Abstract
In recent years the Drosophila heart has become an established model of many different aspects of human cardiac disease. This model has allowed identification of disease-causing mechanisms underlying congenital heart disease and cardiomyopathies and has permitted the study underlying genetic, metabolic and age-related contributions to heart function. In this review we discuss methods currently employed in the analysis of the Drosophila heart structure and function, such as optical methods to infer heart function and performance, electrophysiological and mechanical approaches to characterize cardiac tissue properties, and conclude with histological techniques used in the study of heart development and adult structure.
Keywords: Drosophila, heart, development, cardiomyopathy, congenital heart disease, genetics, optical heart beat analysis, electrophysiology, atomic force microscopy, immunohistochemistry, KCNQ, tinman
Introduction
The Drosophila heart as a tool for investigating cardiomyopathies
The Drosophila heart or dorsal vessel is a linear tube that is reminiscent of the primitive vertebrate embryonic heart tube. Although the final heart structure in Drosophila is very different from that in vertebrates, the basic elements for cardiac development, function and aging are remarkably conserved (Bodmer 1995, Cripps and Olson 2002, Ocorr et al. 2007a). Because of the simplicity in structure and availability of powerful genetic tools, the Drosophila heart has emerged as a pioneering model system for unraveling the basic genetic and molecular mechanisms of cardiac development, function and aging (Bodmer and Frasch 2010, Nishimura et al. 2011). The Drosophila heart model has proven to be a valuable asset to elucidate the etiology of human cardiac disease, including dilated and restricted cardiomyopathy, channelopathies, diabetic and congenital heart disease, as well as cardiac senescence (Birse et al. 2010, Cammarato et al. 2008, Melkani et al. 2013, Na et al. 2013, Ocorr et al. 2007b, Qian et al. 2008, Taghli-Lamallem et al. 2008, Wessells et al. 2004, Wolf et al. 2006). The Drosophila heart has also been used as a tool for the identification of novel genes and pathways potentially involved in heart disease (e.g. (Kim et al. 2010, Kim et al. 2004, Neely et al. 2010, Qian et al. 2011). Of note, certain important ion channel gene functions are conserved between Drosophila and humans to maintain a regular heart rhythm, such as KCNQ (Ocorr et al. 2007b). Interestingly, some of these ion channels do not play a significant role in the (faster beating) adult mouse heart (Nerbonne 2004). This suggests that in some regards the fly heart model may be more informative than the mouse model.
We will first discuss different methods to assess heart function that may change under different genetic and environmental conditions, as well as with age. Then we will summarize tools and markers for structural features of the heart during development, maintenance and aging. An overview of the different methods discussed in this chapter is presented in Table 1.
Table 1.
Comparison of the different methods for analyzing heart function in Drosophila.
| Method | Effect on Animal |
Heart Rate |
Systolic & Diastolic Intervals |
Systolic & Diastolic Diameters |
Rhythmicity | Structure | Stiffness | Stress Response |
|---|---|---|---|---|---|---|---|---|
| Gross Δ Intensity Optical Method | Stressful / in vivo | + | + | |||||
| SOHA Optical Method | Invasive / in situ | + | + | +/− 1µ | + | + | ||
| Atomic Force Microscopy | Invasive / in situ | + | ||||||
| Optical Coherence Tomography | Stressful / in vivo | + | +/− 10µ | + | + | |||
| Electrical Recordings | Invasive / in situ | + | + | + | ||||
| Immuno histochemistry / Electron Microscopy | in situ | ++ | ||||||
| Electrical Pacing | Stressful / in vivo | + |
Optical-based analysis methods to measure heart function and performance
Heart Rate and Pacing
The Drosophila heart is a linear tube with a non-contractile aorta that extends from the posteriorly located heart to the head in both larva and adults. Rudimentary valve-like structures divide the heart into chambers and prevent back flow of hemolymph. In larvae the heart is suspended within the hemocoel and undergoes substantial remodeling during pupal stages prior to forming the four chamber abdominal heart of the adult fly (Molina and Cripps 2001; Zeitouni et al. 2007). Early efforts to examine heart function in intact Drosophila were performed on dissected larva (Gu and Singh 1995) and early pupa, where the cuticle is nearly transparent (Dowse et al. 1995). For larval preparations the heart rate was determined visually. For the pupal preparation, light is passed through the heart and detected by a phototransistor in the microscope eyepiece. Changes in overall light intensity could be recorded and displayed as linear traces. Using custom software, Maximum Entropy Spectral Analysis (MESA) the overall heart rate could be determined (Dowse et al. 1989, Dowse and Ringo 1991). In addition, heartbeat rhythmicity could be quantified by MESA as a correlation coefficient. Using genetic mutants and pharmacological manipulations these techniques provided evidence that the fly heartbeat is myogenic and that the cardiac action potential likely does not have a substantial Na+ current since heartbeats were not affected by tetrodotoxin (TTX; Gu and Singh 1995, Johnson et al. 1998). These studies also showed that reduction of extracellular Ca2+ stopped heart function suggesting that the cardiac action potential is Ca2+ based, although each of these groups came to different conclusions concerning the specific type of Ca2+ channel that was involved. In addition a prominent role for K+ channels was suggested in that channel blockers such as TEA caused dramatic reductions in heart rate (Gu and Singh 1995) and mutations in shaker, ether-a-gogo and slowpoke K+ channels all affected heart rate albeit to different extents (Johnson et al. 1998).
A variation on these optical detection methods has been developed for monitoring the adult hearts expressing GFP (Dulcis and Levine 2005). The GFP signal permits the heart walls to be visualized through the cuticle; however, since the fluorescent signal is usually very low, high frame rate recordings require specialized equipment that is costly. Another approach for optically monitoring heart function in adult makes use of infrared light and an array of sensors oriented along the abdominal cuticle (Wasserthal 2007). Longer wavelengths are better able to penetrate the pigmented cuticle of the adult thus permitting heart rate and rhythmicity to be determined in vivo from tethered adult flies. However, the spatial resolution is limited and frame rate is still relatively low with this method.
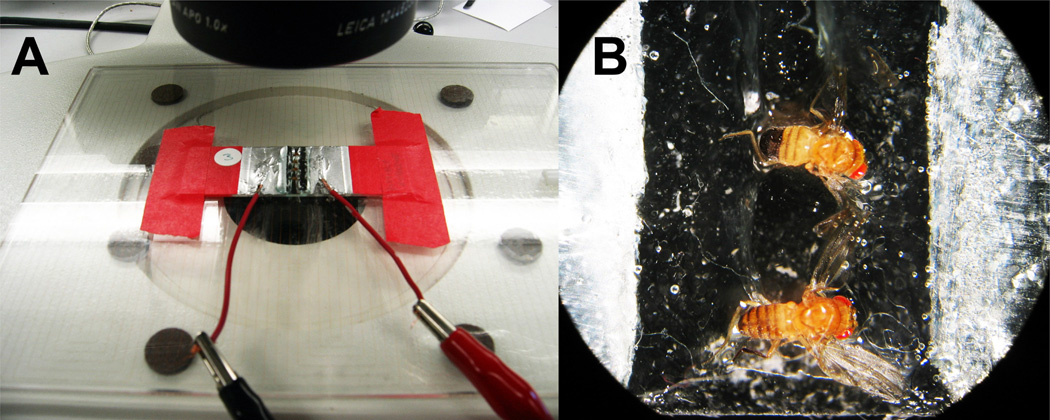
In order to test cardiac performance, cardiac stress tests have been developed based on increasing or ‘pacing’ the resting heart rate. These assays can be used to compare the effects of different mutations as well as the effect of age on heart function. In one such assay, pacing of pupal or adult hearts to higher rates was achieved by increasing temperature (Paternostro et al. 2001, White et al. 1992). In another pacing assay, the head and abdomen of adult flies are placed to touch two strips of electrode jelly (Fig. 1) and a square wave current is passed through the animal to pace the heart to 6 Hz (the upper end of the unperturbed heart rate range, which normally is 4–5 Hz) for a set amount of time (30 sec; (Wessells and Bodmer 2004, Wessells et al. 2004). Immediately after pacing, heart function is monitored visually through the abdominal cuticle (A2–3) and flies are scored depending on whether the heart can still beat following the stimulus or whether it fails to contract or contracts in a spasmodic fashion. This assay has been used to show that insulin signaling plays a heart-autonomous role in cardiac senescence (Wessells et al. 2004) and KCNQ channels are critical for cardiac repolarization in Drosophila (Ocorr et al. 2007b).
Figure 1. Pacing Assay of Cardiac Function.
(A) Electrical pacing setup showing microscope slide with foil electrodes connected to two leads that deliver the square wave pulse from a stimulator (not pictured). (B) View through the microscope of two flies with their heads inserted into one line of electrode gel and the abdomen tip into the other. It is important that the two lines of gel do not touch.
Semi-Automated Optical Heartbeat Analysis (SOHA)
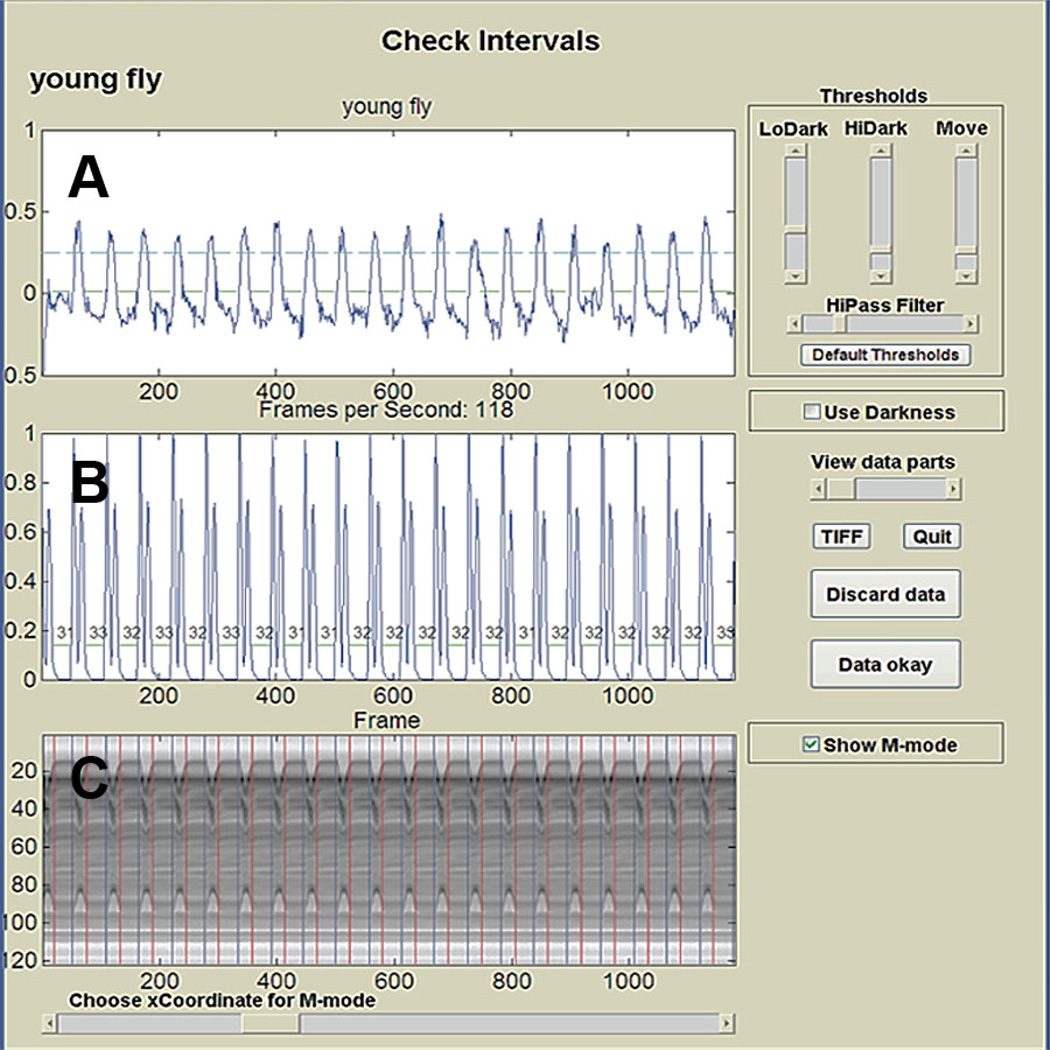
The movement detection methodologies described above all rely on detecting overall changes in the intensity of light passing through the heart while it is beating (‘darkness’ profiles, Fig. 2A). A more sensitive method that relies on the combined use of two different computer algorithms has been developed for use with high speed, high resolution digital cameras (Ocorr et al. 2007b). This Semi-automated Optical Heartbeat Analysis (SOHA; detailed method in Fink et al. 2009) uses two computer algorithms that were developed to combine information about overall darkness changes (Fig. 2A), which denote systolic periods, with a pixel-by-pixel analysis of intensity changes detecting only the regions in the movie frame that are moving (Fig. 2B). This combinatorial approach thus detects the individual inward and outward movements of the heart wall and uses the gross darkness changes to determine the “state” (i.e. contracted v. relaxed); permitting the quantification of large numbers of contractions in relatively long movie clips. This capability becomes significant with respect to rhythmicity measurements (see below). Camera setups that can record images at speeds of 150–500+ fps with pixel resolution in the 1 micron range allow very precise temporal and spatial measurements. In addition, the high spatial resolution enhances the ability to detect even subtle changes in contractility measured as Fractional Shortening (FS, e.g. decreased FS with age; Cammarato et al. 2008). Of particular usefulness, this program produces high-resolution qualitative records of heart wall movements (M-mode, kymograph) by excising a 1 pixel horizontal “slice” of pixels from each frame of the movie and aligning them horizontally to provide an edge trace displaying heart wall movements in the X-axis and time along the Y-axis (Fig. 2C; Fink et al. 2009).
Figure 2.
SOHA output showing the gross darkness changes in (A) the top window, (B) the pixel-by-pixel intensity changes in the middle window, and (C) the corresponding M-mode in the bottom window. Adapted from Fink et al. 2009.
This analysis program can also be used with lower speed and lower resolution movies, such as of GFP fluorescent-labelled hearts (Lalevee et al. 2006, Monnier et al. 2012). However, due to the lower resolution and speed, the output will be limited to heart rate (see discussion below). This analysis method also works with movies of intact (in vivo) preparations of larval, pupal and adult Drosophila hearts (Ocorr et al. 2007b; Senatore et al. 2010), as well as larval zebrafish and day 8–9 embryonic mouse hearts (Fink et al. 2009). However, SOHA is most sensitive when using semi-dissected preparations to expose the beating heart and using direct immersion objectives. Although it takes some time to learn the dissection technique (Vogler and Ocorr 2009), the attainable speed of dissection is roughly equivalent to the time it takes to position the intact organisms and the light source for other optical methods, including OCT (see below). SOHA is not fully automatic because it is desirable to permit user input to verify and correct (when needed) the systolic and diastolic intervals detected by the combined algorithms. It also requires the user to manually mark the heart edges for the diameter and fractional shortening measurements (detailed in Fink et al. 2009, and illustrated in Ocorr et al. 2009).
Using the SOHA method, a number of disease models have been developed, including channelopathy models (Akasaka et al. 2006, Ocorr et al. 2007b), restricted and dilated cardiomyopathy models (Cammarato et al. 2008, Taghli-Lamallem et al. 2008, Cozhimuttam Viswanathan et al. 2013), diabetic cardiomyopathy models (Birse et al. 2010, Na et al. 2013), and a cardiac amyloidosis model (Melkani et al. 2013).
A more fully automated optical methodology for data collection has been also developed for intact adult flies (Feala et al. 2008). In this method the automation is achieved thanks to a customized fly deposition system and software that can recognize and orient the heart for the camera. An automated stage orients the flies such that heart wall movements can then be detected and a single line of pixels is recorded to produce a real time M-mode records from which heart rate can be determined. This methodology was used to characterize the response to acute hypoxia and could be used, with appropriate optimization, to screen for genetic modifiers of this response.
Optical Coherence Tomography (OCT)
Optical coherence tomography (OCT) uses optical wave lengths in the infrared light range to produce subsurface images. Light passing through the fly is scattered but the pattern of scattering can be used to build an image of structure lying 1–2mm below the surface using a technique referred to as interferometry. These images are referred to as A-scans and can be generated over time to provide a movie of the beating heart. These scans can also be used to generate a cross-sectional image called a B-scan. This technique has been used successfully to study the Drosophila heart in anaesthetized flies (Wolf et al. 2006). As proof of principle, the effects of mutations in muscle structural proteins on heart function were examined. Mutations in Troponin I, Tropomyosin 2 and δ-Sarcoglycan all produced a dilated cardiac phenotype in the fly model.
Because of the processing required, the image capture rate is relatively low compared to high speed optical cameras. However, higher speeds up to 100 fps have been recently reported (Tsai et al. 2011, Tsai et al. 2013, Zhou et al. 2013). The advantage of this method is that tissue structure and dynamics can be observed in vivo, however because resolution is limited to a 10 micron range (Yu et al. 2010, Zhou et al. 2013) this method is best suited to determinations of heart rate and cardiac dilation and less so for rhythmicity and contractility measurements such as fractional shortening. Finally, the equipment required for this analytical method is highly specialized and costly.
Comparisons between optical-based analysis methods
All of these optical methodologies have limitations that must be kept in mind. One is the speed of data capture. The adult fly heart beats in vivo at rates of 3–6 Hz while the denervated adult heart preparation as well as the larval heart both beat more slowly at 1–3 Hz. Unlike most other heart models, such as Zebrafish and mouse, the fly has distinct systolic and diastolic intervals (Figure 2C). In denervated hearts the heart contraction event (systolic interval) is typically shorter than the relaxation event (diastolic interval); the average systole is 0.2 – 0.25 sec, whereas the average diastole is 0.3 – 0.6 sec. Thus, the ability to precisely track the contraction event will be dependent on the rate at which the optical images are captured. For example a 30fps movie will have only 3–4 frames covering systole and 11–12 frames covering diastole, whereas a 150fps movie will have 5 times as many frames covering each interval. Thus data captured at low frame rates cannot be used to reliably quantify distinct intervals but will permit rate and crude rhythmicity measurements.
A second limitation is the image resolution. This limitation primarily comes into play when one is using images to determine the contractility based on the diameters of the heart wall during diastole and systole (fractional shortening). The heart tube in adult flies is on average 60 – 80 microns in diameter during diastole and 40 – 50 microns during systole. Ideally one would like to resolve images to at least the 1 micron level for this type of measurement. Even with adequate resolution this measurement could be affected by low frame rates which might make it difficult to identify a frame where the heart is in its most contracted state.
A related consideration concerns the ability to accurately mark the edges of the heart in intact animals for the purpose of measuring diameters. Since fat bodies are closely associated with the heart in most animals and due to the opacity of fat in light microscope, it is not possible to accurately identify the edges thus accurate fractional shortening measurements are not usually possible in intact preparations.
Finally a consideration that is unrelated to the method of data capture is that of rhythmicity measurements. In the fly heart there are two pacemaker regions that control the direction of contraction. One is located in the anterior conical chamber region; the other is located in the posterior terminal chamber. The pacemakers themselves have not been identified and may well be a “network” property of the myocardial cells themselves. Numerous studies of adult flies have documented spontaneous alterations in directionality of the contractions that are also associated with changes in beating frequency. The changes in beating frequency appear to be influenced by neuronal input because removal of the ventral nerve cord, and not just the brain, results in a more consistent and rhythmical beating pattern although directionality can still alternate (Ocorr et al. 2007b). Thus a determination of rhythmicity in intact animals will have to contend with this inherent arrhythmicity. In addition, movement artifacts present in most in vivo manipulations could be interpreted as arrhythmic heart beats so these must be identified and controlled for.
Electrophysiological and mechanical assessments of heart function
Extracellular Electrical Recording
Extracellular signals have been recorded from the exposed fly heart using standard electrical recoding techniques. All electrical recording methodologies necessitate that the heart first be exposed by dissection. Suction electrodes have been used to record extracellular signals from the posterior end of the adult fly heart (Papaefthimiou et al. 2001). This heart preparation involved loosening the posterior tip of the heart from its attachment to the abdominal cuticle and gently sucking the detached portion into a small bore glass capillary. Extracellular recordings were relatively stable over an 8 hour period and the output was used to demonstrate an excitatory effect of octopamine on the heart rate, accompanied by a reduction in the amplitude and duration of the extracellular potential.
A Multi-electrode Array (MEA) apparatus has also been used to record extracellular field potentials from semi-intact fly heart preparations (Ocorr et al. 2007b). A typical MEA plate contains indium-tin oxide or titanium microelectrodes that are typically 10 microns in diameter and spaced in a variety of grid patterns at intervals of 30 or more microns. The dimensions of the adult fly heart are roughly 80–100 microns wide and 800–1000 microns in length. Thus for standard arrays only two or three electrodes are likely to contact the heart at the same time. Nevertheless it is possible to obtain extracellular field potentials. This technique was used to show prolongation of repolarizing potentials in hearts that were mutant the KCNQ K+ channel that underlies cardiac repolarization in both flies and humans (Ocorr et al. 2007b).
Intracellular Electrical Recording
Intracellular recordings have been reported from both larval and adult heart preparations. In the larva the heart is not attached to the body wall and is relatively mobile, thus a floating electrode technique was employed to minimize damage to the heart by the impaled electrode during contractions (Lalevee et al. 2006). In this study, recordings from larval hearts were used to examine a role for the outwardly rectifying K+ channel ork1 in larval hearts. The data indicated that currents through Ork1 likely set the (resting) membrane potential and influenced the rate of spontaneous action potential generation in the heart.
Unlike in larva, the adult heart is somewhat fixed to the abdominal cuticle by the extracellular matrix and standard intracellular recording techniques have been used successfully to record from beating hearts (Dulcis and Levine 2005). In this study the authors state that the recordings were made from the ventral longitudinal muscle (VLM). This ventral layer of muscle, though not myocardial in origin, but made from progenitor cells from larval lymph glands, contains longitudinally oriented myofibrils and thus would not be expected to mechanically contribute to the heart tube contractions, which are preferentially circumferential. This notion is further supported by recent studies using atomic force microscopy that have shown that the VLM layer is much softer than the myocardial layer (Kaushik et al. 2011; see below). Thus, it is likely that the electrode had actually penetrated through the VLM and that they were recording from myocardial cells.
In both larva and adult hearts, the recorded action potentials exhibit pacemaker potentials that would be expected of a myogenic heart. However, both preparations exhibited suspiciously high resting membrane potentials (≥2212;25 mV; Dulcis and Levine 2005, Lalevee et al. 2006). This could be due to un-optimized extracellular medium and recordings or could point to a basic difference between myocardial cells from flies and other organisms. Our unpublished observations and results from a recently published study (Magny et al. 2013) record resting membrane potentials of −40 mV and lower, suggesting that the former is the case. Finally, for all electrical recording methods, damage due to movement and movement based artifacts remain a problem when recording from beating cardiac muscle.
Assessment of Muscle Mechanics
Cardiac disease and aging is often accompanied by diastolic dysfunction that has been attributed to a stiffening of the cardiac tissue, due in part to damaging passive mechanical properties of the cardiomyocytes and their extracellular milieu (Kass et al. 2004). Diastolic dysfunction is also observed in Drosophila under mutant or aging conditions (Cammarato et al. 2008), suggesting that similar changes in mechanical properties could occur in flies as in humans. One method to detect changes in muscle tension has been applied to Drosophila hearts (Ocorr et al. 2007b). This methodology was adapted from a system used to measure tension in mammalian cultured myocytes and uses tiny carbon fibers attached to a force transducer to monitor tension generation across the fly heart tube, and used to show that the ability to generate tension was significantly altered in hearts from KCNQ mutant flies.
Recently, the use of atomic force microscopy (AFM) has been applied to monitor cardiac mechanical properties of the adult Drosophila heart (Kaushik et al. 2011, Kaushik et al. 2012, Cozhimuttam Viswanathan et al. 2013). Using this approach it has been shown that the heart’s stiffness changes with age or with genetic manipulations that perturb the integrity of the contractile apparatus within cardiomyocytes. Interestingly, the age-dependent increase in cardiac stiffness observed in wildtype fly hearts was reversed with genetic manipulations that prolong lifespan and improve cardiac performance with age (Nishimura et al. 2014).
The principle of AFM and its application to the Drosophila heart (using a MFP-3D Bio Atomic Force Microscope) are described in detail in Kaushik et al. (2011). Briefly, AFM uses a nano-indentation approach to measure the stiffness of the Drosophila heart and requires a similar dissection procedure as for SOHA. A silicon nitride cantilever with a 2µm borosilicate sphere tip is placed directly on the heart and stepped down on the heart tube’s centerline. Its deflection is measured with a laser, providing Hertzian plots of force versus indentation depth, whose linearized slope is a relative measure of stiffness. Remarkably, using this technique it has been possible to identify differences in stiffness between individual muscle layers, i.e. between the contractile myocardial layer and the non-cardiac ventral layer beneath the heart (Kaushik et al. 2012).
A caveat for both of these methods of heart mechanical analysis is that the equipment is highly specialized, requires considerable experience and the techniques themselves may be damaging to heart muscle function. Nevertheless, these protocols do provide valuable information concerning some mechanical properties of the cardiac tissue that cannot as of yet be obtained by other methods.
Histological methods to assess heart development and structural integrity
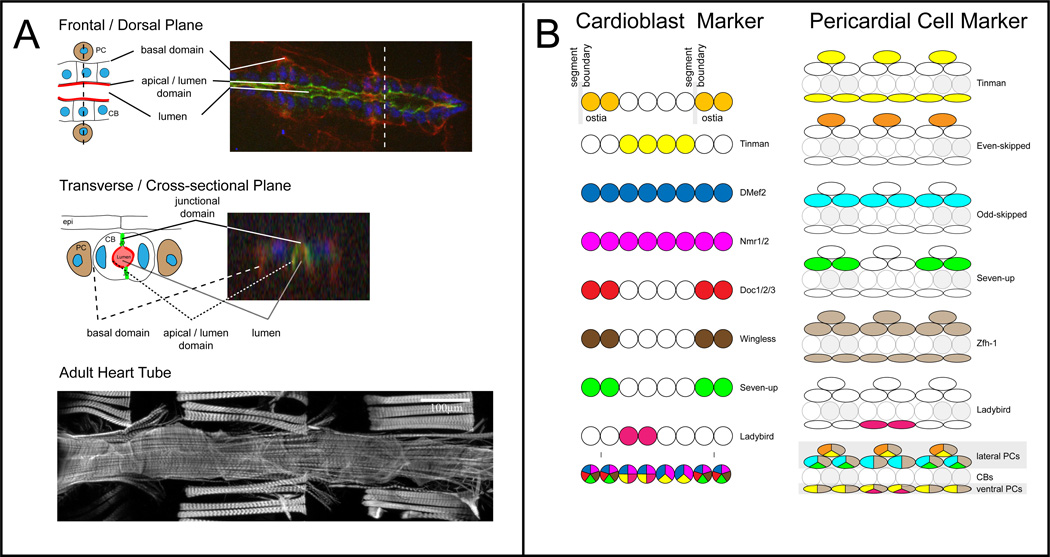
The Drosophila heart develops from cardioblasts (CB) and pericardial cells (PC), which derive from progenitors that are specified in the lateral mesoderm during mid-embryonic stages. From this region both cell types migrate towards the dorsal midline, where they assemble into a linear tube and differentiate into the beating dorsal vessel. Since the identification of the cardiac determinant Tinman, a homeobox transcription factor, additional transcription factors and signaling pathways that underlie cardiac specification have been characterized and have subsequently also been identified as relevant for vertebrate cardiogenesis (for review see Harvey and Rosenthal 2010). Specified heart precursor cells fall into 3 groups, which can be individually identified based on the combination of marker genes they express (Figure 3). Similarly, PCs can also be subdivided into different subtypes most of which likely undergo histolysis at the end of embryogenesis (Sellin et al. 2006); only ~40 PCs persist into larval stages (as nephrocyte-like cells). Antibodies and reporter lines with lacZ and GFP are available for many of the key cardiac transcription factors that control cardiac specification (see Table 2).
Figure 3. Heart Lumen Cross sections, Adult Heart Staining and Cardiac Marker genes.
A. Schematic overview and fluorescent image of the embryonic heart tube (early stage 17), with domains highlighted. Cardiac nuclei are labeled with anti-Neuromancer1 (blue), basal + luminal domains are labeled with anti-Dystroglycan (red), and the luminal domain is marked with anti-Slit (green; for more markers see Table 2). Adult heart tube stained against F-actin. B. Combinatorial code of transcription factors and signaling pathways to identify embryonic cardioblast and pericardial cell types. See Table 2 for references.
Table 2.
Overview of markers for heart specification, polarity and differentiation
| Protein | cardiac cell types |
subcellular localization |
comment | cardiac reference | source (non-cardiac reference) |
|---|---|---|---|---|---|
| Transcription factors/Specification | |||||
| DMef2 | CB | nucleus | Lilly et al. (1995) | ||
| Dorsocross1/2/3 (Doc1/2/3) | CB | nucleus | Ostia cell marker, also in amnioserosa | Reim et al. (2003) | |
| Even-Skipped (Eve) | PC | nucleus | Su et al. (1999) | DSHB clone 3C10 Frasch et al. (1987) | |
| Hand-LacZ | CB + PC | cytoplasm | lacZ reporter | Han and Olson (2005) | |
| Ladybird-early (Lbe) | CB + PC | nucleus | Jagla et al. (1997) | ||
| Nmr1/H15 | CB | nucleus | Leal et al. (2009) | ||
| Odd-Skipped (Odd) | PC | nucleus | Ward and Coulter (2000) | ||
| Seven-up-LacZ | CB | cytoplasm | lacZ reporter, Ostia cell marker | Lo and Frasch (2001) | |
| Tail-up | CB + PC | nucleus | in amnioserosa | Mann et al. (2009) | DSHB clone 40.3A4 |
| Tinman (Tin) | CB + PC | nucleus | Venkatesh et al. (2000), Yin et al. (1997) | ||
| Zfh-1 | CB + PC | nucleus | Bodmer Rolf (1993) | Ruth Lehmann | |
| Wingless (Wg) | Ostia cell marker | Lo et al. (2002) | DSHB clone 4D4 | ||
| Polarity/Cell shape | |||||
| Actin | filopodia, myofilaments | Phalloidin (Life Technologies) | |||
| alpha-Spectrin | membrane marker | Ponzielli et al. (2002) | DSHB clone 3A9 | ||
| Armadillo (Arm) | junctional domain marker | Medioni et al. (2008) | DSHB clone N2 7A1 | ||
| beta-Integrin (beta-PS1) | apical+basal | Vanderploeg et al. (2012) | DSHB clone CF.6G11 | ||
| beta3-Tubulin | Damm et al. (1998) | ||||
| DE-Cadherin | junctional domain | Haag et al. (1999) | DSHB DCAD2 | ||
| Discs-large (Dlg1) | membrane marker | Qian et al. (2005a) | DSHB clone 4F3 | ||
| Dystroglycan (Dg) | apical+basal | Qian et al. (2005a) | Deng et al. (2003) | ||
| Multiplexin | heart proper lumen (st.17) | Harpaz et al. (2013) | |||
| NetrinB | luminal domain | Albrecht et al. (2011) | |||
| Robo | luminal domain | Qian et al. (2005b) | DSHB clone 13C9 | ||
| Robo2 (lea) | PC | Qian et al. (2005b) | Rajagopalan et al. (2000) | ||
| Slit | aorta + heart proper lumen | Rothberg et al. (1990) | DSHB clone C555.6D | ||
| Toll (Tl) | membrane marker | Wang et al. (2005) | Hashimoto et al. (1991) | ||
| Unc5 | luminal domain | Albrecht et al. (2011) | |||
| Differentiation | |||||
| Pericardin (Prc) | Chartier et al. (2002) | DSHB clone EC11 | |||
| Tropomyosin | LaBeau et al. (2009) | Abcam ab50567 | |||
| Myosin-heavy chain (Mhc) | LaBeau et al. (2009) | Kiehart and Feghali (1986) | |||
| DSur | RNA probe | Akasaka et al. (2006) | |||
From stage 13 to 16 of embryonic development, the heart precursors facilitate two key steps of morphogenesis: the migration and alignment of the cardioblasts at the dorsal midline and the formation of the heart lumen. These steps require the activity of cell polarity and cell adhesion proteins and a number of genes that are involved in the formation of most or all tissues (such as Laminins, DE-Cadherin, see Haag et al. 1999, Yarnitzky and Volk 1995), as well as signaling pathways that have a more restricted, tissue-specific pattern of expression, such as Slit/Robo or Net/Unc5 (Albrecht et al. 2011, Macabenta et al. 2013, MacMullin and Jacobs 2006, Medioni et al. 2008, Qian et al. 2005b, Santiago-Martínez et al. 2006). At this stage, the cardioblasts appear columnar, with distinct localization of basement-membrane markers at the apical/dorsal and basal/ventral side as well as cell-junction markers present at the interface between neighboring CBs. Once the CBs have established the dorsal contact between the junctional domains, they undergo cell shape changes and form a second, ventral cell-cell contact that encloses a single lumen (as originally described in Rugendorff et al. 1994). During these steps, the shape, polarity and identity of all CBs can be determined using a battery of antibodies (summarized in Table 2). Finally, CBs differentiate into beating cardiomyocytes, a process that includes the onset of expression of muscle-myosin and its incorporation into myofibrils (Lehmacher et al. 2012).
For the immunohistochemical analysis of protein expression and localization, standard protocols of Drosophila embryo fixation can be applied (Sullivan et al. 2000). In general, embryos from overnight collections are fixed using Formaldehyde/PBS/Heptane/MetOH-based protocols, and embryos of the correct stage are selected during mounting of specimens (staging according to Hartenstein 1993). Alternatively, timed embryo collections can be used to limit the embryos to relevant stages, but care has to be taken to carry over a sufficient number of embryos during all staining and washing steps. While standard fixation works for the majority of antibodies, there are several antibodies that do require an optimized protocol to preserve the localization pattern or epitope. To detect F-actin with fluorescence-labeled phalloidin exposure to methanol needs to be avoided; this can be accomplished by either by hand-devitellinization of embryos or by replacing methanol with 80–90% ethanol in the devitellinization step (Narashima and Brown 2006). Both methods will result in a limited number of usable embryos, which is disadvantageous when an embryo of a rare genotype and specific stage is to be selected. Heatfixation offers an alternative for some antibodies, since it allows for better visualization of cortical proteins such as Armadillo or DE-Cadherin (Müller and Wieschaus 1996).
Because the heart is localized to the dorsal surface it can easily be imaged from wholemounts. While the central part of the dorsal vessel is very close to the epidermis (8–10µm), high quality images can be obtained with high-NA objectives. The posterior heart proper descends another 25µm into the tissue, adding significantly more, but usually acceptable noise. Filleting the embryo along the ventral midline can improve the image quality (Lovato et al. 2002), but this also increases the risk of damaging the specimen. To bring the coverslip as close as possible to the embryo but without compressing it, a bridge can be built with No. 1-sized coverslips as spacers (No. 1.5 coverslips would increase the space between the embryo and the top coverslip, resulting in a reduced image quality at high resolution). This allows to reliably image embryonic hearts to depths of 35µm even when using laser confocal microscopes or epifluorescent microscopes in combination with structured illumination.
For the analysis of cardiac cell specification, migration and alignment whole-mount imaging along the D/V axis is usually sufficient; however the three-dimensional properties of heart assembly are not optimally captured with confocal microscopy due to the lower axial resolution. The diameter of the embryonic heart tube is only 2–5µm, and with a typical axial resolution of only about 0.7µm the luminal properties are poorly reflected by Z-scans. While gross-morphological lumen defects can be identified using whole-mounts, using cross-sections of embryos and imaging along the A/P axis results in images of much higher resolution (Harpaz and Volk 2011, Macabenta et al. 2013). These methods reveal clearer defects of cardioblast shape and heart lumen that would not have been resolved otherwise. However, since heart lumen formation is a dynamic process that does not occur synchronously along the heart but at different time points along A/P positions (aorta versus heart proper), and is presumably different depending on cardioblast cell type (ostia lumen versus cardiomyocyte lumen), it is important to register sections according to their position along the A/P axis for the interpretation of phenotypes.
Due to the dynamic nature of heart morphogenesis, live-imaging offers an alternative to follow cardiac development over time. In Drosophila, many fluorescent reporter lines such as Moesin-RFP/Actin-GFP exist and have been successfully applied to study aspects of heart tube assembly (Knox et al. 2011, Medioni et al. 2008, Vanderploeg et al. 2012). The dorsal, subcutaneous location of the heart allows easily obtainable high-quality confocal images of the heart as it undergoes morphogenesis.
The larval and adult heart can also be characterized using immunohistochemical methods (Alayari et al. 2009). In general, visualizing the structural proteins of the heart such as proteins of the sarcomere, (e.g. phalloidin staining of F-actin, Myosin or the Z-band marker α-Actinin (Mery et al. 2008)), allows reliable detection of cardiac defects in overall morphology and myofibrillar organization. In addition, antibodies against extracellular proteins like the collagen-IV Pericardin provide information about the extracellular matrix surrounding the heart (Na et al. 2013). GFP fusion proteins have been used to detect subcellular localization of sarcomeric proteins (Mery et al. 2008), human genes expressed in the fly heart such as α-B-crystalin (Xie et al. 2013) and amyloid aggregates (Melkani et al. 2013). Finally, electron microscopy has been used to examine the adult heart tube; especially noteworthy is the work from A. Paululat’s group on the ultrastructure of the wildtype fly heart (Lehmacher et al. 2012).
Summary
The Drosophila heart model provides an accessible and easily manipulated system for the analysis of cardiac development as well as function. The ease of genetic manipulation, limited genetic complexity and conservation of gene function with vertebrates makes it an ideal system for the identification of gene candidates and mechanisms involved in vertebrate heart development, function and disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akasaka T, Klinedinst S, Ocorr K, Bustamante EL, Kim SK, Bodmer R. The ATP-sensitive potassium (KATP) channel-encoded dSUR gene is required for Drosophila heart function and is regulated by tinman. Akasaka T, Klinedinst. 2006b;103:11999–12004. doi: 10.1073/pnas.0603098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht S, Altenhein B, Paululat A. The transmembrane receptor Uncoordinated5 (Unc5) is essential for heart lumen formation in Drosophila melanogaster. Dev Biol. 2011;350:89–100. doi: 10.1016/j.ydbio.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Alayari NN, Vogler G, Taghli-Lamallem O, Ocorr K, Bodmer R, Cammarato A. Fluorescent labeling of Drosophila heart structures. J Vis Exp. 2009;pii:1423. doi: 10.3791/1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse RT, Choi J, Reardon K, Rodriguez J, Graham S, Diop S, Ocorr K, Bodmer R, Oldham S. High-fat-diet-induced obesity and heart dysfunction are regulated by the TOR pathway in Drosophila. Cell Metab. 2010;12:533–544. doi: 10.1016/j.cmet.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- Bodmer R. Heart development in Drosophila and its relationship to vertebrate systems. Trends Cardiovasc Med. 1995;5:21–27. doi: 10.1016/1050-1738(94)00032-Q. [DOI] [PubMed] [Google Scholar]
- Bodmer R, Frasch M. Development and Aging of the Drosophila Heart. Pages 47–86. In: Rosenthal N, Harvey R, editors. Heart Development and Regeneration. Vol. 2. Amsterdam: Elsevier; 2010. [Google Scholar]
- Cammarato A, Dambacher CM, Knowles AF, Kronert WA, Bodmer R, Ocorr K, Bernstein SI. Myosin transducer mutations differentially affect motor function, myofibril structure, and the performance of skeletal and cardiac muscles. Mol Biol Cell. 2008;19:553–562. doi: 10.1091/mbc.E07-09-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier A, Zaffran S, Astier M, Sémériva M, Gratecos D. Pericardin, a Drosophila type IV collagen-like protein is involved in the morphogenesis and maintenance of the heart epithelium during dorsal ectoderm closure. Development. 2002;129:3241–3253. doi: 10.1242/dev.129.13.3241. [DOI] [PubMed] [Google Scholar]
- Cozhimuttam Viswanathan M, Kaushik G, Engler AJ, Lehman WJ, Cammarato A. A Drosophila Melanogaster Model of Diastolic Dysfunction and Cardiomyopathy Based on Impaired Troponin-T Function. Circ Res. 2014;114:e6–e17. doi: 10.1161/CIRCRESAHA.114.302028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripps RM, Olson EN. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev Biol. 2002;246:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- Damm C, Wolk A, Buttgereit D, Löher K, Wagner E, Lilly B, Olson EN, Hasenpusch-Theil K, Renkawitz-Pohl R. Independent regulatory elements in the upstream region of the Drosophila beta 3 tubulin gene (beta Tub60D) guide expression in the dorsal vessel and the somatic muscles. Dev Biol. 1998;199:138–149. doi: 10.1006/dbio.1998.8916. [DOI] [PubMed] [Google Scholar]
- Deng W-M, Schneider M, Frock R, Castillejo-Lopez C, Gaman EA, Baumgartner S, Ruohola-Baker H. Dystroglycan is required for polarizing the epithelial cells and the oocyte in Drosophila. Development. 2003;130:173–184. doi: 10.1242/dev.00199. [DOI] [PubMed] [Google Scholar]
- Dowse H, Ringo J, Power J, Johnson E, Kinney K, White L. A congenital heart defect in Drosophila caused by an action-potential mutation. J Neurogenet. 1995;10:153–168. doi: 10.3109/01677069509083461. [DOI] [PubMed] [Google Scholar]
- Dowse HB, Dushay MS, Hall JC, Ringo JM. High-resolution analysis of locomotor activity rhythms in disconnected, a visual-system mutant of Drosophila melanogaster. Behav Genet. 1989;19:529–542. doi: 10.1007/BF01066252. [DOI] [PubMed] [Google Scholar]
- Dowse HB, Ringo JM. Comparisons between“periodograms” and spectral analysis: apples are apples after all. J Theor Biol. 1991;148:139–144. doi: 10.1016/s0022-5193(05)80468-0. [DOI] [PubMed] [Google Scholar]
- Dulcis D, Levine RB. Glutamatergic innervation of the heart initiates retrograde contractions in adult Drosophila melanogaster. J Neurosci. 2005;25:271–280. doi: 10.1523/JNEUROSCI.2906-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feala JD, Omens JH, Paternostro G, McCulloch AD. Discovering regulators of the Drosophila cardiac hypoxia response using automated phenotyping technology. Ann N Y Acad Sci. 2008;1123:169–177. doi: 10.1196/annals.1420.019. [DOI] [PubMed] [Google Scholar]
- Fink M, Callol-Massot C, Chu A, Ruiz-Lozano P, Izpisua Belmonte JC, Giles W, Bodmer R, Ocorr K. A new method for detection and quantification of heartbeat parameters in Drosophila, zebrafish, and embryonic mouse hearts. BioTechniques. 2009;46:101–113. doi: 10.2144/000113078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M, Hoey T, Rushlow C, Doyle H, Levine M. Characterization and localization of the even-skipped protein of Drosophila. The EMBO Journal. 1987;6:749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu GG, Singh S. Pharmacological analysis of heartbeat in Drosophila. J Neurobiol. 1995;28:269–280. doi: 10.1002/neu.480280302. [DOI] [PubMed] [Google Scholar]
- Haag TA, Haag NP, Lekven AC, Hartenstein V. The role of cell adhesion molecules in Drosophila heart morphogenesis: faint sausage, shotgun/DE-cadherin, and laminin A are required for discrete stages in heart development. Dev Biol. 1999;208:56–69. doi: 10.1006/dbio.1998.9188. [DOI] [PubMed] [Google Scholar]
- Han Z, Olson EN. Hand is a direct target of Tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Development. 2005;132:3525–3536. doi: 10.1242/dev.01899. [DOI] [PubMed] [Google Scholar]
- Harpaz N, Ordan E, Ocorr K, Bodmer R, Volk T. Multiplexin Promotes Heart but Not Aorta Morphogenesis by Polarized Enhancement of Slit/Robo Activity at the Heart Lumen. PLoS Genetics. 2013;9:e1003597. doi: 10.1371/journal.pgen.1003597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpaz N, Volk T. A novel method for obtaining semi-thin cross sections of the Drosophila heart and their labeling with multiple antibodies. Methods. 2012;56:63–68. doi: 10.1016/j.ymeth.2011.09.017. [DOI] [PubMed] [Google Scholar]
- Hartenstein V. Atlas of Drosophila development. Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Harvey RP, Rosenthal N, editors. Heart Development and Regeneration. 1 edition. Academic Press; 2010. (June 18, 2010) [Google Scholar]
- Hashimoto C, Gerttula S, Anderson KV. Plasma membrane localization of the Toll protein in the syncytial Drosophila embryo: importance of transmembrane signaling for dorsal-ventral pattern formation. Development. 1991;111:1021–1028. doi: 10.1242/dev.111.4.1021. [DOI] [PubMed] [Google Scholar]
- Jagla K, Jagla T, Heitzler P, Dretzen G, Bellard F, Bellard M. ladybird, a tandem of homeobox genes that maintain late wingless expression in terminal and dorsal epidermis of the Drosophila embryo. Development. 1997;124:91–100. doi: 10.1242/dev.124.1.91. [DOI] [PubMed] [Google Scholar]
- Johnson E, Ringo J, Bray N, Dowse H. Genetic and pharmacological identification of ion channels central to the Drosophila cardiac pacemaker. J Neurogenet. 1998;12:1–24. doi: 10.3109/01677069809108552. [DOI] [PubMed] [Google Scholar]
- Kass DA, Bronzwaer JG, Paulus WJ. What mechanisms underlie diastolic dysfunction in heart failure? Circ Res. 2004;94:1533–1542. doi: 10.1161/01.RES.0000129254.25507.d6. [DOI] [PubMed] [Google Scholar]
- Kaushik G, Fuhrmann A, Cammarato A, Engler AJ. In situ mechanical analysis of myofibrillar perturbation and aging on soft, bilayered Drosophila myocardium. Biophys J. 2011;101:2629–2637. doi: 10.1016/j.bpj.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik G, Zambon AC, Fuhrmann A, Bernstein SI, Bodmer R, Engler AJ, Cammarato A, Mercola M. Measuring passive myocardial stiffness in Drosophila melanogaster to investigate diastolic dysfunction. J Cell Mol Med. 2012;16:1656–1662. doi: 10.1111/j.1582-4934.2011.01517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Feghali R. Cytoplasmic myosin from Drosophila melanogaster. The J Cell Biol. 1986;103:1517–1525. doi: 10.1083/jcb.103.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IM, Wolf MJ, Rockman HA. Gene deletion screen for cardiomyopathy in adult Drosophila identifies a new notch ligand. Circ Res. 2010;106:1233–1243. doi: 10.1161/CIRCRESAHA.109.213785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YO, Park SJ, Balaban RS, Nirenberg M, Kim Y. A functional genomic screen for cardiogenic genes using RNA interference in developing Drosophila embryos. Proc Natl Acad Sci U S A. 2004;101:159–164. doi: 10.1073/pnas.0307205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox J, Moyer K, Yacoub N, Soldaat C, Komosa M, Vassilieva K, Wilk R, Hu J, Vasquez Pas LdL, Syed Q, Krause HM, Georgescu M, Jacobs JR. Syndecan contributes to heart cell specification and lumen formation during Drosophila cardiogenesis. Dev Biol. 2011;356:279–290. doi: 10.1016/j.ydbio.2011.04.006. [DOI] [PubMed] [Google Scholar]
- LaBeau EM, Trujillo DL, Cripps RM. Bithorax complex genes control alary muscle patterning along the cardiac tube of Drosophila. Mech Dev. 2009;126:478–486. doi: 10.1016/j.mod.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalevee N, Monier B, Senatore S, Perrin L, Semeriva M. Control of cardiac rhythm by ORK1, a Drosophila two-pore domain potassium channel. Curr Biol. 2006;16:1502–1508. doi: 10.1016/j.cub.2006.05.064. [DOI] [PubMed] [Google Scholar]
- Leal SM, Qian L, Lacin H, Bodmer R, Skeath JB. Neuromancer1 and Neuromancer2 regulate cell fate specification in the developing embryonic CNS of Drosophila melanogaster. Dev Biol. 2009;325:138–150. doi: 10.1016/j.ydbio.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmacher C, Abeln B, Paululat A. The ultrastructure of Drosophila heart cells. Arthropod Struct Dev. 2012;41:459–474. doi: 10.1016/j.asd.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- Lo PCH, Frasch M. A role for the COUP-TF-related gene seven-up in the diversification of cardioblast identities in the dorsal vessel of Drosophila. Mech Dev. 2001;104:49–60. doi: 10.1016/s0925-4773(01)00361-6. [DOI] [PubMed] [Google Scholar]
- Lo PCH, Skeath JB, Gajewski K, Schulz RA, Frasch M. Homeotic genes autonomously specify the anteroposterior subdivision of the Drosophila dorsal vessel into aorta and heart. Dev Biol. 2002;251:307–319. doi: 10.1006/dbio.2002.0839. [DOI] [PubMed] [Google Scholar]
- Lovato TL, Nguyen TP, Molina MR, Cripps RM. The Hox gene abdominal-A specifies heart cell fate in the Drosophila dorsal vessel. Development. 2002;129:5019–5027. doi: 10.1242/dev.129.21.5019. [DOI] [PubMed] [Google Scholar]
- Macabenta FD, Jensen AG, Cheng Y-S, Kramer JJ, Kramer SG. Frazzled/DCC facilitates cardiac cell outgrowth and attachment during Drosophila dorsal vessel formation. Dev Biol. 2013;380:233–242. doi: 10.1016/j.ydbio.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMullin A, Jacobs JR. Slit coordinates cardiac morphogenesis in Drosophila. Dev Biol. 2006;293:154–164. doi: 10.1016/j.ydbio.2006.01.027. [DOI] [PubMed] [Google Scholar]
- Magny EG, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA, Couso JP. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013;341:1116–1120. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- Mann T, Bodmer R, Pandur P. The Drosophila homolog of vertebrate Islet1 is a key component in early cardiogenesis. Development. 2009;136:317–326. doi: 10.1242/dev.022533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medioni C, Astier M, Zmojdzian M, Jagla K, Semeriva M. Genetic control of cell morphogenesis during Drosophila melanogaster cardiac tube formation. J Cell Biol. 2008;182:249–261. doi: 10.1083/jcb.200801100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkani GC, Trujillo AS, Ramos R, Bodmer R, Bernstein SI, Ocorr K. Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Drosophila Heart. PLoS Genet. 2013;9:e1004024. doi: 10.1371/journal.pgen.1004024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mery A, Taghli-Lamallem O, Clark KA, Beckerle MC, Wu X, Ocorr K, Bodmer R. The Drosophila muscle LIM protein, Mlp84B, is essential for cardiac function. J Exp Biol. 2008;211:15–23. doi: 10.1242/jeb.012435. [DOI] [PubMed] [Google Scholar]
- Molina MR, Cripps RM. Ostia, the inflow tracts of the Drosophila heart, develop from a genetically distinct subset of cardial cells. Mech Dev. 2001;109:51–59. doi: 10.1016/s0925-4773(01)00509-3. [DOI] [PubMed] [Google Scholar]
- Monnier V, Iche-Torres M, Rera M, Contremoulins V, Guichard C, Lalevee N, Tricoire H, Perrin L. dJun and Vri/dNFIL3 are major regulators of cardiac aging in Drosophila. PLoS Genet. 2012;8:e1003081. doi: 10.1371/journal.pgen.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller HA, Wieschaus E. armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. The Journal of Cell Biology. 1996;134:149–163. doi: 10.1083/jcb.134.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na J, Musselman LP, Pendse J, Baranski TJ, Bodmer R, Ocorr K, Cagan R. A Drosophila model of high sugar diet-induced cardiomyopathy. PLoS Genet. 2013;9:e1003175. doi: 10.1371/journal.pgen.1003175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narashima M, Brown NH. In: Confocal Microscopy of Drosophila Embryos. Celis JE, editor. Vol. 1. Academic Press; 2006. [Google Scholar]
- Neely GG, Kuba K, Cammarato A, Isobe K, Amann S, Zhang L, Murata M, Elmén L, Gupta V, Arora S, Sarangi R, Dan D, Fujisawa S, Usami8 D, Xia C-P, Keene AC, Alayari NN, Yamakawa H, Elling U, Berger C, Novatchkova M, Koglgruber R, Fukuda K, Nishina H, Isobe M, Pospisilik JA, Imai Y, Pfeufer A, Hicks AA, Pramstaller PP, Subramaniam S, Kimura A, Ocorr K, Bodmer R, Penninger JM. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 2010;141:142–153. doi: 10.1016/j.cell.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerbonne JM. Studying cardiac arrhythmias in the mouse--a reasonable model for probing mechanisms? Trends Cardiovasc Med. 2004;14:83–93. doi: 10.1016/j.tcm.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Kumsta C, Kaushik G, Diop SB, Ding Y, Bisharat-Kernizan J, Catan H, Cammarato A, Ross RS, Engler AJ, Bodmer R, Hansen M, Ocorr K. A dual role for integrin-linked kinase and beta1-integrin in modulating cardiac aging. Aging Cell. 2014 doi: 10.1111/acel.12193. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M, Ocorr K, Bodmer R, Cartry J. Drosophila as a model to study cardiac aging. Exp Gerontol. 2011;46:326–330. doi: 10.1016/j.exger.2010.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K, Akasaka T, Bodmer R. Age-related cardiac disease model of Drosophila. Mech Ageing Dev. 2007a;128:112–116. doi: 10.1016/j.mad.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K, Fink M, Cammarato A, Bernstein S, Bodmer R. Semi-automated Optical Heartbeat Analysis of small hearts. J Vis Exp. 2009;pii:1435. doi: 10.3791/1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K, Reeves NL, Wessells RJ, Fink M, Chen HS, Akasaka T, Yasuda S, Metzger JM, Giles W, Posakony JW, Bodmer R. KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc Natl Acad Sci U S A. 2007b;104:3943–3948. doi: 10.1073/pnas.0609278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaefthimiou I, Hamilton A, Denti M, Baulcombe D, Tsagris M, Tabler M. Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res. 2001;29:2395–2400. doi: 10.1093/nar/29.11.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternostro G, Vignola C, Bartsch DU, Omens JH, McCulloch AD, Reed JC. Age-associated cardiac dysfunction in Drosophila melanogaster. Circ Res. 2001;88:1053–1058. doi: 10.1161/hh1001.090857. [DOI] [PubMed] [Google Scholar]
- Ponzielli R, Astier M, Chartier A, Gallet A, Thérond P, Sémériva M. Heart tube patterning in Drosophila requires integration of axial and segmental information provided by the Bithorax Complex genes and hedgehog signaling. Development. 2002;129:4509–4521. doi: 10.1242/dev.129.19.4509. [DOI] [PubMed] [Google Scholar]
- Qian L, Liu J, Bodmer R. Neuromancer Tbx20-related genes (H15/midline) promote cell fate specification and morphogenesis of the Drosophila heart. Dev Biol. 2005a;279:509–524. doi: 10.1016/j.ydbio.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Qian L, Liu J, Bodmer R. Slit and Robo control cardiac cell polarity and morphogenesis. Curr Biol. 2005b;15:2271–2278. doi: 10.1016/j.cub.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Qian L, Mohapatra B, Akasaka T, Liu J, Ocorr K, Towbin JA, Bodmer R. Transcription factor neuromancer/TBX20 is required for cardiac function in Drosophila with implications for human heart disease. Proc Natl Acad Sci U S A. 2008;105:19833–19838. doi: 10.1073/pnas.0808705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Wythe JD, Liu J, Cartry J, Vogler G, Mohapatra B, Otway RT, Huang Y, KIng J, Maillet M, Zheng Y, Crawley T, Taghli-Lamallem O, Semsarian C, Dunwoodie S, Winlaw D, Harvey RP, Fatkin DA, Towbin JA, Molkentin JD, Srivastava D, Ocorr K, Bruneau B, Bodmer R. Tinman/Nkx2–5 acts via miR-1 and upstream of Cdc42 to regulate heart function across species J. Cell Biol. 2011;193:1181–1196. doi: 10.1083/jcb.201006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Vivancos V, Nicolas E, Dickson BJ. Selecting a longitudinal pathway: Robo receptors specify the lateral position of axons in the Drosophila CNS. Cell. 2000;103:1033–1045. doi: 10.1016/s0092-8674(00)00207-5. [DOI] [PubMed] [Google Scholar]
- Reim I, Lee H-H, Frasch M. The T-box-encoding Dorsocross genes function in amnioserosa development and the patterning of the dorsolateral germ band downstream of Dpp. Development. 2003;130:3187–3204. doi: 10.1242/dev.00548. [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Jacobs JR, Goodman CS, Artavanis-Tsakonas S. Slit: an extracellular protein necessary for development of midline glia and commissural axon pathways contains both EGF and LRR domains. Genes Dev. 1990;4:2169–2187. doi: 10.1101/gad.4.12a.2169. [DOI] [PubMed] [Google Scholar]
- Rugendorff A, Younossi-Hartenstein A, Hartenstein V. Embryonic origin and differentiation of the Drosophila heart. Roux’s Arch Dev Biol. 1994;203:266–280. doi: 10.1007/BF00360522. [DOI] [PubMed] [Google Scholar]
- Santiago-Martínez E, Soplop NH, Kramer SG. Lateral positioning at the dorsal midline: Slit and Roundabout receptors guide Drosophila heart cell migration. Proc Natl Acad Sci U S A. 2006;103:12441–12446. doi: 10.1073/pnas.0605284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin J, Albrecht S, Kölsch V, Paululat A. Dynamics of heart differentiation, visualized utilizing heart enhancer elements of the Drosophila melanogaster bHLH transcription factor Hand. Gene Expr Patterns. 2006;6:360–375. doi: 10.1016/j.modgep.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Senatore S, Rami Reddy V, Semeriva M, Perrin L, Lalevee N. Response to mechanical stress is mediated by the TRPA channel painless in the Drosophila heart. PLoS Genet. 2010;6:e1001088. doi: 10.1371/journal.pgen.1001088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su MT, Fujioka M, Goto T, Bodmer R. The Drosophila homeobox genes zfh-1 and even-skipped are required for cardiac-specific differentiation of a numb-dependent lineage decision. Development. 1999;126:3241–3251. doi: 10.1242/dev.126.14.3241. [DOI] [PubMed] [Google Scholar]
- Sullivan W, Ashburner M, Hawley RS. Drosophila protocols. Cold Spring Harbor Laboratory Pr; 2000. [Google Scholar]
- Taghli-Lamallem O, Akasaka T, Hogg G, Nudel U, Yaffe D, Chamberlain JS, Ocorr K, Bodmer R. Dystrophin deficiency in Drosophila reduces lifespan and causes a dilated cardiomyopathy phenotype. Aging Cell. 2008;7:237–249. doi: 10.1111/j.1474-9726.2008.00367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MT, Chang FY, Lee CK, Chi TT, Yang KM, Lin LY, Wu JT, Yang CC. Observations of cardiac beating behaviors of wild-type and mutant Drosophilae with optical coherence tomography. J Biophotonics. 2011;4:610–618. doi: 10.1002/jbio.201100009. [DOI] [PubMed] [Google Scholar]
- Tsai MT, Lee CK, Chang FY, Wu JT, Wu CP, Chi TT, Yang CC. Noninvasive imaging of heart chamber in Drosophila with dual-beam optical coherence tomography. J Biophotonics. 2013;6:708–717. doi: 10.1002/jbio.201200164. [DOI] [PubMed] [Google Scholar]
- Vanderploeg J, Vazquez Paz LL, MacMullin A, Jacobs JR. Integrins are required for cardioblast polarisation in Drosophila. BMC Dev Biol. 2012;12:8. doi: 10.1186/1471-213X-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh TV, Park M, Ocorr KA, Nemaceck J, Golden K, Wemple M, Bodmer R. Cardiac enhancer activity of the homeobox gene tinman depends on CREB consensus binding sites in Drosophila. Genesis. 2000;26:55–66. [PubMed] [Google Scholar]
- Vogler G, Ocorr K. Visualizing the beating heart in Drosophila. J Vis Exp. 2009;pii:1425. doi: 10.3791/1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tao Y, Reim I, Gajewski K, Frasch M, Schulz RA. Expression, regulation, and requirement of the toll transmembrane protein during dorsal vessel formation in Drosophila melanogaster. Mol Cell Biol. 2005;25:4200–4210. doi: 10.1128/MCB.25.10.4200-4210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward EJ, Coulter DE. odd-skipped is expressed in multiple tissues during Drosophila embryogenesis. Mechanisms of development. 2000;96:233–236. doi: 10.1016/s0925-4773(00)00389-0. [DOI] [PubMed] [Google Scholar]
- Wasserthal LT. Drosophila flies combine periodic heartbeat reversal with a circulation in the anterior body mediated by a newly discovered anterior pair of ostial valves and 'venous' channels. J Exp Biol. 2007;210:3707–3719. doi: 10.1242/jeb.007864. [DOI] [PubMed] [Google Scholar]
- Wessells RJ, Bodmer R. Screening assays for heart function mutants in Drosophila. BioTechniques. 2004;37:58–60. doi: 10.2144/04371ST01. [DOI] [PubMed] [Google Scholar]
- Wessells RJ, Fitzgerald E, Cypser JR, Tatar M, Bodmer R. Insulin regulation of heart function in aging fruit flies. Nat Genet. 2004;36:1275–1281. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- White L, Ringo J, Dowse H. A circadian clock of Drosophila: effects of deuterium oxide and mutations at the period locus. Chronobiol Int. 1992;9:250–259. doi: 10.3109/07420529209064534. [DOI] [PubMed] [Google Scholar]
- Wolf MJ, Amrein H, Izatt JA, Choma MA, Reedy MC, Rockman HA. Drosophila as a model for the identification of genes causing adult human heart disease. Proc Natl Acad Sci U S A. 2006;103:1394–1399. doi: 10.1073/pnas.0507359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie HB, Cammarato A, Rajasekaran NS, Zhang H, Suggs JA, Lin H-C, Berstein SI, Benjamin IJ, Golic KG. The NADPH Metabolic Network Regulates Human aBcrystallin Cardiomyopathy and Reductive Stress in Drosophila melanogaster. PLoS Genet. 2013;9:e1003544. doi: 10.1371/journal.pgen.1003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnitzky T, Volk T. Laminin is required for heart, somatic muscles, and gut development in the Drosophila embryo. Dev Biol. 1995;169:609–618. doi: 10.1006/dbio.1995.1173. [DOI] [PubMed] [Google Scholar]
- Yin Z, Xu XL, Frasch M. Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Development. 1997;124:4971–4982. doi: 10.1242/dev.124.24.4971. [DOI] [PubMed] [Google Scholar]
- Yu L, Lee T, Lin N, Wolf MJ. Affecting Rhomboid-3 function causes a dilated heart in adult Drosophila. PLoS Genet. 2010;6:e1000969. doi: 10.1371/journal.pgen.1000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitouni B, Senatore S, Severac D, Aknin C, Semeriva M, Perrin L. Signalling pathways involved in adult heart formation revealed by gene expression profiling in Drosophila. PLoS Genet. 2007;3:1907–1921. doi: 10.1371/journal.pgen.0030174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Alex A, Rasakanthan J, Ma Y. Space-division multiplexing optical coherence tomography. Opt Express. 2013;21:19219–19227. doi: 10.1364/OE.21.019219. [DOI] [PMC free article] [PubMed] [Google Scholar]