastic changes in release kinetics, we expect that other parameters that influence network structure as related to the hydrogel correlation length, such as polymer weight % and crosslinking density, can be systematically varied to obtain desired release kinetics. Flexibility in protein encapsulation, as well as the ability to simultaneously control therapeutic release kinetics and particle size, render this encapsulation platform ideal for a wide range of protein delivery applications.
Having shown the ability to exclude high molecular weight proteins such as IgG with minimal impact on the transport of critical molecules such as glucose and insulin, we next examined cell encapsulation applications of this microgel system using clinically relevant cell types. Human pancreatic islets were encapsulated with high efficiency (>99% of islets loaded into microfluidic device were encapsulated, and 80% of microgels produced contain at least one islet), in microgels made from PEG-4MAL, a polymer that has been shown to support islet engraftment and function.[13] Encapsulated islets were maintained in culture for 8 days with no decrease in viability (Figure 3c,d), demonstrating the capacity of this synthetic hydrogel network to support high viability of these sensitive human cells. This result also shows that any potentially cytotoxic effects of hydrogel precursor constituents (e.g., DTT) prior to network formation are mitigated by the short residence time of cells in crosslinker emulsion. The device used for islet encapsulation was scaled to have a 600 µm nozzle, and produced microgels from 300–800 µm in diameter. As a further demonstration of the versatility of this platform, human mesenchymal stem cells (hMSCs), currently under investigation for various biomedical applications due to their regenerative and immunomodulatory properties, were encapsulated in PEG-4MAL microgels of either 400 µm or 90 µm diameter. These microgels were precisely functionalized with a cell adhesive RGD peptide (2.0 mM) by simply reacting maleimide groups in the macromer with this peptide prior to cell encapsulation and hydrogel crosslinking. This RGD peptide supports cell adhesion, survival and function when incorporated into the PEG-4MAL network.[12] After encapsulation, hMSCs encapsulated in both microgel sizes exhibited high viability (Figure 3e), and hMSCs in 400 µm diameter microgels were maintained in suspension culture for 7 days with no loss in viability (Figure 3f). Therefore, controlled presentation of adhesive peptides to cells encapsulated using a cytocompatible crosslinking reaction provides an environment amenable to long-term cell viability. Such a microenvironment presenting defined bioactive peptides may be suitable not only for cell encapsulation and delivery,[20] but also for directing stem cell behavior and fate.[21] Additionally, control of microgel size facilitates optimization for cell delivery applications.
A crucial consideration in the engineering of microgels for cell encapsulation is that critical cell functions are not negatively impacted following encapsulation. To this end, we performed a glucose-stimulated insulin secretion (GSIS) assay to evaluate the function of encapsulated human islets. Bare or encapsulated islets were challenged with either 1.67 mM or 16.7 mM glucose for 30 minutes, and the normalized insulin content from each group was assayed using ELISA. The stimulation index (SI), or ratio of normalized insulin secreted in high glucose group to that of low glucose group, was calculated for both bare and encapsulated islets. No significant difference was found between the groups (Figure 3f), demonstrating that microfluidic-based encapsulation in PEG-4MAL has no deleterious effects on human islet function or viability, and that mass transfer of molecules relevant to islet function is not significantly affected by microencapsulation.
The high potential of synthetic hydrogel microencapsulation for cell and protein therapeutics has been limited by the lack of synthetic polymer systems with tunable capsule size, cytocompatible crosslinking reactions, rapid crosslinking rates, adequate biomolecule permeability, and ease of functionalization with bioactive molecules (e.g., adhesive peptides). Using a synthetic hydrogel system with tunable network and crosslinking characteristics and a microfluidics encapsulation platform, we have created an integrated and robust strategy for microencapsulation of cells in which we can control capsule size and local cellular microenvironment. Additionally, microgel network structure can be tuned to optimize permeability of the capsule to molecules of various sizes. We have demonstrated proof of concept with two different clinically relevant human cell types, but the versatility of this strategy will allow it to be tailored to fit diverse engineering applications.
Experimental Section
Microfluidic device preparation
PDMS microfluidic flow focusing devices were cast using soft lithography from silicon and SU8 masters that were fabricated by the Stanford Microfluidics Foundry. Devices with 300 µm nozzles were bonded directly to glass slides after treatment with air plasma. 600 µm nozzle devices were manufactured by first bonding mirror-image PDMS channels, each with 300 µm depth, together to create a channel with 600 µm depth.
PEG-4MAL microgel formation and particle encapsulation
Flow-focusing microfluidic geometry was utilized to form polymer droplets. Both shielding and crosslinker phases consisted of light mineral oil (Sigma) with 2% SPAN80 (Sigma). The crosslinker phase also contained an emulsion, at a ratio of 1:15, of 20 mg/mL dithiothreitol (DTT) (Sigma) in PBS. A co-flowing shielding phase protected the macromer solution – a 5% PEG-4MAL (10 kDa or 20 kDa, Laysan Bio) solution containing molecules or cells to be encapsulated – from the crosslinker phase until droplets of the macromer solution were formed. DTT rapidly diffused into macromer droplets, forming crosslinked microgels. To functionalize hydrogel with GRGDSPC (‘RGD’, AAPPTec), macromer was reacted for 20 minutes before encapsulation with 2.0 mM RGD in buffer solution containing 4 mM triethanolamine (Sigma). After formation, microgels were washed 5 times by centrifugation to remove mineral oil and excess DTT.
Microgel size control
To characterize the relationship between microgel size and the various macromer solution and continuous phase flow rates, hydrogel droplets were generated using computer-controlled syringe pumps, and were measured while still in the microfluidic chip. Harvard Apparatus Elite syringe pumps were computer controlled using FlowControl software to pump inlet solutions at various flow rates. Video was recorded during droplet generation using a Hammamatsu ORCA-ERA 1394 camera connected to a Nikon TE300 microscope. Droplet diameter was measured using ImageJ analysis software. The coefficient of variation (CV) was calculated for each flow rate combination by dividing the standard deviation of the sample by its mean. At least 30 microgels were measured for each flow rate combination.
Protein encapsulation and release
AlexaFluor488-labeled IgG (goat anti-rabbit IgG, Life Technologies), bovine serum albumin-AlexaFluor488 conjugate (Life Technologies), 2-NBD-glucose (Life Technologies) or insulin (Sigma) tagged with AlexaFluor488 was dissolved in a 5% PEG-4MAL (10 kDa or 20 kDa) solution before being microencapsulated by macromer droplet gelation. To prevent proteins from being crosslinked by the macromer, thiols were capped using aminoethylate reagent (Thermo Scientific) according to product instructions. Particles were washed and resuspended in PBS and divided into 5 replicates containing 2 mL total volume. 50 µL samples were taken of supernatant alone, as well as of supernatant containing well-mixed, protein-laden microgels. These samples were placed in a 96 well plate, and their fluorescent intensity was measured using a Perkin Elmer HTS 7000 plate reader. To generate release curves, supernatant samples were collected over the course of 3 days, and their fluorescent intensity was measured. Protein release was normalized by setting fluorescent intensity of the supernatant alone correspond to 0% protein released, and fluorescent intensity of the buffer/microgel mixture correspond to 100% protein released. This data was plotted using GraphPad Prism, and exponential best fit curves were calculated from normalized data.
Human MSC encapsulation and viability assay
Passage 3 hMSCs (Texas A&M Health Science Center College of Medicine) were trypsinized and washed 3 times with PBS before being suspended in RGD-functionalized macromer solution (5% wt macromer) at a concentration of 5 × 106 cells/mL. Generation and subsequent gelation of cell-laden macromer solution droplets, using a microfluidic device with a 300 µm nozzle, resulted in microencapsulated hMSCs. These cells were maintained under static culture conditions in chemically defined MSC media (Lonza) for 7 days, with media changes every 2 days. On days 1, 2, 3, 4, and 7, microencapsulated cells were removed from culture, stained with Calcein AM and TOTO-3 iodide (Life Technologies) for 15 minutes, washed, and resuspended in fresh media. At least 200 cells were imaged each day using a Nikon Eclipse Ti microscope, and their viabilities were assessed based on fluorescent signal. ANOVA analysis was performed using GraphPad Prism software. The percent viability was calculated by taking the ratio of live cells to total cells. Viability data was plotted using GraphPad Prism. ANOVA analysis between the groups found no significant difference in viability, and a student’s t-test between days 1 and 7 also found no significant difference in viability.
Human islet encapsulation and in vitro characterization
Human pancreatic islets (PRODO Laboratories and the Integrated Islet Distribution Program) were suspended at a concentration of 2 × 104 IEQ/mL in culture media containing 5% (w/v) macromer. A microfluidic device with a 600 µm nozzle was used for droplet generation and subsequent crosslinking of the macromer solution, resulting in microencapsulated islets. After microencapsulation, islets were washed 5 times with media (PRODO labs PIM(S)), placed in fresh media, and allowed to recover overnight. On days 1, 2, 5, and 8 after encapsulation, islets were stained with Calcein AM and TOTO-3 iodide (Life Technologies) for 15 minutes, washed, and resuspended in fresh media. At least 74 islets were imaged each day using a Nikon Eclipse Ti microscope, and their viabilities were assessed based on fluorescent signal. For each islet, dead cell area to total islet area was computed, and this fraction was subtracted from 100% to obtain percent viability. ANOVA analysis was performed using GraphPad Prism software, and no significant difference in viability was found. On day 1 following encapsulation, a glucose-stimulated insulin secretion assay was performed. Islets were washed and were equilibrated using 1.67 mM glucose in Hanks Buffered Salt Solution for 30 minutes. Two groups, containing 5 replicates of approximately 10 islets each, were collected from both microencapsulated and bare islets. One group from each treatment was incubated with high (16.7 mM) glucose HBSS, and the other group was incubated with low (1.67 mM) glucose HBSS for one hour. Supernatant from each sample was collected, and insulin content was quantified using human insulin ELISA (Sigma). DNA from each sample was then quantified using a Quant-iT PicoGreen kit (Invitrogen). Insulin secretion was normalized to DNA content for each well. The Stimulation Index for each replicate was calculated by taking the ratio of normalized high glucose insulin secretion to normalized low glucose insulin secretion (n=5). Groups were compared using a student’s t-test in GraphPad Prism. Human islets and MSCs were obtained by third party distributors, and consent was provided by donors or next of kin.
Acknowledgements
Funding provided by the Juvenile Diabetes Research Foundation (17-2013-277), the National Institutes of Health (DP3 DK094346, P20 HL113451), and the National Science Foundation-sponsored Stem Cell Biomanufacturing IGERT (NSF DGE 0965945). The authors thank E.A. Phelps for helpful suggestions. Human islets were provided by the Integrated Islet Distribution Program and PRODO Laboratories. hMSCs were provided by the Texas A&M Health Science Center College of Medicine Institute for Regenerative Medicine at Scott & White through a grant from NCRR of the NIH (P40RR017447).
References
- 1.Lim F, Sun AM. Science. 1980;210:908. doi: 10.1126/science.6776628. [DOI] [PubMed] [Google Scholar]
- 2.Wang T, Lacik I, Brissova M, Anilkumar AV, Prokop A, Hunkeler D, Green R, Shahrokhi K, Powers AC. Nature Biotechnology. 1997;15:358. doi: 10.1038/nbt0497-358. [DOI] [PubMed] [Google Scholar]
- 3.Santos E, Pedraz JL, Hernandez RM, Orive G. J. Control. Release. 2013;170:1. doi: 10.1016/j.jconrel.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 4.Lutolf MP, Hubbell JA. Nat Biotechnol. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 5.Peppas NA. Langer, Science. 1994;263:1715. doi: 10.1126/science.8134835. [DOI] [PubMed] [Google Scholar]; Langer R, Tirrell DA. Nature. 2004;428:487. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- 6.a) Onoe H, Okitsu T, Itou A, Kato-Negishi M, Gojo R, Kiriya D, Sato K, Miura S, Iwanaga S, Kuribayashi-Shigetomi K, Matsunaga YT, Shimoyama Y, Takeuchi S. Nature Materials. 2013;12:584. doi: 10.1038/nmat3606. [DOI] [PubMed] [Google Scholar]; b) Jun Y, Kim MJ, Hwang YH, Jeon EA, Kang AR, Lee SH, Lee DY. Biomaterials. 2013;34:8122. doi: 10.1016/j.biomaterials.2013.07.079. [DOI] [PubMed] [Google Scholar]
- 7.Goosen MFA, AlGhafri AS, ElMardi O, AlBelushi MIJ, AlHajri HA, Mahmoud ESE, Consolacion EC. Biotechnology Progress. 1997;13:497. [Google Scholar]
- 8.a) Choi C-H, Jung J-H, Rhee Y, Kim D-P, Shim S-E, Lee C-S. Biomed Microdevices. 2007;9:855. doi: 10.1007/s10544-007-9098-7. [DOI] [PubMed] [Google Scholar]; b) Tan WH, Takeuchi S. Advanced Materials. 2007;19:2696. [Google Scholar]; c) Um E, Lee D-S, Pyo H-B, Park J-K. Microfluid Nanofluid. 2008;5:541. [Google Scholar]
- 9.Rossow T, Heyman JA, Ehrlicher AJ, Langhoff A, Weitz DA, Haag R, Seiffert S. Journal of the American Chemical Society. 2012;134:4983. doi: 10.1021/ja300460p. [DOI] [PubMed] [Google Scholar]
- 10.Velasco D, Tumarkin E, Kumacheva E. Small. 2012;8:1633. doi: 10.1002/smll.201102464. [DOI] [PubMed] [Google Scholar]
- 11.a) Allazetta S, Hausherr TC, Lutolf MP. Biomacromolecules. 2013;14:1122. doi: 10.1021/bm4000162. [DOI] [PubMed] [Google Scholar]; b) Panda P, Ali S, Lo E, Chung BG, Hatton TA, Khademhosseini A, Doyle PS. Lab Chip. 2008;8:1056. doi: 10.1039/b804234a. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chung SE, Park W, Park H, Yu K, Park N, Kwon S. Applied Physics Letters. 2007;91:041106. [Google Scholar]
- 12.Phelps EA, Enemchukwu NO, Fiore VF, Sy JC, Murthy N, Sulchek TA, Barker TH, Garcia AJ. Advanced Materials. 2012;24:64. doi: 10.1002/adma.201103574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phelps EA, Headen DM, Taylor WR, Thule PM, Garcia AJ. Biomaterial. 2013;34:4602. doi: 10.1016/j.biomaterials.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorsen T, Roberts RW, Arnold FH, Quake SR. Physical review letters. 2001;86:4163. doi: 10.1103/PhysRevLett.86.4163. [DOI] [PubMed] [Google Scholar]
- 15.a) Anna SL, Bontoux N, Stone HA. Applied Physics Letters. 2003;82:364. [Google Scholar]; b) Wan J, Shi L, Benson B, Bruzek MJ, Anthony JE, Sinko PJ, Prudhomme RK, Stone HA. Langmuir. 2012;28:13143. doi: 10.1021/la3025952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koster S, Angile FE, Duan H, Agresti JJ, Wintner A, Schmitz C, Rowat AC, Merten CA, Pisignano D, Griffiths AD, Weitz DA. Lab Chip. 2008;8:1110. doi: 10.1039/b802941e. [DOI] [PubMed] [Google Scholar]
- 17.Kesselman LRB, Shinwary S, Selvaganapathy PR, Hoare T. Small. 2012;8:1092. doi: 10.1002/smll.201102113. [DOI] [PubMed] [Google Scholar]
- 18.Tumarkin E, Kumacheva E. Chemical Society Reviews. 2009;38:2161. doi: 10.1039/b809915b. [DOI] [PubMed] [Google Scholar]
- 19.a) Blasi P, Luca G, Mancuso F, Schoubben A, Calvitti M, Giovagnoli S, Basta G, Becchetti E, Ricci M, Calafiore R. International Journal of Pharmaceutics. 2013;440:141. doi: 10.1016/j.ijpharm.2012.10.010. [DOI] [PubMed] [Google Scholar]; b) Teramura Y, Oommen OP, Olerud J, Hilborn J, Nilsson B. Biomaterials. 2013;34:2683. doi: 10.1016/j.biomaterials.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 20.Ranganath Sudhir H, Levy O, Inamdar Maneesha S, Karp Jeffrey M. Cell Stem Cell. 2012;10:244. doi: 10.1016/j.stem.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.a) Lutolf MP, Gilbert PM, Blau HM. Nature. 2009;462:433. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lienemann PS, Karlsson M, Sala A, Wischhusen HM, Weber FE, Zimmermann R, Weber W, Lutolf MP, Ehrbar M. Advanced Healthcare Materials. 2013;2:292. doi: 10.1002/adhm.201200280. [DOI] [PubMed] [Google Scholar]