Abstract
Chromatin immunoprecipitation coupled with ultra high-throughput sequencing (ChIP-seq) is a widely used method for mapping the interactions of proteins with DNA. However, the requirements for ChIP-grade antibody impedes wider application of this method and variations can be high due to differences in affinity and cross-reactivity of antibodies. Therefore, we developed Chromatin Tandem Affinity Purification (ChTAP) as an effective alternative. Through the use of identical affinity reagents, ChTAP enables the direct comparison of binding between different transcription factors and the direct assessment of background in control experiments. Thus, ChTAP can be used to rapidly map the genome-wide binding of multiple DNA-binding proteins in a wide range of cell types. Notably, performing ChTAP with matching affinity controls has demonstrated that the use of affinity-based control is superior to input DNA to assess background noise. ChTAP takes three to four days for completion starting from cross-linking of chromatin to purification of ChIP DNA.
Keywords: Chromatin immunoprecipitation, Tandem affinity purification, ChIP-seq, Next generation sequencing, data analysis
INTRODUCTION
Approaches to mapping transcription factor binding sites generally involve cross-linking of DNA–protein complexes followed by immunoprecipitation of DNA-bound protein with a ChIP-grade antibody. Analysis of enriched genomic sites is then carried out by various means, such as quantitative PCR (qPCR) using locus-specific primers on the purified ChIP DNA, hybridization to microarray 1–3, high-throughput sequencing of ChIP DNA (ChIP-seq) 4–7 or chromatin interaction analysis using paired-end tag sequencing (Chia-PET) 8,9.
A significant limitation of these approaches to study many DNA-binding proteins and chromatin modifiers has been the lack of ChIP-grade antibodies. Additionally, the presence of various isoform of the same protein recognized by the antibody can also complicate the analysis of its binding sites 10. Cross-comparison of multiple ChIP-seq datasets can also be complicated by differences in affinity and cross-reactivity of different antibodies. We have used chromatin tandem affinity purification (ChTAP) to compare chromatin binding of Pax3 to that of Pax7 11and that of MyoD to Snail1 11. Here, we detail the ChTAP-seq protocol, that we employed to comparatively assess binding between transcription factors using identical affinity reagents in all experimental and control conditions 11,12.
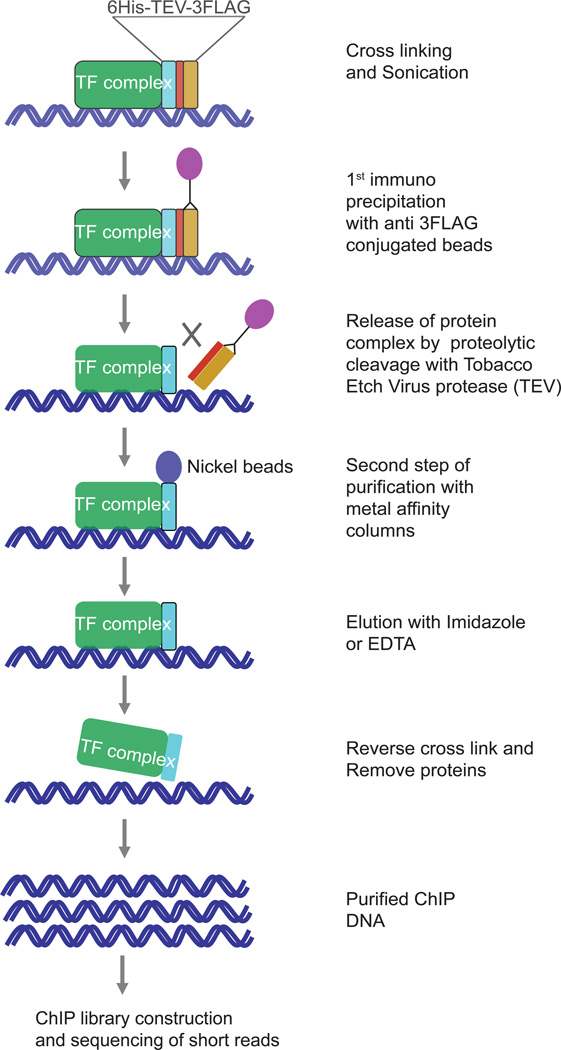
ChTAP combines tandem affinity purification (TAP, Fig. 1) — a widely used technique for protein purification 13–16 — with ultra high-throughput sequencing to map the global targets of any DNA-binding protein. We have used this technique to map genome-wide targets of diverse transcription factors and chromatin modifiers in muscle cells and fibroblasts 11,12 as well as in mouse embryonic stem cells (Soleimani et al., unpublished results). A similar approach can be used to create numerous other stable cell lines expressing TAP-tagged proteins. Therefore, ChTAP can be used to study a wide range of cells types, including primary cells derived from various tissues and organs.
Figure 1. Chromatin Tandem Affinity Purification.
The protein of interest is expressed as a C-terminal, TAP-tagged fusion protein at near-physiological levels. Cells stably expressing the fusion protein are subjected to a process by which proteins attached to chromatin are cross-linked to DNA by formaldehyde (using a 1%(vol/vol) formaldehyde solution in 1X PBS). After cell lysis and chromatin shearing, the DNA-bound protein complex of interest is purified using anti FLAG antibody conjugated to agarose beads. Following three sequential washes, the complex is eluted from the beads by either 3XFLAG peptide competition or proteolytic cleavage with tobacco etch virus (TEV) protease. The eluted material is used for a second step of purification using nickel affinity gel. Further contaminants are removed by three washes and chromatin is eluted from the beads with imidazole. The protein–DNA complex is dissociated by reverse cross-linking and DNA is purified by phenol/chloroform extraction.
ChTAP bypasses the requirement for a ChIP-grade antibody, which is a major problem for the study of many DNA-binding proteins. It also allows for a quantitative assessment of background noise due to the use of identical affinity reagents in the control and ChIP experiment. A direct comparison between ChTAP and traditional endogenous ChIP with a ChIP-grade MyoD antibody performed to identify MyoD binding sites showed that ChTAP could robustly identify bona fide MyoD binding sites as efficiently as the ChIP-grade antibody 11. Importantly, ChTAP robustly identified genome-wide binding sites of Snai1, for which no commercially available ChIP-grade antibody exists 12.
Although ChTAP can be routinely applied to any cells, such as transformed cells and cells derived from various tissues and organs, its application to tissue samples and cells within the endogenous niche is limited. Furthermore, insertion of a TAP tag has the potential to alter the biological function of some proteins. This possibility requires that additional assays for functional validation of the tagged protein are performed, a need that should be taken into consideration when deciding whether or not to implement this protocol. The procedure takes three to four days for completion starting from cross-linking of chromatin of the cells to the purification of ChIP DNA.
Protocol workflow
Generation of TAP-tagged constructs
An important consideration in the ChTAP protocol is to aim for near-physiological levels of TAP-tagged protein expression by retroviral-mediated integration of TAP-tagged expression cassette with low multiplicity of infection (MOI) to minimize multiple retroviral integrations into target cells. In this protocol, the full-length Open Reading Frame (ORF) of the gene of interest is fused in-frame with TAP tag (6xHIS-TEV-3FLAG) sequence at its multiple cloning site (MCS) as described previously 12. We produced a GateWay compatible version of this vector by inserting a GateWay reading frame RfC.1 at SalI site at MCS using GateWay conversion kit (Invitrogen). By incorporating the GateWay cloning strategy into ChTAP-seq pipeline, we have streamlined this protocol for high-throughput data acquisition and analysis.
Validation assay for TAP-tagged fusion construct
Studies published in 2003 on the global analyses of yeast proteome by protein tagging have shown that almost 80% of proteins tagged at their C-terminus retain their function and correct subcellular localization. However, a minority of proteins may require an intact C-terminus for correct localization and biological function17,18. Although we have demonstrated that the C-terminal TAP tag did not interfere with the biological function of any of the myogenic factors and chromatin modifiers we studied previously11,14, we recommend that an appropriate functional validation assay be carried out for any new TAP-tagged fusion construct as a first step in the ChTAP-seq pipeline. The validation steps include Western blot analysis for the expression of the TAP tagged construct using an antibody against FLAG epitope, DNA binding of the tagged protein with electrophoretic shift assay (EMSA) and correct subcellular localization as described previously 11,12.
Generation of stable cell lines expressing TAP tagged protein
To generate a stable cell line expressing the TAP-tagged construct of interest, retroviral vectors harboring the gene of interest fused with a C-terminal TAP-tag (6HIS-TEV-3FLAG) 11,14 are created. In our studies, retroviral particles were generated by transfection of Phoenix helper-free retrovirus producer cell line (a kind gift from Dr. Garry Nolan, http://www.stanford.edu/group/nolan/retroviral_systems/phx.html), also available from ATCC as described previously 11,14. Other vectors such as lenti- and adeno-viral systems can be equally useful in the production of stable cell lines expressing the TAP-tagged protein. In addition, tetracycline-inducible vectors or other alternatives can be utilized in ChTAP pipeline for the temporally controlled expression of the protein of interest.
ChTAP
The next stage of the protocol involves the implementation of ChTAP, which is a derivative of TAP purification, first applied to the identification of yeast protein complexes 13,15 and later to the study of diverse organisms14,19,20. TAP was designed for two primary reasons: Firstly, to eliminate the requirement for a specific antibody for immunoprecipitation and, secondly, to purify protein complexes to near-homogeneity through sequential immunoprecipitations with two independent epitopes. Both of these qualities also apply to ChTAP.
Direct comparison of ChTAP with ChIP
To undertake a direct comparison between ChTAP with TAP tag purification and ChIP with endogenous antibody, we first identified genome-wide target sites of key myogenic factors such as Pax3, Pax7, Myf5 and MyoD and others using ChTAP-seq 11,12. We selected the MyoD dataset for direct comparison due to the commercial availability of a ChIP-grade MyoD antibody. Furthermore, genome-wide MyoD binding sites using an endogenous MyoD antibody have been previously reported 21,22.
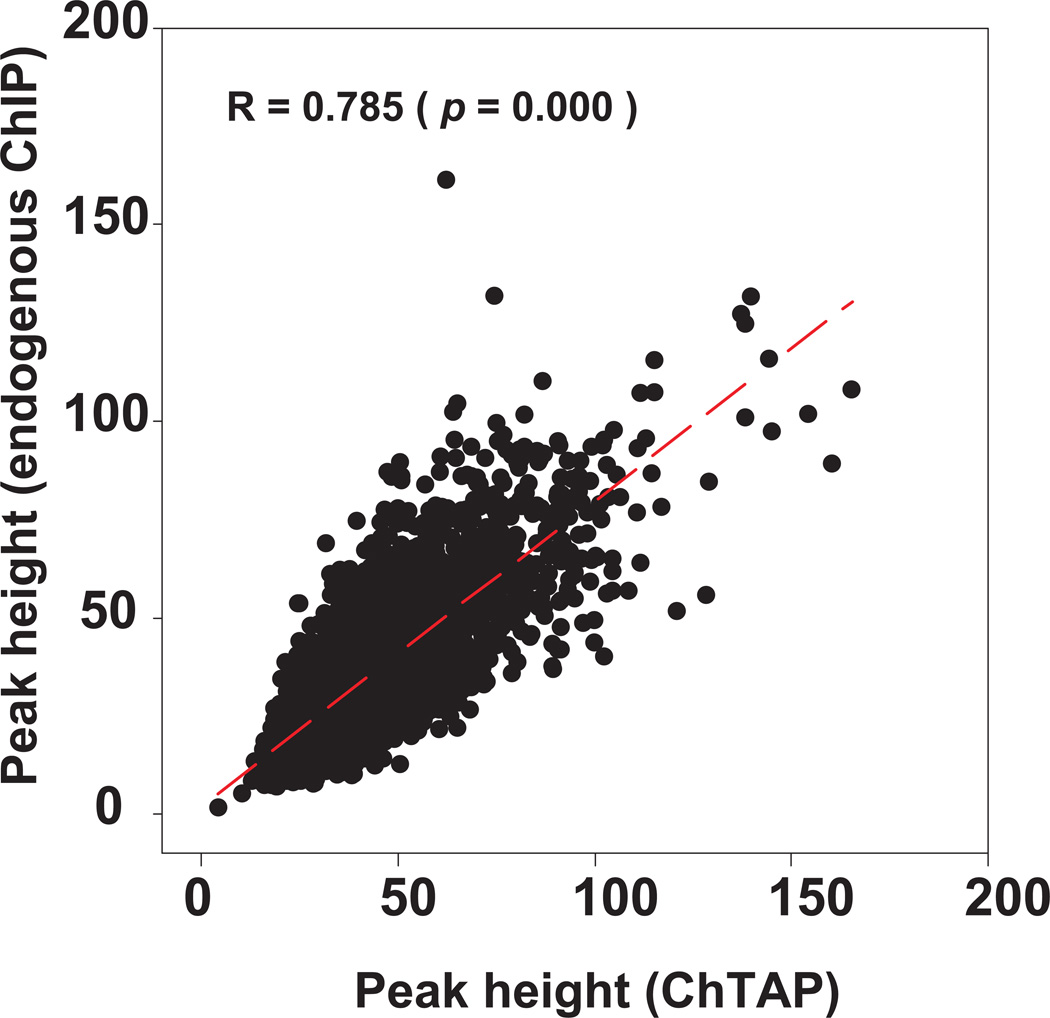
We selected 20 random sites from the MyoD ChTAP-seq dataset in primary myotubes 12 and performed quantitative ChIP PCR (qPCR) analysis of these target sites using an antibody against the endogenous MyoD. This analysis revealed that ChTAP is a robust method for identification of bona fide MyoD binding sites12. Furthermore, in a direct comparison between ChTAP-qPCR (i.e., TAP purification of chromatin) and ChIP-qPCR with the endogenous MyoD antibody, our data shows that ChTAP-qPCR is as efficient as ChIP-qPCR with a ChIP-grade antibody 12. Finally, a direct correlation analysis for tag enrichment on ~5,800 common sites between our MyoD ChTAP-seq in primary myotubes and an independent MyoD ChIP-seq in C2C12 myotubes 21 showed a significant positive correlation (Fig. 2). Together, these results indicate that ChTAP can applied for robust identification of bona fide binding site of any DNA binding protein without the requirement for a ChIP-grade antibody.
Figure 2. Direct comparison between endogenous ChIP-seq and ChTAP-seq.
There is a significant positive correlation for peak height for the ~5,800 common binding sites between endogenous ChIP-seq in C2C12 myotubes 21 and MyoD ChTAP-seq MyoD in primary myotubes 12. The figure shows a correlation analysis for the enrichment of sequence reads from an endogenous MyoD ChIP-seq and ChTAP-seq on the common binding sites between the two independent datasets.
Assessment of background noise and the choice of control for ChIP-seq data analysis
Like for most other high-throughput, genome-wide approaches, the ‘signal’ obtained after ChIP-seq tends to be characterized by high background noise level. The background noise could result from structural differences in chromatin, mappability or systematic biases of immunoprecipitation for certain genomic landmarks 23,24. By far, the most widely used controls for peak calling are input DNA (purified sheared genomic DNA) or a generic null model of the read distribution, such as a Poisson or negative binomial distribution 7,21,22,25,26.
Importantly, the approach we have taken to assess background by performing cell-type specific affinity control experiments follows the recommendations of existing best practices as set forth by the ENCODE/modENCODE consortia 27. Input DNA is prepared from sonicated chromatin without any selection or enrichment for specific genomic regions. Another control that has been used is a mock IP (mock ChIP). Mock ChIP goes through the same process of selection as the experimental ChIP; however, this is done in the absence of the antibody. Other controls include performing ChIP on cells or tissue in which the gene encoding the protein being studied has been knocked out or performing ChIP in the presence of a blocking peptide, which binds the antibody against which they are designed and blocks further antibody binding to the target protein.
To perform the experimental and control ChIP under identical conditions we used empty vector ChIP, a control that is similar to a mock ChIP. However, unlike the mock ChIP, in our control empty vector ChIP we use the same antibody and beads as the experimental ChIP. In this control the TAP tag is not fused with the protein of interest therefore any enrichment arising from it most likely due to non-specific binding to beads, tag or other factors during the ChIP process.
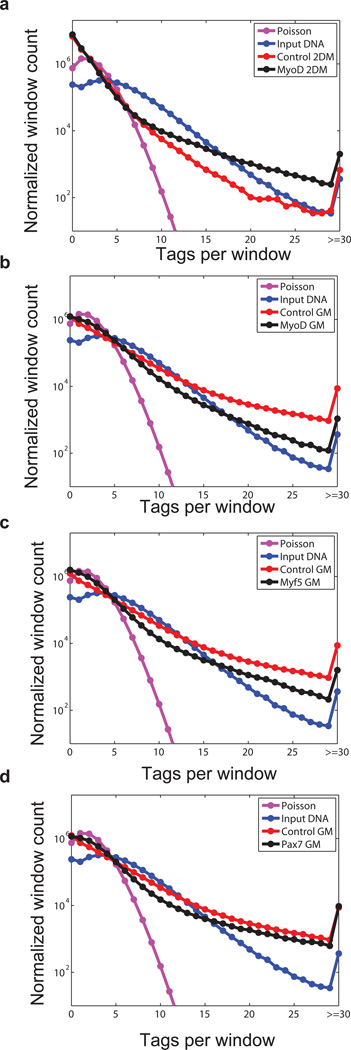
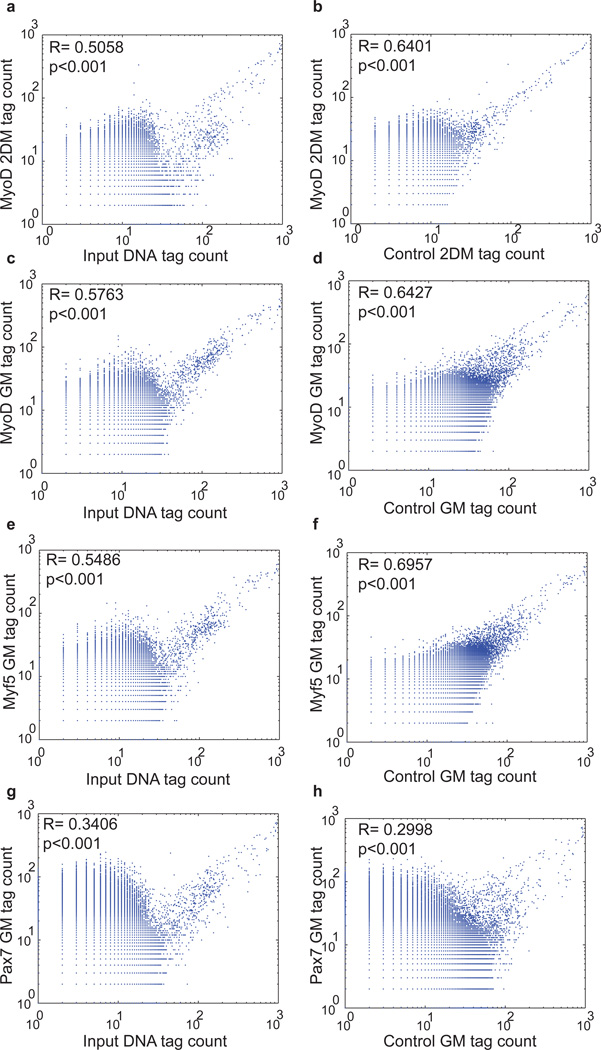
To provide a quantitative estimate of the extent of background noise in ChIP-seq data and assess the read distribution we analyzed the distribution of reads in 500 base pair windows across genome for ChTAP-seq, empty vector control and the input data. The 500 base pair window approximately matches the average peak length in our datasets. Importantly, our analysis shows that read distributions in all of these datasets are poor fit to a Poisson model (Fig. 3, Fig. S1a and see Supplementary Discussion). Hence the background noise in the ChIP-seq data cannot be modeled based on a Poisson model of reads distribution. Pair wise correlation analysis of read distributions between ChTAP-seq, affinity control and input DNA showed that ChTAP signal is more correlated with the empty vector affinity control signal than to input DNA (Fig. 4, Fig. 5a, Fig. S1b and Supplementary Discussion). This observation is consistent with previous findings that signal from mock IP is more related to experimental ChIP than is the input sheared genomic DNA 27.
Figure 3. Read distribution in ChIP-seq, control and input sample.
(a) The genome was divided into 500 bp windows and, for each data set; the number of reads per window was calculated. These counts were used to derive the frequency of windows with 0, 1, 2, …, 29 reads. Windows with reads ≥ 30 were also counted. Data was normalized to 10 million reads to adjust for differences in read counts among datasets. The expected number of reads within each window was computed under a Poisson model. Pink lines (Poisson) represent read distribution of normalized data under a uniform model in which 10 million tags are uniformly distributed across the genome. Blue lines represent read distribution from input DNA sample, while red and black lines represent read distribution of control- and MyoD ChIP-seq datasets respectively. (b-d) Similar analysis as above for MyoD GM, Myf5 GM and Pax7 GM ChIP-seq datasets. GM: myoblasts in growth media, 2DM: myotubes 2 days in differentiation media.
Figure 4. ChIP-seq reads distribution has significant similarities to control reads distribution.
(a) Correlation analysis between the read distributions of input DNA and MyoD ChIP-seq and (b) the same analysis for control versus MyoD ChIP-seq, as described in supplementary information. (c-h) Similar analysis for additional three independent datasets of MyoD GM, Myf5 GM and Pax7 GM shows similar results. GM: myoblasts in growth media, 2DM: myotubes 2 days in differentiation media.
Figure 5. Non uniform distribution of reads across genome in control datasets.
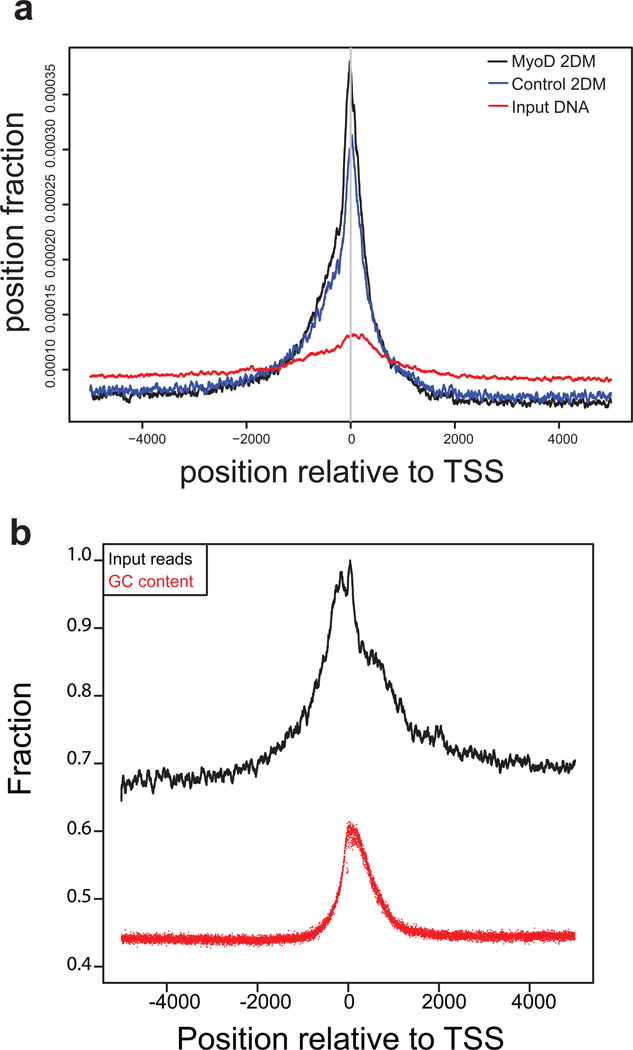
(a) Pileup analysis of tag distributions around transcription start sites (TSS) for MyoD 2DM, control (empty vector ChTAP) 2DM and input DNA. All ENSEMBL protein-coding genes are included in the analysis irrespective of their expression status. The graph shows reads piled up (not extended) divided by total non-duplicated mapped reads plotted around TSS. (b) Pileup analysis of input DNA reads around TSS. Input reads pileup (not extended) scaled to max in window being 1.0 and a corresponding relationship to average positional GC percentage (x-axis) suggests that there is a correlation between increased input reads and GC content around the TSS. GM: myoblasts in growth media.
To make a direct comparison between input DNA and empty vector affinity control for peak calling, we applied MACS to separately call MyoD peaks using either of the two controls. We show that there is a large discrepancy between the numbers of peaks called between the two controls. For example, using input DNA for background subtraction we obtained 28, 788 MyoD peaks. However, when we use affinity control (i.e., empty vector ChIP) data for background subtraction the number of MyoD peaks in myotubes is reduced to 10,705. Importantly, 10,156 out of 10,705 (94%) of peaks identified using affinity control are also called when we use input DNA control. However, we observe additional 18,083 peaks that are called when we use input DNA but not affinity control (empty vector ChIP).
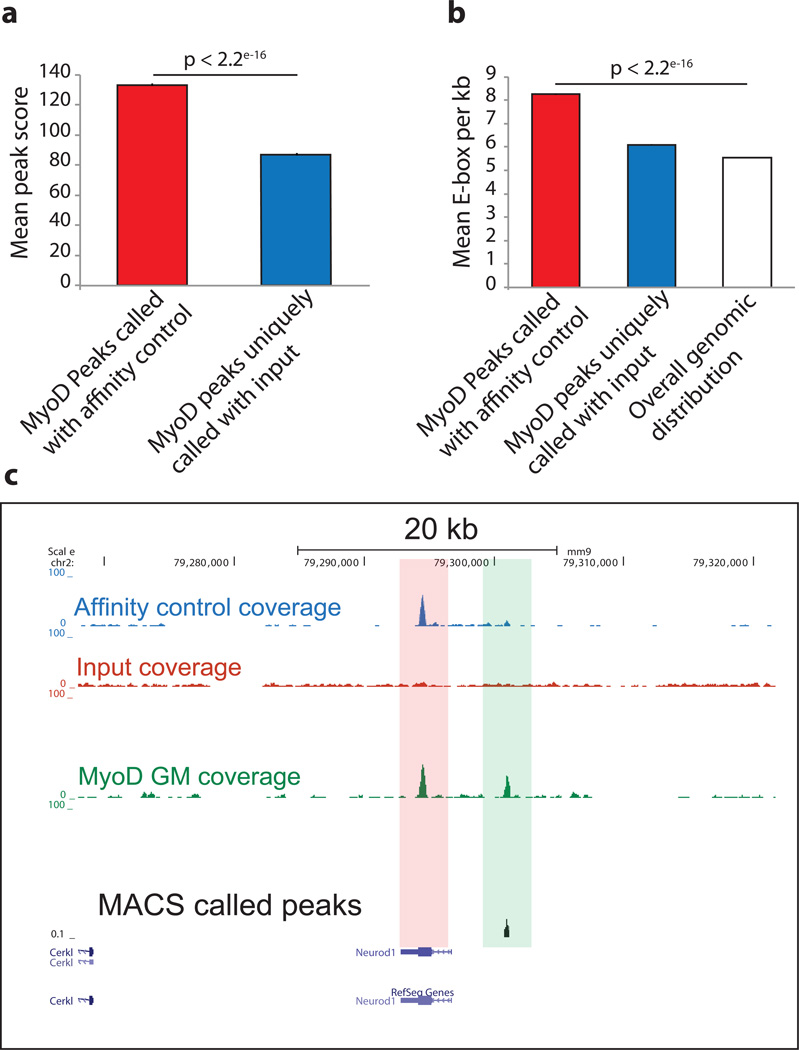
To determine if the extra set of 18,083 peaks, which are called by input but not an empty vector affinity control during background normalization, represent real binding sites we measured their average peak score (calculated as -10log10 p-value) (Fig. 6a). Additionally, since MyoD peaks are significantly more enriched for E-box motif than random 12 we measured the average number of E-boxes per kilo base of peaks and showed that this set of peaks have lower peak score and E-box density comparable to genome wide distribution (Fig. 6b). This analysis suggests that a substantial number of peaks called after background subtraction with input DNA may be false positives. Input DNA is valuable for assessing GC compositional bias (Fig. 5b and Supplementary Discussion). Furthermore, it can also be used to flag certain genomic regions that are highly enriched in all ChIP-seq, control and input data sets (Fig. S2, Table S1) from further analyses. These regions, with unknown etiology are likely the result of mapping artifacts, which can confound downstream analysis. A complete list of these flagged regions of mouse genome, which is an intersection of input, affinity control and ChIP-seq datasets reported here is provided in Table S1. Importantly, many of these regions are equally enriched in other independent datasets (Fig. S2).
Figure 6. Use of input DNA for background subtraction during peak calling results in substantial increase in the number of peaks with low quality scores.
(a) MyoD binding sites in myotubes were determine by ChTAP-seq using either input or an empty vector control for background subtraction. Peaks called using empty vector control have high quality score (−10 × log10 p-value) compared to peaks uniquely called using input as control. (b) Average number of E-boxes per kilo base of peaks for the two peak sets from above were calculated and compared to genome-wide distribution of E-boxes. P-values were calculated by pair wise t-test between the datasets. Error bars represent standard error of the mean (SEM). (c) A USCS browser snapshot showing a representative enriched site in affinity control and the ChIP but not in the input. Bottom track shows the location of peaks called by MACS using an affinity control for peak calling.
Our analysis has clearly shown that there are hundreds of genomic regions that are highly enriched in both affinity control and ChIP datasets but not in the input. Therefore, the use of input for background subtraction can erroneously identify numerous genomic loci as bona fide transcription factor binding site and confound downstream analysis as shown in the representative (Fig. 6c). Our similar observation with other independent and publically available ChIP-seq datasets (Fig. S1 and Supplementary Discussion) strongly suggests that a substantial portion of ChIP-seq signal constitutes background noise and can only be capture by an appropriate affinity control but not by the use of input DNA.
In conclusion, we show that using a cell-type specific matching affinity control is critical for objective assessment of background noise in ChIP-seq data. Furthermore, we provide a methodology and a detailed protocol to map genome-wide binding sites of any DNA binding protein without requirement for a ChIP-grade antibody. Finally, our method of background subtraction can be broadly applied to the analysis of any ChIP-seq dataset.
MATERIALS
REAGENTS
Agarose gel
Anti FLAG M2 affinity gel (Sigma A2220)
Dithiothreitol (DTT, Sigma D0632)
DNA ladder, 100 bp (New England Biolabs N3231)
DNA loading buffer (Life technologies, SKU 10816-015)
Ethanol (anhydrous and 95% solution)
Ethidium Bromide (Sigma E1510)
FLAG peptide (Sigma F3290)
Formaldehyde (37% (vol/vol) solution, Sigma 252549)
Glycine (Sigma G8898)
Glycoblue (AM 9516 Life Technologies)
HAM’s F10 cell culture media (Life technologies 11550-043)
HEK293T cell line (ATCC CRL-11268)
Phoenix Helper-Free Retrovirus Producer Line (ATCC CRL-3214)
His-Select HF Nickel Affinity Gel (Sigma H0537)
Hydrochloric Acid (Fisher A508SK-212)
Illumina ChIP-Seq Sample Preparation Kit (Illumina IP-102–1001)
Imidazole (Sigma-Aldrich I5513)
MinElute PCR Purification Kit (Qiagen 28004)
NaCl (5M) from salt (Fisher S271)
Phenol/Chloroform (Fisher BP1752I)
Phosphate Buffered Saline (PBS)
Protease inhibitors (Aprotinin, A1153; Leupeptin, L2884; Pepstatin A, P5318; Phenylmethanesulfonyl fluoride (PMSF), P7626 all available from Sigma)
Protein loading buffer (Laemmli 2X, Sigma-Aldrich S3401)
Proteinase K (New England Biolabs P8102)
Puromycin (Life Technologies A11138)
Quant-iT Pico Green (Invitrogen P11496)
Slide-A-Lyser Dialysis Cassette (Thermo Scientific 6630)
Sodium acetate (Sigma S2889)
Sodium dodecyl sulfate (SDS) (Sigma L4390)
Tobacco Etch Virus (TEV) Protease (Invitrogen 12575-015)
Tris-acetate-EDTA (TAE buffer, 10X, Fisher BP13354)
Tris Base (Fisher BP152)
Triton X-100 (Sigma X100)
EQUIPMENT
Agilent Technologies 2100 Bioanalyzer
Cell scraper (BD Bioscience 353085)
Centrifuges
Falcon tube (5 ml round bottom polypropylene tube for FACS analysis VWR)
Falcon tubes (15 ml, 50 ml VWR)
Fixed rotation mixer (Fisher Scientific 14-059-346Q)
Fluorescent plate reader or a qPCR machine
Freezer and refrigerators (various sources)
Heat blocks
Horizontal shaker
Illumina Genome Analyzer (Illumina)
Low-retention microcentrifuge tubes (Fisher 02-681-320)
Needles, 25G 5/8 (Becton Dickinson 305122)
Poly-Prep chromatography columns (Bio RAD 731–1550)
Tissue culture dish (6 cm, 10 cm, 15 cm)
Water bath sonication device
REAGENT SETUP
ChIP dilution buffer: 40 mM Tris-HCl, pH 8.0; 4 mM EDTA, pH 8.0. Buffer can be stored at 4 °C for up to 6 moths.
ChIP fixation buffer: 1% (vol/vol) formaldehyde in 1X PBS. This solution needs to be prepared fresh each time.
ChIP lysis buffer: 40 mM Tris-HCl, pH 8.0; 1% (vol/vol) Triton X-100, 4 mM EDTA, pH, 8.0; 300 mM NaCl. Store at 4 °C for up to 4 weeks. Add protease inhibitors to each batch before use. Dialysis buffer: 20 mM Tris-HCl, pH 8.0; 150 mM NaCl, 0.1 mM EDTA, pH 8.0; 10% glycerol EDTA solution: EDTA 0.5 M, pH 8.0. Adjust pH by adding HCl. CAUTION! HCl is a hazardous and corrosive material use safety goggle and gloves during handling.
M2 wash buffer: 10 mM Tris-HCl, pH 8.0; 150 mM NaCl, 0.1% (vol/vol) Triton X-100. Can be stored at 4 °C for up to 6 months.
Quenching (glycine) buffer: 0.125 M glycine in 1X PBS 500 ml, make fresh solution each time TBS buffer: 50 mM Tris-HCl, pH 7.4; 150 mM NaCl
TEV protease buffer 10X: 500 mM Tris-HCl, pH 8.0; 5 mM EDTA, pH, 8.0; 10 mM DTT Tris-EDTA (TE buffer): 10 mM Tris-HCl, pH 8.0; 1 mM EDTA, pH, 8.0.
PROCEDURE
This protocol has been used to identify genome-wide binding sites of Pax3 and Pax7 in myoblasts 11, MyoD, Myf5, E47, Snai1, HDAC1 and HDAC2 in myoblasts, myotubes or C3H10T1/2 fibroblasts 12.
Generation of stable cell lines expressing TAP tagged construct • TIMING ~1 week
-
1
Generate C-terminal TAP tagged fusion construct by removing the stop codon from the Open Reading Frame (ORF) of the gene of interests and fuse the ORF with 6XHIS-TEV-3XFLAG tag as described previously 14.
-
2
Validate the construct by sequencing, Western blot analysis after transfection of the plasmid into a mammalian cell line such as HEK 293T cells and by Electrophoretic mobility shift assay EMSA as describe previously 11,12.
-
3
Generate retroviral particle by transfecting the plasmid into helper-free Pheonix –ECO or HEK 293T cells as described previously 14.
-
4
Generate stable cells over expressing low level of TAP tagged protein by infecting cells of interest with low (<10) multiplicity of infection (MOI) as described previously 11,12,14. A parallel cell line over expressing TAP tag not fused to any protein in same cell line, as the experimental ChIP is required as a cell-type specific affinity control. This cell line should be generated concurrently.
Cell harvesting, lysis and chromatin shearing • TIMING ~3 h
This protocol describes the purification of chromatin using two independent epitopes. Starting material is 100–150 million cells.
CAUTION: All work involving retroviruses should be performed in class II biosafety chamber. Personal protective equipment such as gloves and lab coat should be worn at all time. The waste containing retrovirus vectors should be disposed of according to approved biosafety guidelines.
-
5
Aspirate media from growing cells murine cell and rinse the cells with 1X PBS solution.
CRITICAL STEP To minimize cell loss due to possible dislodging from the plate, add the PBS solution gently. Step 5 to 10 of this procedure apply to adherent cells. The procedure can easily be adapted to non-adherent cells with minimum effort and no extra reagents or equipment.
-
6
Add ChIP fixation buffer. The amount of fixation buffer depends on the plate size. Add up to 15 ml per 15 cm plate or enough to cover the entire surface of the plate and place the plates on horizontal shaker at room temperature (22 °C) for 10 min with gentle rotation.
-
7
Gently pour off the fixation buffer into a labeled container for waste disposal. Add quenching (glycine buffer) and place the plates on a horizontal shaker for 5 min at room temperature (22 °C) with gentle rotation.
-
8
Wash cells with 1X ice-cold PBS solution containing protease inhibitors (Leupeptin, 100 µM; Pepstatin A, 100 µM; Aprotinin, 10 µg/ml; PMSF, 1mM). Aspirate the wash buffer and replenish the plates with an additional 5 ml of ice-cold 1X PBS.
-
9
Harvest cells using a cell scraper and collect the cell–PBS mixture into a 50 ml falcon tube. Combine all cell material per experiment in one tube.
-
10
Collect cell pellet using a refrigerated centrifuge by spinning at 700 ×g for 5 min at 4 °C
-
11
Carefully pour off the PBS without losing the cell pellet.
-
12
Add to the cell pellets ChIP lysis buffer containing working concentration of protease inhibitors and place the tubes on ice for 15–20 min. Follow 2:1 (vol/wt) ratio for the amount of lysis buffer to cell pellet.
-
13
Dispense cell lysate in 0.5-ml aliquots to 1.5 ml eppendorf tubes
-
14
Place the tubes from step 8 into a water bath sonicator/bioruptor, with circulating cold water. Sonication time will need to be empirically determined for each cell/tissue type. Typically, to get chromatin shearing within the range of 100–300 bp for ChIP-seq, we sonicate for 30 cycles (30 s on 60 s off) using a Diagenode bioruptor on high power setting.
-
15
Assess the quality of DNA shearing implementing the procedure detailed in Box 1. Prepare also a sample of input DNA control as described in Box 2. Once the shearing of chromatin has achieved optimal fragmentation size, typically between 100 to 300 base pairs as assessed by agarose gel and ethidium bromide visualization, take 2% of input and proceed to the preparation of input DNA for sequencing. While implementing that procedure, incubate the sample from step 10 at −20 °C
Box 1 | Assessing the shearing efficiency of chromatin during sonication • TIMING ~1 d.
Collect 10 µl of sheared chromatin from step 10 and place it into a 1.5-ml eppendorf tube.
Bring the volume to 200 µl by adding TE buffer. Add 10 µl of NaCl 5 M solution and incubate in a heat block at 65 °C for at least 4 h or up to overnight.
Add 5 µl of proteinase K 5 mg/ml solution and incubate at 40 °C for 1 h.
Bring the volume up to 500 µl by adding TE buffer.
Add 500 µl of phenol/chloroform/isoamyl alcohol.
Shake vigorously or vortex briefly until a homogenous milky color mixture is observed.
Centrifuge the sample at 20K ×g for 3 min in a refrigerated microcentrifuge at 4 °C.
Take the upper phase (aqueous) and place it in a new 1.5-ml eppendorf tube and add 1:10 vol/vol of 2.5 M sodium acetate, pH 5.2 followed by 2:1 vol/vol of anhydrous ethanol.
Incubate the sample at −80 °C for at least 1 h.
(OPTIONAL) Add 2 µl of GlycoBlue to ease visualization of the pellet.
Centrifuge the sample at maximum speed (>20K ×g for 15 min at 4 °C.
Pour off the supernatant. Be careful not to loose the pellet, which is usually located where the Glyco Blue pellet is located at the bottom of the tube (if step 11 has been implemented).
Wash the pellet twice with 70% ethanol.
Air-dry the pellet for up to 1 h and dissolve the pellet in 20 µl of TE buffer.
Analyze the shearing efficiency by agarose gel and ethidium bromide staining.
(OPTIONAL) Most of the sheared DNA should be in the 100–300 (average 200) base pairs (bp) length range. If the average fragment length is substantially higher than 200 bp, perform an additional round of sonication by repeating step 14 of the main procedure.
Take samples from step 14 of the procedure and proceed with input library preparation and immunoprecipitation.
Box 2 | Preparation of input DNA • TIMING 1d.
Take 2% of sample from step 14 of the main procedure (the sonicated cell lysate).
Bring the volume to 200 µl by adding TE buffer.
Add 10 µl of NaCl (5M) and incubate sample at 65 °C for 4 to 6 h.
Add 5 µl of proteinase K and incubate sample at 37 °C for 1 h.
Add 200 µl of phenol/chloroform, shake vigorously and centrifuge at 20K ×g for 3 minutes.
Take the upper aqueous phase and place it in a labeled 1.5 ml eppendorf tube.
Add 2 vol/vol of anhydrous ethanol and incubate the sample at −80 °C for 1 h
Centrifuge the sample at 20K ×g for 20 minutes.
Carefully pour off the supernatant without loosing the pellet, which is barely visible.
Wash the pellet with 1 ml of 70% ethanol.
Air dry the pellet by leaving on the bench upside down with open cap.
Dissolve the pellet in 20 µl H2O.
Measure DNA concentration and quality with Bioanalyzer.
Take 10 n of input DNA for library construction.
Please note that sonication produces double-stranded DNA with various configurations of fragment ends, including blunt ends, 3’ overhangs, or 5’ overhangs. To prepare input DNA library for sequencing, blunt the fragment ends with T4 DNA polymerase followed by incorporation of “A” overhang and adapter ligation as described in the Illumina library preparation protocol (http://www.illumina.com). The resulting libraries are sequenced on the Illumina Genome Analyzer.
Chromatin tandem affinity purification • TIMING ~3–4 d
-
16
Transfer the cell lysate from step 10 and centrifuge at 20K ×g for 10 min in a refrigerated microfuge at 4 °C Transfer the supernatant to a freshly labeled 15-ml eppendorf tube. Be careful to avoid carrying over the insoluble material and pellet.
-
17
Add an equal volume to the supernatant from step 12 of ChIP dilution buffer. Typically at this stage one should have 5–6 ml of diluted lysate.
-
18
Place 150 µl of the anti FLAG affinity gel in a 1.5-ml eppendorf tube and add 1,350 µl of M2 wash buffer.
-
19
Place the samples on a fixed rotation mixer in a refrigerated room at 4 °C for 5 min with constant rotation to wash beads.
-
20
Collect beads by centrifugation at 400 ×g for 1–2 min in a refrigerated centrifuge at 4 °C. Carefully aspirate off the supernatants and dissolve the beads in M2 wash buffer again as described in step 15.
-
21
Repeat step 16 two more times
-
22
After the final wash, mix the anti FLAG M2 affinity gel (beads) in 150 µl of M2 wash buffer and directly add to the cell lysate from step 13 and incubate on a fixed rotation mixer at 4 °C for 3 h.
-
23
Pellet beads by centrifugation at 400 ×g at 4 °C for 1–2 min. Carefully transfer the supernatant to an eppendorf tube and label it as flow-through #1.
-
24
(OPTIONAL) We advise using the supernatant from step 23 western blot analysis to determine the efficiency of binding (capturing antigen), a procedure that will help troubleshoot the experiment in case of failure of the experiment.
-
25
Wash beads from step 19 with up to 10 volumes of M2 wash buffer and transfer the slurry to a freshly labeled low-retention (siliconized) 1.5 ml eppendorf tube.
-
26
Repeat step 21 two more times.
Carefully aspirate the wash buffer without disturbing the pellet and proceed with elution of chromatin from beads.
-
27
Elute chromatin using one of the two following independent options:
Option 1 Chromatin elution by competition with 3XFLAG peptide. TIMING 3–4 h
Dissolve the lyophilized 3XFLAG (5 mg) provided by the supplier in in 1 ml of TBS buffer to bring the concentration to 5 mg/ml.
Add 250 µl of 3XFLAG peptide elution buffer by diluting the stock solution 3XFLAG peptide from above to 200 to 400 g/ml in TBS and add directly to the beads. The final concentration of the 3XFLAG peptide should be between 200 µg/ml and 400 µg/ml in TBS for efficient elution.
Incubate the sample at 4 °C for at least 1 h on a fixed rotation mixer with rotation.
Set up a Poly-prep chromatography column with a 25G 5/8 needle attached to the bottom.
Add 5 ml of TBS buffer to the column at let drain under gravity.
Place a labeled 1.5-ml eppendorf low-retention, siliconized tube at the bottom of the column to collect the flow-through.
Take the slurry (agarose-conjugated beads with the protein complex) and add directly to the column and let drain into the tube under gravity.
Prepare another 250 µl of elution buffer (3XFLAG peptide in TBS) and add it directly to the column. At this stage the total volume of eluate is ~500 µl.
At this stage most of the protein–DNA complex is eluted, however, to check the efficiency of elution, boil agarose beads in protein-loading buffer.
Option 2 Chromatin elution by photolytic cleavage using TEV protease • TIMING 1 d
-
Add the following components to the beads from step 22: 25 µl of 10X TEV protease buffer, 2.5 µl of 0.1 M DTT, 1.5 µl TEV protease, and ddH2O to a total volume of 250 µl.
CRITICAL STEP TEV protease functions optimally under reducing condition. Therefore, this step requires a significant amount of DTT, which is a strong reducing agent and must be removed prior to purification with nickel (Ni-NTA) beads as described below.
Incubate overnight at 4 °C on a fixed rotation mixer with constant rotation.
Centrifuge beads at 400 ×g for 1–2 min in a refrigerated centrifuge at 4 °C.
Carefully remove the supernatant with a pipette and place it into a 1.5-ml, low-retention eppendorf tube.
Repeat step i one more time and combine the supernatant.
Keep the digested beads at −20 °C while performing western blots analysis to determine the efficiency of TEV digest.
Remove DTT by dialysis using Slide-A-Lyser Dialysis Cassette 3,500 MWCO in dialysis buffer. Load the sample into the dialysis cassette according to the manufacturer’s recommendation and dialyze at 4 °C overnight.
-
Recover the sample from dialysis cassette with a syringe and place it in a 1.5 ml siliconized, low retention eppendorf tube.
PAUSE POINT. The eluted product can be stored at −20 °C for a few days or at −80 °C for a few months.
Final purification of chromatin by Ni-NTA affinity chromatography (6XHIS tag purification) • TIMING 0.5 d
-
28
Take 150 µl of HIS-Select Nickel Affinity Gel and place it into a 1.5-ml eppendorf tube. Add to it 1,350 µl of nickel wash buffer.
-
29
Place the tubes containing the beads on a fixed rotation mixer at 4 °C for 5 min with constant rotation.
-
30
Centrifuge the bead slurry at 400 ×g for 1 min in a refrigerated centrifuge at 4 °C. Carefully pour off the supernatants without disturbing beads.
-
31
Repeat step 26 (washing) two more times and pour off the wash buffer.
-
32
Dissolve beads in an additional 150 µl of nickel wash buffer and directly add the beads slurry to the sample eluted by 3FLAG or TEV digest.
-
33
Incubate the samples on a fixed rotation mixer with rotation at 4 °C for 2–3 h.
-
34
Centrifuge the tubes at 400 ×g for 1–2 min at 4 °C and carefully pour off supernatant.
-
35
Wash beads, containing chromatin with 1,350 µl of nickel wash buffer as described in step 26.
-
36
Centrifuge the tubes at 400 ×g in a refrigerated centrifuge at 4 °C and pour off the supernatant.
-
37
Add 250 µl of elution buffer (250–400 mM imidazole in H2O). Incubate samples on a fixed rotation mixer at room temperature for at least 1 h with continuous rotation. Please note that longer incubation times with elution buffer usually increase the elution efficiency.
-
38
Set up Poly-Prep chromatography columns. Attack a 25G 5/8 needle to the bottom of the column.
-
39
Pour 5 ml of nickel wash buffer directly into the columns and let the column drain completely under gravity.
-
40
After the column has completely drained, place a labeled 1.5-ml low-retention (siliconized) eppendorf tube at the bottom of the column beneath the needle to collect the eluate from the next step.
-
41
Pour the slurry from step 10 directly onto the column and let it drain into the collection tube under gravity. Please note that occasionally, clogging occurs and the flow stops. When this happens, apply a slight pressure to the top of the column with the palm of your gloved hand, which will restart the flow. The sample is generally drained in 2 to 5 min.
-
42
Add an additional 250 µl of elution buffer to the beads and collect the eluate into the same siliconized tube. Keep the beads at −20 °C while performing western blot analysis to determine the efficiency of elution. Please note that at this stage the purified chromatin is highly enriched for the transcription factor binding site. The control sample contains non-specific chromatin, which is pulled down by non-specific interaction with agarose and nickel affinity beads.
-
43
To the 500 µl of eluted sample, add 20 µl of NaCl 5 M and incubate in a heat block at 65 °C for a minimum of 4 h and up to overnight.
-
44
Add 5 µl of proteinase K (5 mg/ml) and incubate at 37 °C for 1 h.
-
45
Proceed with DNA extraction using phenol/chloroform
-
46
Quantify ChIP DNA using Quant-iT PicoGreen ds DNA reagent and kits following manufacturer’s instructions. Alternatively, the quality and quantity of DNA can be assessed with Agilent Technologies 2100 Bioanalyzer.
Anticipated results
The yield of ChIP DNA is typically in the range of 5–50 ng and it varies depending of the protein under investigation. Control ChIPs yield is substantially less than the yield of the experimental ChIP. In cases where the quantity of ChIP DNA is substantially lower than expected, the chromatin may have been lost during recovery throughout the pipeline. Western blot analysis of samples from step 23, 42 and step vi of option 2 should be performed to find out the step(s) that require troubleshooting.
It is also advised that in cases where there are known binding sites for the protein of interest, to do quick quality control test by quantitative Real-time PCR using locus-specific primers to ascertain that the ChIP DNA is enriched for the bona fide binding sites, prior to proceeding with ChIP library construction and sequencing.
ChTAP library construction and sequencing TIMING 2–3 d
-
47
Proceed with ChTAP library construction and sequencing. At this point, ChTAP samples, empty vector control and the input DNA (Box 2) can be processed in parallel using Illumina protocol for ChIP-DNA library construction.
Supplementary Material
Table 1.
Troubleshooting
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 2 | Tagged protein not expressed |
Cloning and mutation issues |
Verify the construct by sequencing |
| 4 | Expression level of the tagged protein is too high relative to the endogenous protein |
Inefficient viral infection |
Titer the virus prior to infection, use a GFP control virus |
| Viral infection with high multiplicity of infection (MOI) |
Infect with low MOI, use a vector with weaker promoter |
||
| 14 | Under/over shearing of DNA |
Inappropriate sonication |
Extend/reduce sonication time, keep the concentration of input sample constant |
| 27 | Inefficient elution from M2 beads |
Inefficient TEV cleavage |
Extend incubation with TEV to overnight at 4 °C |
| Inefficient 3xFLAG peptide competition |
Increase the concentration of 3xFLAG peptide to ~400 µg/ml |
||
| 28 | Weak binding to Ni-NTA |
Presence of reducing agents, such as DTT |
Remove interfering compounds with dialysis prior to purification with Ni-NTA. |
| 40– 43 |
Inefficient elution from Ni-NTA beads |
Short elution and/or low imidazole concentration |
Perform elution with imidazole for extended time (>3 h) at room temperature |
| 44– 46 |
Insufficient DNA recovery |
Tagged protein non- Functional Protein folding masks TAP tag |
Functional validation and correct cellular localization of the tagged protein Assay the efficiency of IP with anti FLAG antibody |
| Washes were too stringent or too extended |
Each washing step should be ~5 min. The ratio of wash buffer to beads should be ~10:1 |
||
| Use molecular biology grade and nuclease- free reagents |
|||
| Cross-linking sub optimal |
Make fresh fixation buffer prior to ChIP. | ||
| 47 | ChIP-seq data is suboptimal |
Chromatin fragmentation not appropriate |
Size exclusion during ChIP library construction results in chromatin fragment between 100–300 base pairs. Assess the efficiency of chromatin fragmentation prior to ChIP experiment |
| Insufficient antibody quantity used |
Increase the amount of antibody. We typically use 150 µl of M2 agarose beads slurry for ChIP with 100–150 million cells |
||
| Insufficient sequencing depth |
Run additional lane on the flow cell or switch to another platform such as HI-SEQ2000 |
||
| No enrichment on the expected genomic regions |
TAP tag epitope may have been masked due to cross-linking or protein folding. Place TAP tag on the opposite end of protein or switch to a different tag |
TIMING
Steps 1–4 Generation of stable cell lines expressing TAP tagged construct ~1 week
Steps 5–14 Cell harvesting, lysis and chromatin shearing ~3 h
Steps 16–22 Chromatin tandem affinity purification: ~3–4 days.
Step 27 Elution of chromatin from anti FLAG M2 affinity beads: option1, 3–4 h; option 2, 1 d
Steps 28–42 Final purification of chromatin by Ni-NTA affinity chromatography: 0.5 days.
Steps 43–46 Reverse cross-linking of chromatin and ChIP DNA purification: 1d
Step 47 ChTAP library construction and sequencing: ~2–3 days.
ACKNOWLEDGEMENTS
We thank Kathy Sheikheleslamy at the StemCore laboratory, Ottawa Hospital Research Institute for discussions on Illumina library preparation and sequencing. VDS is supported by a Computational Regulomics Training Program (CRTP) fellowship from Ontario Research Funds. M.A.R. holds the Canada Research Chair in Molecular Genetics and is an International Research Scholar of the Howard Hughes Medical Institute. This work was supported by the Canadian Institutes of Health Research grant MOP12080, NIH/NIAMS grant R01AR044031, and Ontario Research Fund grants to M.A.R and by a Natural Sciences and Engineering Council of Canada (NSERC) Discovery Grant 328154-2009 to T.J.P.
Footnotes
This protocol has been used in the following two research articles:
Soleimani et al. Developmental Cell 2012 Jun 12;22(6):1208-20. doi: 10.1016/j.devcel.2012.03.014. Epub 2012 May 17.
Soleimani et al. Molecular Cell 2012 Aug 10;47(3):457-68. doi: 10.1016/j.molcel.2012.05.046.Epub 2012 Jul 5.
AUTHOR CONTRIBUTIONS VDS and MAR conceived the project and designed the experiments. VDS performed the experiments and together with TJP and GAP analyzed the data. VDS wrote the manuscript with help from TJP, GAP and MAR.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
REFERENCES
- 1.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 2.Horak CE, et al. GATA-1 binding sites mapped in the beta-globin locus by using mammalian chIp-chip analysis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:2924–2929. doi: 10.1073/pnas.052706999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blais A, et al. An initial blueprint for myogenic differentiation. Genes & development. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mardis ER. ChIP-seq: welcome to the new frontier. Nature methods. 2007;4:613–614. doi: 10.1038/nmeth0807-613. [DOI] [PubMed] [Google Scholar]
- 5.Schmid CD, Bucher P. ChIP-Seq data reveal nucleosome architecture of human promoters. Cell. 2007;131:831–832. doi: 10.1016/j.cell.2007.11.017. author reply 832–833. [DOI] [PubMed] [Google Scholar]
- 6.Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nature methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harada R, Nepveu A. Chromatin affinity purification. Methods Mol Biol. 2012;809:237–253. doi: 10.1007/978-1-61779-376-9_16. [DOI] [PubMed] [Google Scholar]
- 11.Soleimani VD, et al. Transcriptional dominance of Pax7 in adult myogenesis is due to high-affinity recognition of homeodomain motifs. Developmental cell. 2012;22:1208–1220. doi: 10.1016/j.devcel.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soleimani VD, et al. Snail regulates MyoD binding-site occupancy to direct enhancer switching and differentiation-specific transcription in myogenesis. Molecular cell. 2012;47:457–468. doi: 10.1016/j.molcel.2012.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puig O, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 14.McKinnell IW, et al. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nature cell biology. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nature biotechnology. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 16.Burckstummer T, et al. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nature methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- 17.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 18.Ghaemmaghami F, et al. First-line chemotherapy with 5-FU and platinum for advanced and recurrent cancer of the cervix: a phase II study. Journal of obstetrics and gynaecology : the journal of the Institute of Obstetrics and Gynaecology. 2003;23:422–425. doi: 10.1080/0144361031000120969. [DOI] [PubMed] [Google Scholar]
- 19.Rohila JS, Chen M, Cerny R, Fromm ME. Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. The Plant journal : for cell and molecular biology. 2004;38:172–181. doi: 10.1111/j.1365-313X.2004.02031.x. [DOI] [PubMed] [Google Scholar]
- 20.Abe M, Fujiwara M, Kurotani K, Yokoi S, Shimamoto K. Identification of dynamin as an interactor of rice GIGANTEA by tandem affinity purification (TAP) Plant & cell physiology. 2008;49:420–432. doi: 10.1093/pcp/pcn019. [DOI] [PubMed] [Google Scholar]
- 21.Cao Y, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Developmental cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mullen AC, et al. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auerbach RK, et al. Mapping accessible chromatin regions using Sono-Seq. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14926–14931. doi: 10.1073/pnas.0905443106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vega VB, Cheung E, Palanisamy N, Sung WK. Inherent signals in sequencing-based Chromatin-ImmunoPrecipitation control libraries. PloS one. 2009;4:e5241. doi: 10.1371/journal.pone.0005241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 26.Rozowsky J, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nature biotechnology. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome research. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.