Abstract
Background:
Lung cancer is a leading cause of mortality, and patients often present at a late stage. More recently, advances in screening, diagnosing, and treating lung cancer have been made. For instance, greater numbers of minimally invasive procedures are being performed, and identification of lung adenocarcinoma driver mutations has led to the implementation of targeted therapies. Advances in molecular techniques enable use of scant tissue, including cytology specimens. In addition, per recently published consensus guidelines, cytology-derived cell blocks (CBs) are preferred over direct smears. Yet, limited comparison of molecular testing of fine-needle aspiration (FNA) CBs and corresponding histology specimens has been performed. This study aimed to establish concordance of epidermal growth factor receptor (EGFR) and Kirsten rat sarcoma (KRAS) virus homolog testing between FNA CBs and histology samples from the same patients.
Materials and Methods:
Patients for whom molecular testing for EGFR or KRAS was performed on both FNA CBs and histology samples containing lung adenocarcinoma were identified retrospectively. Following microdissection, when necessary, concordance of EGFR and KRAS molecular testing results between FNA CBs and histology samples was evaluated.
Results:
EGFR and/or KRAS testing was performed on samples obtained from 26 patients. Concordant results were obtained for all EGFR (22/22) and KRAS (17/17) mutation analyses performed.
Conclusions:
Identification of mutations in lung adenocarcinomas affects clinical decision-making, and it is important that results from small samples be accurate. This study demonstrates that molecular testing on cytology CBs is as sensitive and specific as that on histology.
Keywords: Cell blocks, cytopathology, endobronchial ultrasound-guided, epidermal growth factor receptor, fine needle aspiration, Kirsten rat sarcoma, lung adenocarcinoma, molecular testing
INTRODUCTION
Lung carcinoma is the most common cause of cancer mortality world-wide.[1] Because patients often present at a late stage, average 5-year survival is approximately 15%. Non-small cell lung carcinoma (NSCLC) pathology predominates, representing approximately 80% of all lung carcinomas. Recently, adenocarcinoma surpassed squamous cell carcinoma as the most common form of NSCLC in the United States.[2,3] Because of differences in therapy, distinguishing adenocarcinoma from squamous cell carcinoma by morphology and/or immunohistochemistry has become crucial. Furthermore, the introduction of tyrosine kinase inhibitors (TKIs) heralds entry into an era of personalized medicine, where genotype identification and not solely phenotype is critical to optimizing therapy.[4,5] Identification of mutations in adenocarcinomas, including in epidermal growth factor receptor (EGFR) or Kirsten rat sarcoma (KRAS) virus oncogene homolog, among others, determines eligibility for treatment with targeted therapies, clinical trials, and/or ruling out TKI-therapy.[6,7,8,9,10] Evaluating mutation status before initiating therapy is important because TKI administration to patients with unknown mutation status is, overall, detrimental rather than beneficial.[11]
Introduction of target-specific therapy for lung adenocarcinoma has coincided with the performance of greater numbers of minimally invasive procedures, including computed tomography-guided (CT-guided) fine-needle aspiration (FNA) biopsy, endobronchial ultrasound-guided (EBUS) FNA biopsy, and navigational bronchoscopic FNA biopsy for diagnosis and/or staging of pulmonary and mediastinal lesions or serial testing to monitor therapeutic response and/or drug resistance.[11,12,13] Because resection of locally advanced or metastatic disease as first line therapy is discouraged, fewer than 30% of patients are surgical candidates at diagnosis.[14]
In contrast to histopathologic resection specimens, cytologic specimens, including FNA biopsies and exfoliative specimens, have relatively lower cellular volume and may represent the only tissue available for immunohistochemical and/or molecular testing. There is concern by some as to whether cytology specimens are acceptable for molecular testing.[4,5] Routine use of procedures that provide higher quantities of tissue has been proposed because low tumor volume or proportion may theoretically cause mutant alleles to fall below the level of detection and be masked by non-neoplastic cells resulting in reporting of false negative results.[5,15] Alternatively, a potential concern is that cytology specimens may not be representative since carcinomas demonstrate intratumoral histologic and genetic heterogeneity, and a mutated clone may be missed.[16]
According to recently published guidelines from the College of American Pathologists, International Association for the Study of Lung Cancer (IASLC), and Association for Molecular Pathology, cytologic samples are suitable for EGFR testing, and cell blocks (CBs) are the preferred medium.[17]
Multiple studies have illustrated the successful use of cytology specimens for molecular testing.[18,19,20,21,22,23,24,25,26] However, while aspirate cytology-derived CB material is more convenient than and generally preferred to direct smears for mutation status evaluation,[17,27,28,29] direct comparison of specific mutations between matched histology and cytology specimens has been predominantly reported for analysis of smear derived material.[25,30,31,32] In this study, molecular testing results of FNA cytology CB specimens are evaluated and compared with corresponding histologic specimens to validate the expert consensus guidelines.
MATERIALS AND METHODS
Patient specimens
Patients with available histology and cytology samples that both contained lung adenocarcinoma were identified retrospectively over a period of 122 months. Patient age at the time of diagnosis, sex of the patient, interval between acquisition of the cytology and histology specimens and their respective locations were recorded. Targets of histopathologic (biopsy and resection) specimens and cytologic FNA biopsies included lung lesions, lymph nodes (LNs), and distant metastases. Histology specimens were collected by CT-guided, transthoracic, spring-loaded, core needle (20-gauge) biopsy (n = 3), endoscopic, transbronchial biopsy (n = 3), video-assisted, thoracic surgical resection (n = 8) or open surgical resections (n = 16, from 15 patients). One punch biopsy of a metastasis to skin was also collected. Cytology specimens were collected by CT-guided-FNA (22-gauge) biopsy (n = 8) or EBUS-FNA (21-gauge) biopsy (n = 18) with rapid on-site evaluation performed by a cytopathologist and/or cytotechnologist. FNA cytology specimens were processed according to routine cytopathology procedures including CB preparation. CBs were prepared by allowing the specimen to clot and placing it directly into 10% neutral buffered formalin[33] and/or fixing it in the needle rinse placed in CytoLyt (Hologic). Following centrifugation of the specimen in a 50 ml tube for 5 min, the supernatant was removed. Well-formed clots were placed directly in Bio-Wrap® (Leica Biosystems, Buffalo Grove, IL) and fixed in 10% neutral buffered paraffin before paraffin embedding. For the remainder, HistoGel™ (Thermo Fisher Scientific, Waltham, MA) was added to the pellet and solidified in the refrigerator at 4°C. Solidified pellets were then placed in Bio-Wrap®, fixed in 10% neutral buffered formalin, and embedded in paraffin.
Histologic and cytologic interpretation
Histologic and cytologic specimens were evaluated using the 2004 World Health Organization Classification for lung tumors and the small biopsy and cytology classification proposed by the IASLC, American Thoracic Society and European Respiratory Society.[34]
EGFR and KRAS mutation status
At our institution, diagnosis of lung adenocarcinoma or adenosquamous carcinoma prompts reflex mutational analysis of EGFR and KRAS. Initially, the reflex testing was for KRAS, subsequently it was for both EGFR and KRAS. Depending on the available deoxyribonucleic acid (DNA) and/or the test result, one or both tests were performed. (Fluorescence in situ hybridization to detect rearrangement of the anaplastic lymphoma kinase (ALK) gene is also performed and mutational analysis of BRAF is subsequently undertaken if sufficient material remains). Except when a specimen has no or few isolated cells on each slide, there are no strict criteria for the minimum number of cells for molecular testing. Multiple[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20] serial sections of the block are utilized and stained with cresyl violet to identify and collect the neoplastic cells. When necessary, the carcinoma is microdissected manually or with laser capture, depending on the tumor content and its relationship to the surrounding non-neoplastic cells, to enrich the sample for molecular analysis.
Polymerase chain reactions (PCR) with flanking intronic primers[35] were performed to amplify regions of interest and identify all mutations in EGFR exons 18-21. DNA was extracted from paraffin-embedded histology specimens and CBs using QIAmp® (Qiagen, Inc., Valencia, CA) spin columns per the manufacturer's instructions. Cycle dideoxy terminator sequencing of the PCR amplicons was performed using the ABI BigDye™ Terminator (Applied Biosystems, Carlsbad, CA) kit V1.1 per the manufacturer's instructions. Raw sequence data were analyzed and aligned using SeqScape (Life Technologies™, Grand Island, NY) software.
Common KRAS mutations were detected with the KRAS codon 12/13 amplification-refractory mutation system-scorpions assay™ (Qiagen) per the manufacturer's instructions. Briefly, real-time PCR with allele-specific primers covalently linked to fluorophores with signal quenchers was performed to amplify regions potentially containing seven common KRAS mutations (listed in the supplementary data). The fluorophores and quenchers separate upon binding to amplified sequences, resulting in increased fluorescence in the reaction tubes. The number of cycles necessary to detect fluorescent signal above background indicated presence or absence of mutation.
Beginning in 2012, a PCR-based method for identifying KRAS mutations was employed. Briefly, PCR with flanking intronic primers[35] were performed to amplify regions of interest in KRAS exon 2. DNA was extracted from paraffin-embedded histology specimens and CBs using QIAmp® (Qiagen, Inc., Valencia, CA) spin columns per the manufacturer's instructions. Cycle dideoxy terminator sequencing of the PCR amplicons was performed using the ABI BigDye™ Terminator (Applied Biosystems, Carlsbad, CA) kit V1.1 per the manufacturer's instructions. Raw sequence data were analyzed and aligned using SeqScape (Life Technologies™) software.
RESULTS
EGFR and/or KRAS testing were performed on histology and cytology samples from 26 patients, 18 female and 8 male (mean age = 69 years, median age = 72 years). Of the 26 samples, EGFR and KRAS testing was performed on 13, EGFR only on 9, and KRAS only on 4. Mutation of more than one molecular marker was not detected in any sample pair.
Evaluation of EGFR mutation status
EGFR mutation testing was performed on both histology and cytology specimens from 22 separate patients, including 5 men and 17 women with median age 73.5 [Table 1]. Evaluation of all sample pairs yielded concordant results - 7 positive and 15 negative for EGFR mutation. Interestingly, 6/7 pairs harboring EGFR mutation were obtained from female patients. Five of the 15 pairs negative for EGFR mutation were subsequently shown to harbor KRAS mutation (patients 5, 9, 13, 17 and 22). Eleven concordant sample pairs consisted of a primary tumor and its LN or distant metastasis (patients 1, 4, 5, 6, 9, 12, 16, 21, 23, 24, and 26). EGFR mutation was identified in 5 of these 11 cancers (patients 4, 6, 12, 23, and 24).
Table 1.
Concordant results of molecular testing on cytology and histology specimens
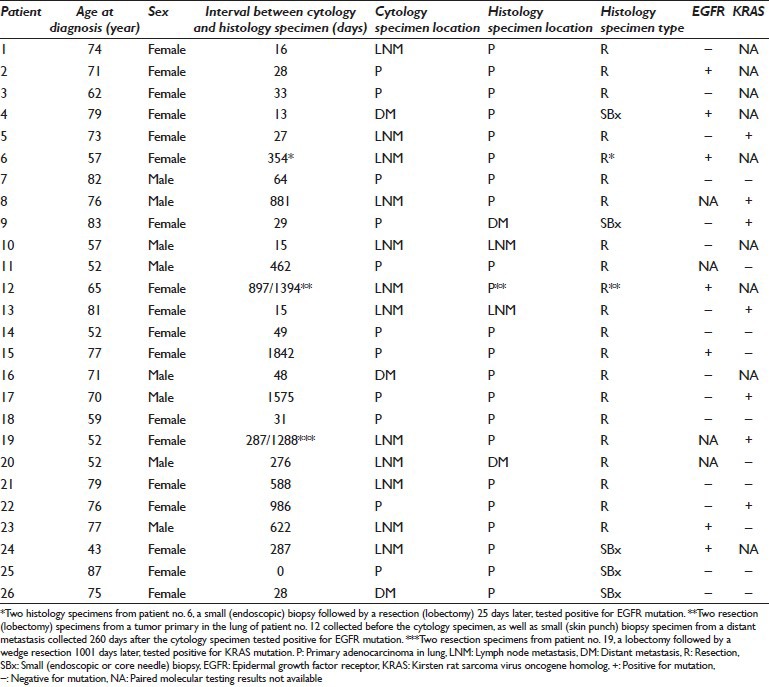
The histology specimen in the majority of patients consisted of a tumor resection. However, the histology specimen from 5 patients consisted of a small (endoscopic snare or transthoracic CT-guided core needle) biopsy (patients 4, 9, 24, 25, and 26). EGFR mutation was identified in 2 of these 5 cancers (patients 4 and 24). In two additional patients (6 and 12) for whom EGFR was evaluated on more than one histology specimen, one of the histology specimens consisted of a small biopsy. EGFR mutation was identified in both of these cancers.
Evaluation of KRAS mutation status
KRAS mutation testing was performed on both histology and cytology specimens from 17 separate patients, including 6 men and 11 women with median age 76 [Table 1]. Evaluation of all sample pairs yielded concordant results - 7 positive and 10 negative for KRAS mutation. Two of the 10 pairs negative for KRAS mutation had been previously shown to harbor EGFR mutation. Seven sample pairs consisted of a primary tumor and its metastasis (patients 5, 8, 9, 19, 21, 23, and 26). KRAS mutation was identified in 4 of these 7 cancers (patients 5, 8, 9, and 19). The histology specimen from 3 patients consisted of a small biopsy (patients 9, 25, and 26). KRAS mutation was identified in 1 of these 3 cancers (patient 9).
DISCUSSION
In recent years, a significant proportion of lung carcinomas have been diagnosed on cytology specimens obtained with minimally invasive techniques. Recent trends demonstrate that use of minimally invasive procedures such as FNAs for diagnosis of lung carcinoma is increasing.[36] However, adequacy of cytology specimens for molecular testing is controversial because of contamination by non-neoplastic cells, lack of standardized cytological preparations, and relatively small size.[4,37] The possibility of false negative results, particularly in specimens with scant tumor, may contribute to the reluctance to the use of cytology specimens for molecular testing.[20] Accordingly, in one study, <5% of 239 samples tested for EGFR mutation were obtained by FNA.[36] However, others have argued that FNA is suitable and underutilized for molecular testing.[15,17] The current study demonstrates that CBs prepared from EBUS and CT-guided FNAs provide sufficient material for EGFR and/or KRAS mutation analysis. Similar outcomes have been described elsewhere.[11,20,21,26,38,39,40] In fact, Fukui et al. achieved better results with cytological preparations than histological ones regardless of tumor volume.[41] Smouse et al. also demonstrated that mutation detection in cytological specimens is equally sensitive to that in histological specimens, and no significant difference exists between the frequency of inconclusive results observed in cytology and surgical pathology specimens.[36]
The results of the current study also illustrate that EGFR and KRAS molecular testing results of cytology specimens are concordant with those of corresponding histology specimens in 39/39 tests performed - 22/22 EGFR tests (including 7/7 positive results) and 17/17 KRAS tests (including 7/7 positive results). Previous studies have directly compared the results of mutational analysis predominantly between matched histology specimens and direct cytology smears; we compare both EGFR and KRAS results on matched histology specimens and aspirate cytology-derived CBs. Though the published guidelines recommend prioritizing testing for EGFR and ALK, we test for KRAS mutation as part of a reflex protocol when sufficient tissue is available. This is by far the largest such study of lung adenocarcinoma, and the first to include comparison between both EGFR and KRAS on cytological aspirate CB material and corresponding surgical pathology material.[25,26,30,31,32] The concordant results indicate that CBs provide sufficient tissue for analysis beyond the recommended EGFR molecular testing. Khode et al., performed only EGFR analysis on 37 pairs of cytology smears and surgical pathology specimens of lung adenocarcinoma/NSCLC.[25] Of those 37 pairs, only one showed discrepant EGFR mutation results between the cytology smear and the biopsy specimen (97% concordance). Aisner et al. have demonstrated the presence of the same EGFR mutation in three cases for which resection and cytology evaluation had been performed on the same site in the same patient and a cytology CB was available.[27] Similarly, van Eijk et al. in their study have reported complete concordance of the results of mutation analysis in a series of 17 CBs from NSCLC patients with corresponding histology specimens.[30] However, only four of their study patients received a diagnosis of adenocarcinoma, and two of them were found to harbor a KRAS mutation. EGFR mutation was detected in none of the 17 patients. Sun et al. demonstrated an overall EGFR concordance of 91.7% between histologic and cytologic specimens; however, the cytologic specimens were not all obtained from the primary site from which the histologic specimens were obtained. They also performed no KRAS and only EGFR analysis on both CBs and scrapings from smears.
Molecular testing results were concordant among all 13 sample pairs consisting of a primary tumor and its metastasis. Of these, six sample pairs were evaluated for the presence of EGFR mutation alone, and two tested positive. Two pairs were evaluated for the presence of KRAS mutation alone, and both tested positive. Of the five pairs in which both genes were evaluated, EGFR mutation was identified in one pair and KRAS mutation was identified in two pairs. Such perfect concordance is interesting as lung tumors are often heterogeneous, and phenotypic and genotypic differences between primary tumors and their metastases have been described.[25,26,35,42] Indeed, Kalikaki et al. described discordance in 28% (7/25) and 24% (6/25) of primary and metastatic sites for EGFR and KRAS, respectively.[43] The 23 patients with matched pairs of cytology smears and surgical specimens from different anatomic sites showed a concordance rate of 82%. The cytological specimens from their study included pleural fluids, bronchial washings, and bronchial brushings in addition to aspirate material. However, for cases in which a diagnostic sample is available from only one site, primary or metastatic, performance of molecular testing on that lone sample may suffice to appropriately guide treatment.
Despite the small size of cytology samples, they provide material for molecular testing of equal or possibly greater quality than that of larger histology samples. The NSCLC working group recommends using tissue blocks rich in neoplastic cells for molecular testing,[4,37] and effective methods to enrich cytology samples for tumor currently exist. These include use of fixative containing a hemolytic agent,[44] rapid on-site evaluation,[45,46] or inclusion of a beacon or marker to visualize the level at which cells are concentrated.[47] Either formalin or alcohol is suitable for EGFR testing, but other fixatives, including heavy metal fixatives (e.g., B5, acid zinc formalin, Zenker) and acidic solutions (e.g., decalcifying solution, Bouin), should be avoided because they interfere with testing.[17] A high ratio of neoplastic to non-neoplastic cells predicts the ability of molecular testing to detect mutations. Though reliable results with as few as 10-20% neoplastic cells have been reported, >40-50% neoplastic cells are considered optimal for evaluation.[4,38] The rarity of mutated cells in a specimen might exceed the sensitivity of the assay. Macro-or micro-dissection can enrich the sample by increasing the ratio of neoplastic to non-neoplastic cells, and these techniques can be performed on cytology preparations. Results of direct sequencing mutation detection with laser capture microdissection may increase yield from 21% to 40%.[12,48] Boldrini et al. analyzed 23 archived specimens, including FNA biopsies and exfoliative specimens. Microdissection using a 25-gauge needle resulted in adequate specimens for molecular testing.[19] Chowdhuri et al. demonstrated that 50 tumor cells microdissected with laser capture from cytology specimens were sufficient for EGFR and KRAS mutation detection.[12] Adequate results from other preparations, including liquid-based cytology preparations such as ThinPrep®[19,48] and scrapings from archival slides stained with Romanowsky or Papaniocolaou stains, have been reported.[20,25,26,38,49] However, yields from cells in Cytolyt™ are lower when compared to those obtained with microdissection of cells on a slide.[48]
CONCLUSIONS
Cytology specimens pose challenges for pathologists, who must perform ancillary tests, including immunophenotyping and/or molecular testing, and for oncologists, who rely on results for selection of therapy. The data from the current study indicate high concordance between results of molecular testing performed on cytology CBs and histology specimens and between primary and metastatic sites. Therefore, it is likely that results obtained from molecular testing of cytology specimens in lung adenocarcinoma are valid, and FNA-derived CBs provide a valuable medium.
COMPETING INTERESTS STATEMENT BY ALL AUTHORS
The authors declare that they have no competing interests.
AUTHORSHIP STATEMENT BY ALL AUTHORS
Each author has participated sufficiently in the work and takes public responsibility for appropriate portions of the content of this article. All authors read and approved the final manuscript. Each author acknowledges that this final version was read and approved.
ETHICS STATEMENT BY ALL AUTHORS
This study was conducted with approval from Institutional Review Board (IRB) of the institution associated with this study (Columbia University Medical Center). Authors take responsibility to maintain relevant documentation in this respect.
EDITORIAL/PEER-REVIEW STATEMENT
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double blind model (authors are blinded for reviewers and vice versa) through automatic online system.
Contributor Information
Jonas J. Heymann, Email: jjh2110@cumc.columbia.edu.
William A. Bulman, Email: wab10@cumc.columbia.edu.
Roger A. Maxfield, Email: ram7@cumc.columbia.edu.
Charles A. Powell, Email: charles.powell@mssm.edu.
Balazs Halmos, Email: bh2376@cumc.columbia.edu.
Joshua Sonett, Email: js2106@cumc.columbia.edu.
Nike T. Beaubier, Email: nikebeaubier@gmail.com.
John P. Crapanzano, Email: jpc2141@cumc.columbia.edu.
Mahesh M. Mansukhani, Email: mm322@cumc.columbia.edu.
Anjali Saqi, Email: aas177@cumc.columbia.edu.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Toh CK. The changing epidemiology of lung cancer. Methods Mol Biol. 2009;472:397–411. doi: 10.1007/978-1-60327-492-0_19. [DOI] [PubMed] [Google Scholar]
- 3.Thun MJ, Lally CA, Flannery JT, Calle EE, Flanders WD, Heath CW., Jr Cigarette smoking and changes in the histopathology of lung cancer. J Natl Cancer Inst. 1997;89:1580–6. doi: 10.1093/jnci/89.21.1580. [DOI] [PubMed] [Google Scholar]
- 4.Pirker R, Herth FJ, Kerr KM, Filipits M, Taron M, Gandara D, et al. Consensus for EGFR mutation testing in non-small cell lung cancer: Results from a European workshop. J Thorac Oncol. 2010;5:1706–13. doi: 10.1097/JTO.0b013e3181f1c8de. [DOI] [PubMed] [Google Scholar]
- 5.Sequist LV, Bell DW, Lynch TJ, Haber DA. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol. 2007;25:587–95. doi: 10.1200/JCO.2006.07.3585. [DOI] [PubMed] [Google Scholar]
- 6.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 7.Fukuoka M, Wu YL, Thongprasert S, Sunpaweravong P, Leong SS, Sriuranpong V, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS) J Clin Oncol. 2011;29:2866–74. doi: 10.1200/JCO.2010.33.4235. [DOI] [PubMed] [Google Scholar]
- 8.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–42. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 9.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–46. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 10.Miller VA, Hirsh V, Cadranel J, Chen YM, Park K, Kim SW, et al. Afatinib versus placebo for patients with advanced, metastatic non-small-cell lung cancer after failure of erlotinib, gefitinib, or both, and one or two lines of chemotherapy (LUX-Lung 1): A phase 2b/3 randomised trial. Lancet Oncol. 2012;13:528–38. doi: 10.1016/S1470-2045(12)70087-6. [DOI] [PubMed] [Google Scholar]
- 11.Nicholson AG, Gonzalez D, Shah P, Pynegar MJ, Deshmukh M, Rice A, et al. Refining the diagnosis and EGFR status of non-small cell lung carcinoma in biopsy and cytologic material, using a panel of mucin staining, TTF-1, cytokeratin 5/6, and P63, and EGFR mutation analysis. J Thorac Oncol. 2010;5:436–41. doi: 10.1097/JTO.0b013e3181c6ed9b. [DOI] [PubMed] [Google Scholar]
- 12.Chowdhuri SR, Xi L, Pham TH, Hanson J, Rodriguez-Canales J, Berman A, et al. EGFR and KRAS mutation analysis in cytologic samples of lung adenocarcinoma enabled by laser capture microdissection. Mod Pathol. 2012;25:548–55. doi: 10.1038/modpathol.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah PL, Singh S, Bower M, Livni N, Padley S, Nicholson AG. The role of transbronchial fine needle aspiration in an integrated care pathway for the assessment of patients with suspected lung cancer. J Thorac Oncol. 2006;1:324–7. [PubMed] [Google Scholar]
- 14.Little AG, Rusch VW, Bonner JA, Gaspar LE, Green MR, Webb WR, et al. Patterns of surgical care of lung cancer patients. Ann Thorac Surg. 2005;80:2051–6. doi: 10.1016/j.athoracsur.2005.06.071. [DOI] [PubMed] [Google Scholar]
- 15.Clark DP. Seize the opportunity: Underutilization of fine-needle aspiration biopsy to inform targeted cancer therapy decisions. Cancer. 2009;117:289–97. doi: 10.1002/cncy.20045. [DOI] [PubMed] [Google Scholar]
- 16.Taniguchi K, Okami J, Kodama K, Higashiyama M, Kato K. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci. 2008;99:929–35. doi: 10.1111/j.1349-7006.2008.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med. 2013;137:828–60. doi: 10.5858/arpa.2012-0720-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.da Cunha Santos G, Saieg MA, Geddie W, Leighl N. EGFR gene status in cytological samples of nonsmall cell lung carcinoma: Controversies and opportunities. Cancer Cytopathol. 2011;119:80–91. doi: 10.1002/cncy.20150. [DOI] [PubMed] [Google Scholar]
- 19.Boldrini L, Gisfredi S, Ursino S, Camacci T, Baldini E, Melfi F, et al. Mutational analysis in cytological specimens of advanced lung adenocarcinoma: A sensitive method for molecular diagnosis. J Thorac Oncol. 2007;2:1086–90. doi: 10.1097/JTO.0b013e31815ba1fa. [DOI] [PubMed] [Google Scholar]
- 20.Billah S, Stewart J, Staerkel G, Chen S, Gong Y, Guo M. EGFR and KRAS mutations in lung carcinoma: Molecular testing by using cytology specimens. Cancer Cytopathol. 2011;119:111–7. doi: 10.1002/cncy.20151. [DOI] [PubMed] [Google Scholar]
- 21.Cai G, Wong R, Chhieng D, Levy GH, Gettinger SN, Herbst RS, et al. Identification of EGFR mutation, KRAS mutation, and ALK gene rearrangement in cytological specimens of primary and metastatic lung adenocarcinoma. Cancer Cytopathol. 2013;121:500–7. doi: 10.1002/cncy.21288. [DOI] [PubMed] [Google Scholar]
- 22.Abele JS, Miller TR, Goodson WH, 3rd, Hunt TK, Hohn DC. Fine-needle aspiration of palpable breast masses.A program for staged implementation. Arch Surg. 1983;118:859–63. doi: 10.1001/archsurg.1983.01390070067013. [DOI] [PubMed] [Google Scholar]
- 23.Saglam A, Can B. Coexistence of lactating adenoma and invasive ductal adenocarcinoma of the breast in a pregnant woman. J Clin Pathol. 2005;58:87–9. doi: 10.1136/jcp.2004.018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veneti S, Daskalopoulou D, Zervoudis S, Papasotiriou E, Ioannidou-Mouzaka L. Liquid-based cytology in breast fine needle aspiration.Comparison with the conventional smear. Acta Cytol. 2003;47:188–92. doi: 10.1159/000326502. [DOI] [PubMed] [Google Scholar]
- 25.Khode R, Larsen DA, Culbreath BC, Parrish S, Walker KL, Sayage-Rabie L, et al. Comparative study of epidermal growth factor receptor mutation analysis on cytology smears and surgical pathology specimens from primary and metastatic lung carcinomas. Cancer Cytopathol. 2013;121:361–9. doi: 10.1002/cncy.21273. [DOI] [PubMed] [Google Scholar]
- 26.Dey P, Luthra UK, George J, Zuhairy F, George SS, Haji BI. Comparison of ThinPrep and conventional preparations on fine needle aspiration cytology material. Acta Cytol. 2000;44:46–50. doi: 10.1159/000326224. [DOI] [PubMed] [Google Scholar]
- 27.Aisner DL, Deshpande C, Baloch Z, Watt CD, Litzky LA, Malhotra B, et al. Evaluation of EGFR mutation status in cytology specimens: An institutional experience. Diagn Cytopathol. 2013;41:316–23. doi: 10.1002/dc.21851. [DOI] [PubMed] [Google Scholar]
- 28.Sanz-Santos J, Serra P, Andreo F, Llatjós M, Castellà E, Monsó E. Contribution of cell blocks obtained through endobronchial ultrasound-guided transbronchial needle aspiration to the diagnosis of lung cancer. BMC Cancer. 2012;12:34. doi: 10.1186/1471-2407-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navani N, Brown JM, Nankivell M, Woolhouse I, Harrison RN, Jeebun V, et al. Suitability of endobronchial ultrasound-guided transbronchial needle aspiration specimens for subtyping and genotyping of non-small cell lung cancer: A multicenter study of 774 patients. Am J Respir Crit Care Med. 2012;185:1316–22. doi: 10.1164/rccm.201202-0294OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Eijk R, Licht J, Schrumpf M, Talebian Yazdi M, Ruano D, Forte GI, et al. Rapid KRAS, EGFR, BRAF and PIK3CA mutation analysis of fine needle aspirates from non-small-cell lung cancer using allele-specific qPCR. PLoS One. 2011;6:e17791. doi: 10.1371/journal.pone.0017791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruno P, Mariotta S, Ricci A, Duranti E, Scozzi D, Noto A, et al. Reliability of direct sequencing of EGFR: Comparison between cytological and histological samples from the same patient. Anticancer Res. 2011;31:4207–10. [PubMed] [Google Scholar]
- 32.Bozzetti C, Negri FV, Azzoni C, Naldi N, Nizzoli R, Bortesi B, et al. Epidermal growth factor receptor and Kras gene expression: Reliability of mutational analysis on cytological samples. Diagn Cytopathol. 2013;41:595–8. doi: 10.1002/dc.22905. [DOI] [PubMed] [Google Scholar]
- 33.Bulman W, Saqi A, Powell CA. Acquisition and processing of endobronchial ultrasound-guided transbronchial needle aspiration specimens in the era of targeted lung cancer chemotherapy. Am J Respir Crit Care Med. 2012;185:606–11. doi: 10.1164/rccm.201107-1199CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bukhari MH, Arshad M, Jamal S, Niazi S, Bashir S, Bakhshi IM, et al. Use of fine-needle aspiration in the evaluation of breast lumps. Patholog Res Int. 2011;2011:689521. doi: 10.4061/2011/689521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–9. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 36.Smouse JH, Cibas ES, Jänne PA, Joshi VA, Zou KH, Lindeman NI. EGFR mutations are detected comparably in cytologic and surgical pathology specimens of nonsmall cell lung cancer. Cancer. 2009;117:67–72. doi: 10.1002/cncy.20011. [DOI] [PubMed] [Google Scholar]
- 37.Eberhard DA, Giaccone G, Johnson BE Non-Small-Cell Lung Cancer Working Group. Biomarkers of response to epidermal growth factor receptor inhibitors in Non-Small-Cell Lung Cancer Working Group: Standardization for use in the clinical trial setting. J Clin Oncol. 2008;26:983–94. doi: 10.1200/JCO.2007.12.9858. [DOI] [PubMed] [Google Scholar]
- 38.Schuurbiers OC, Looijen-Salamon MG, Ligtenberg MJ, van der Heijden HF. A brief retrospective report on the feasibility of epidermal growth factor receptor and KRAS mutation analysis in transesophageal ultrasound- and endobronchial ultrasound-guided fine needle cytological aspirates. J Thorac Oncol. 2010;5:1664–7. doi: 10.1097/JTO.0b013e3181f0bd93. [DOI] [PubMed] [Google Scholar]
- 39.Khazai L, Kundu UR, Jacob B, Patel S, Sneige N, Eapen GA, et al. Endobronchial ultrasound-guided transbronchial needle aspiration biopsy is useful evaluating mediastinal lymphadenopathy in a cancer center. Cytojournal. 2011;8:10. doi: 10.4103/1742-6413.82022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nunez AL, Jhala NC, Carroll AJ, Mikhail FM, Reddy VV, Xian RR, et al. Endoscopic ultrasound and endobronchial ultrasound-guided fine-needle aspiration of deep-seated lymphadenopathy: Analysis of 1338 cases. Cytojournal. 2012;9:14. doi: 10.4103/1742-6413.95845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukui T, Ohe Y, Tsuta K, Furuta K, Sakamoto H, Takano T, et al. Prospective study of the accuracy of EGFR mutational analysis by high-resolution melting analysis in small samples obtained from patients with non-small cell lung cancer. Clin Cancer Res. 2008;14:4751–7. doi: 10.1158/1078-0432.CCR-07-5207. [DOI] [PubMed] [Google Scholar]
- 42.Italiano A, Vandenbos FB, Otto J, Mouroux J, Fontaine D, Marcy PY, et al. Comparison of the epidermal growth factor receptor gene and protein in primary non-small-cell-lung cancer and metastatic sites: Implications for treatment with EGFR-inhibitors. Ann Oncol. 2006;17:981–5. doi: 10.1093/annonc/mdl038. [DOI] [PubMed] [Google Scholar]
- 43.Kalikaki A, Koutsopoulos A, Trypaki M, Souglakos J, Stathopoulos E, Georgoulias V, et al. Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Br J Cancer. 2008;99:923–9. doi: 10.1038/sj.bjc.6604629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weidmann J, Chaubal A, Bibbo M. Cellular fixation.A study of CytoRich Red and cytospin collection fluid. Acta Cytol. 1997;41:182–7. doi: 10.1159/000332321. [DOI] [PubMed] [Google Scholar]
- 45.Monaco SE, Pantanowitz L, Khalbuss WE. Comparing endobronchial ultrasound-guided fine needle aspiration specimens with and without rapid on-site evaluation. Cytojournal. 2012;9:2. doi: 10.4103/1742-6413.92414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Griffin AC, Schwartz LE, Baloch ZW. Utility of on-site evaluation of endobronchial ultrasound-guided transbronchial needle aspiration specimens. Cytojournal. 2011;8:20. doi: 10.4103/1742-6413.90081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varsegi GM, Shidham V. Cell block preparation from cytology specimen with predominance of individually scattered cells. J Vis Exp. 9 doi: 10.3791/1316. Cell block preparation from cytology specimen with predominance of individually scattered cells J Vis Exp 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malapelle U, de Rosa N, Rocco D, Bellevicine C, Crispino C, Illiano A, et al. EGFR and KRAS mutations detection on lung cancer liquid-based cytology: A pilot study. J Clin Pathol. 2012;65:87–91. doi: 10.1136/jclinpath-2011-200296. [DOI] [PubMed] [Google Scholar]
- 49.Smith GD, Chadwick BE, Willmore-Payne C, Bentz JS. Detection of epidermal growth factor receptor gene mutations in cytology specimens from patients with non-small cell lung cancer utilising high-resolution melting amplicon analysis. J Clin Pathol. 2008;61:487–93. doi: 10.1136/jcp.2007.051425. [DOI] [PubMed] [Google Scholar]


