Abstract
Motivation: Alternative splicing (AS) is a regulated process that directs the generation of different transcripts from single genes. A computational model that can accurately predict splicing patterns based on genomic features and cellular context is highly desirable, both in understanding this widespread phenomenon, and in exploring the effects of genetic variations on AS.
Methods: Using a deep neural network, we developed a model inferred from mouse RNA-Seq data that can predict splicing patterns in individual tissues and differences in splicing patterns across tissues. Our architecture uses hidden variables that jointly represent features in genomic sequences and tissue types when making predictions. A graphics processing unit was used to greatly reduce the training time of our models with millions of parameters.
Results: We show that the deep architecture surpasses the performance of the previous Bayesian method for predicting AS patterns. With the proper optimization procedure and selection of hyperparameters, we demonstrate that deep architectures can be beneficial, even with a moderately sparse dataset. An analysis of what the model has learned in terms of the genomic features is presented.
Contact: frey@psi.toronto.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Alternative splicing (AS) is a process whereby the exons of a primary transcript may be connected in different ways during pre-mRNA splicing. This enables the same gene to give rise to splicing isoforms containing different combinations of exons, and as a result different protein products, contributing to the cellular diversity of an organism (Wang and Burge, 2008). Furthermore, AS is regulated during development and is often tissue dependent, so a single gene can have multiple tissue-specific functions. The importance of AS lies in the evidence that at least 95% of human multi-exon genes are alternatively spliced and that the frequency of AS increases with species complexity (Barbosa-Morais et al., 2012; Pan et al., 2008).
One mechanism of splicing regulation occurs at the level of the sequences of the transcript. The presence or absence of certain regulatory elements can influence which exons are kept, while others are removed, before a primary transcript is translated into proteins. Computational models that take into account the combinatorial effects of these regulatory elements have been successful in predicting the outcome of splicing (Barash et al., 2010).
Previously, a ‘splicing code’ that uses a Bayesian neural network (BNN) was developed to infer a model that can predict the outcome of AS from sequence information in different cellular contexts (Xiong et al., 2011). One advantage of Bayesian methods is that they protect against overfitting by integrating over models. When the training data are sparse, as is the case for many datasets in the life sciences, the Bayesian approach can be beneficial. It was shown that the BNN outperforms several common machine learning algorithms, such as multinomial logistic regression (MLR) and support vector machines, for AS prediction in mouse trained using microarray data.
There are several practical considerations when using BNNs. They often rely on methods like Markov Chain Monte Carlo (MCMC) to sample models from a posterior distribution, which can be difficult to speed up and scale up to a large number of hidden variables and a large volume of training data. Furthermore, computation-wise, it is relatively expensive to get predictions from a BNN, which requires computing the average predictions of many models.
Recently, deep learning methods have surpassed the state-of-the-art performance for many tasks (Bengio et al., 2013). Deep learning generally refers to methods that map data through multiple levels of abstraction, where higher levels represent more abstract entities. The goal is for an algorithm to automatically learn complex functions that map inputs to outputs, without using hand-crafted features or rules (Bengio, 2009). One implementation of deep learning comes in the form of feedforward neural networks, where levels of abstraction are modeled by multiple non-linear hidden layers.
With the increasingly rapid growth in the volume of ‘omic’ data (e.g. genomics, transcriptomics, proteomics), deep learning has the potential to produce meaningful and hierarchical representations that can efficiently be used to describe complex biological phenomena. For example, deep networks may be useful for modeling multiple stages of a regulatory network at the sequence level and at higher levels of abstraction.
Ensemble methods are a class of algorithms that are popular owing to their generally good performance (Caruana and Niculescu-Mizil, 2006), and are often used in the life sciences (Touw et al., 2013). The strength of ensemble methods comes from combining the predictions of many models. Random forests is an example, as is the Bayesian model averaging method previously used to model the regulation of splicing. Recently, neural network learning has been improved using a technique called dropout, which makes neural networks behave like an ensemble method (Hinton and Srivastava, 2012). Dropout works by randomly removing hidden neurons during the presentation of each training example. The outcome is that instead of training a single model with N hidden variables, it approximates the training of 2N different networks, each on a different subset of the training data. It is described as an ‘extreme form of bagging’ and is a computationally efficient way of doing model averaging (Hinton and Srivastava, 2012).
With large datasets, learning with MCMC methods can be slow and can be outperformed by stochastic optimization methods in practice (Ahn et al., 2012). These algorithms process small subsets (minibatches) of data at each iteration, and update model parameters by taking small steps in the direction of the gradient to optimize the cost function. It is common to use stochastic gradient descent to train feedforward neural networks. The learning algorithm (backpropagation) is also conceptually simple, involving for the most part matrix multiplications, which makes them suitable for speedup using graphics processing units (GPU).
Here, we show that the use of large (many hidden variables) and deep (multiple hidden layers) neural networks can improve the predictive performances of the splicing code compared with previous work. We also provide an evaluation method for researchers to improve and extend computational models for predicting AS. Another goal is to describe the procedure for training and optimizing a deep neural network (DNN) on a sparse and unbalanced biological dataset. Furthermore, we show how such a DNN can be analyzed in terms of its inputs. To date, aside from a small number of works (Di Lena et al., 2012; Eickholt and Cheng, 2012), deep learning methods have not been applied in the life sciences, even though they show tremendous promise.
We show results supporting that DNN with dropout can be a competitive algorithm for doing learning and prediction on biological datasets, with the advantage that they can be trained quickly, have enough capacity to model complex relationships and scale well with the number of hidden variables and volume of data, making them potentially highly suitable for ‘omic’ datasets.
Different from the previous BNN, which used 30 hidden units, our architecture has thousands of hidden units with multiple non-linear layers and millions of model parameters (Supplementary Table S2). We also explored a different connection architecture compared with previous work. Before, each tissue type was considered as a different output of the neural network. Here, tissues are treated as an input, requiring that the complexity of the splicing machinery in response to the cellular environment be represented by a set of hidden variables that jointly represent both the genomic features and tissue context.
Besides a different model architecture, we also extended the code's prediction capability. In previous work, the splicing code infers the direction of change of the percentage of transcripts with an exon spliced in (PSI) (Katz et al., 2010), relative to all other tissues. Here, we perform absolute PSI prediction for each tissue individually without the need for a baseline averaged across tissues. We also predict the difference in PSI (ΔPSI) between pairs of tissues to evaluate the model's tissue-specificity. We show how these two prediction tasks can be trained simultaneously, where the learned hidden variables are useful for both tasks.
We compare the splicing code's performance trained with the DNN with the previous BNN and additionally optimized a MLR classifier on the same task for a baseline comparison. A GPU was used to accelerate training of the DNN, which made it feasible to perform hyperparameter search to optimize prediction performance with cross validation.
2 METHODS
2.1 Dataset
The dataset consists of 11 019 mouse alternative exons profiled from RNA-Seq data prepared by (Brawand et al., 2011). The exons are based on a set of cassette exons derived from EST/cDNA sequences (Barash et al., 2010, 2013). Five tissue types are available, including whole brain, heart, kidney, liver and testis. To estimate the splicing level for each exon and tissue, we mapped the 75 nucleotide reads to splice junctions, requiring a minimum overhang of 8 nucleotides on each side of the junction, giving 60 mappable positions. A bootstrap method that takes into account position-dependent read biases in RNA-Seq was then used to obtain an estimate of PSI that reflects its uncertainty (Kakaradov et al., 2012). This allows the generation of a distribution of PSI for each exon and tissue. To obtain the distribution denoting the difference in PSI, or ΔPSI, the difference between bootstrap samples was calculated for all pairs of tissues to generate a distribution in the same manner. The possible values range from 0 to 1 for the PSI distribution, and −1 to 1 in the ΔPSI distribution. Additional information about the dataset can be found in Section 1 of the Supplementary Material. To test whether the model generalizes to a different dataset, RNA-Seq data from (Barbosa-Morais et al., 2012) was processed in the same manner for brain and heart, which was used only for testing.
For each exon, a set of intronic, exonic and structural features was derived from sequences in the alternative exon (A), flanking constitutive exons (C1 and C2) and introns between C1 and A (I1) and A and C2 (I2), forming a feature vector of length 1393. These features include those originally described in (Barash et al., 2010) and the extended feature set from (Barash et al., 2013). Features related to the premature termination codon have been removed because they rely on knowing the splicing pattern a priori and cannot be computed by just the local genomic sequences. Instead, four binary ‘translatability’ features are introduced, which describe whether exons can be translated without a stop codon in one of three possible reading frames. The features are summarized in Section 4 of the Supplementary Material.
2.2 The model
We formulate splicing prediction as a classification problem with multiple classes. Figure 1 shows the architecture of the DNN. The parameters of the model are summarized in Section 2 of the Supplementary Material. The nodes of the neural network are fully connected, where each connection is parameterized by a real-valued weight θ. The DNN has multiple layers of non-linearity consisting of hidden units. The output activation a of each hidden unit v in layer l processes a sum of weighted outputs from the previous layer, using a non-linear function f:
| (1) |
where Ml represents the number of hidden units in layer l, and a0 and M0 are the input into the model and its dimensionality, respectively. We used two different activation functions for the hidden units, namely the hyperbolic tangent (TANH) function, and the rectified linear unit (RELU), which is defined as (Glorot et al., 2011):
| (2) |
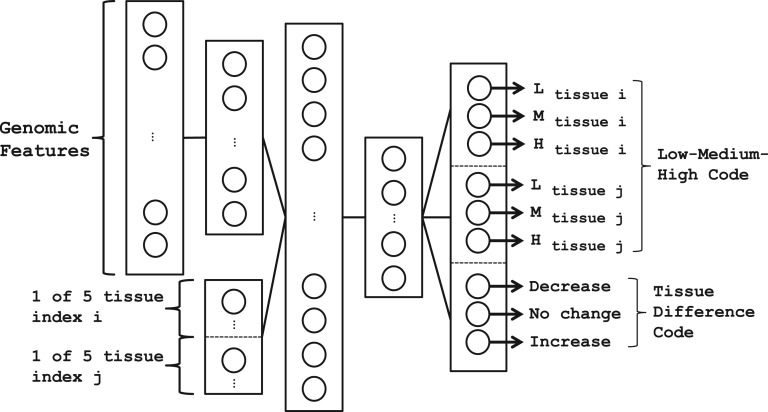
Fig. 1.
Architecture of the DNN used to predict AS patterns. It contains three hidden layers, with hidden variables that jointly represent genomic features and cellular context (tissue types)
The RELU unit was used for all hidden units except for the first hidden layer, which used TANH units, based on empirical performance on validation data.
Inputs into the first hidden layer consist of F = 1393 genomic features xf=1…F describing an exon, neighboring introns and adjacent exons. To improve learning, the features were normalized by the maximum of the absolute value across all exons. The purpose of this hidden layer is to reduce the dimensionality of the input and learn a better representation of the feature space.
The identity of two tissues, which consists of two 1-of-T binary variables ti=1…T and tj=1…T, are then appended to the vector of outputs of the first hidden layer, together forming the input into the second hidden layer. For this work, T = 5 for the five tissues available in the RNA-Seq data. We added a third hidden layer as we found it improved the model's performance. The weighted outputs from the last hidden layer is used as input into a softmax function for classification in the prediction hk(x,t,θ), which represents the probability of each splicing pattern k:
| (3) |
To learn a set of model parameters θ, we used the cross-entropy cost function E on predictions h(x,t,θ) given targets y(x,t), which is minimized during training:
| (4) |
where n denotes the training examples, and k indexes C classes.
We are interested in two types of predictions. The first task is to predict the PSI value given a particular tissue type and a set of genomic features. To generate the targets for training, we created C = 3 classes, which we label as low, medium and high categories. Each class contains a real-value variable obtained by summing the probability mass of the PSI distribution over equally split intervals of 0–0.33, 0.33–0.66 and 0.66–1. They represent the probability that a given exon and tissue type has a PSI value ranging from these corresponding intervals, hence are soft class labels. We will refer this as the ‘low, medium, high’ (LMH) code, with targets .
The second task describes the ΔPSI between two tissues for a particular exon. We again generate three classes, and call them decreased inclusion, no change and increased inclusion, which are similarly generated, but from the ΔPSI distributions. We chose an interval that more finely differentiates tissue-specific AS for this task, where a difference of >0.15 would be labeled as a change in PSI levels. We summed the probability mass over the intervals of −1 to −0.15 for decreased inclusion, −0.15 to 0.15 for no change and 0.15 to 1 for increased inclusion. The purpose of this target is to learn a model that is independent of the chosen PSI class intervals in the LMH code. For example, the expected PSI of two tissues ti and tj for an exon could be 0.40 and 0.60. The LMH code would be trained to predict medium for both tissues, whereas this tissue difference code would predict that tj has increased inclusion relative to ti. We will refer to this as the ‘decrease, no change, increase’ (DNI) code, with targets .
Both the LMH and DNI codes are trained jointly, reusing the same hidden representations learned by the model. For the LMH code, two softmax classification outputs predict the PSI for each of the two tissues that are given as input into the DNN. A third softmax classification function predicts the ΔPSI for the two tissues. We note that two PSI predictions are included in the model's output so we have a complete set of predictions that use the full input features. The total cost of the model used during optimization is the sum of the cross-entropy functions (4) for both prediction tasks.
The BNN architecture used for comparison is the same as previously described (Xiong et al., 2011), but trained on RNA-Seq data with the expanded feature set and LMH as targets. Although hidden variables were shared across tissues in both the BNN and DNN, a different set of weights was used following the single hidden layer to predict the splicing pattern for each tissue separately in the BNN (Supplementary Fig. S3). In the current DNN, the tissue identities are inputs and are jointly represented by hidden variables together with genomic features. For the BNN to make tissue difference predictions in the same manner as the DNI code, we fitted a MLR on the predicted LMH outputs for each tissue pair (Supplementary Fig. S4).
2.3 Training the model
The first hidden layer was trained as an autoencoder to reduce the dimensionality of the feature space in an unsupervised manner. An autoencoder is trained by supplying the input through a non-linear hidden layer, and reconstructing the input, with tied weights going into and out of the hidden layer. This method of pretraining the network has been used in deep architectures to initialize learning near a good local minimum (Erhan et al., 2010; Hinton and Salakhutdinov, 2006). We used an autoencoder instead of other dimensionality reduction techniques like principle component analysis because it naturally fits into the DNN architecture, and that a non-linear technique may discover a better and more compact representation of the features.
In the second stage of training, the weights from the input layer to the first hidden layer (learned from the autoencoder) are fixed, and 10 additional inputs corresponding to tissues are appended. A one-hot encoding representation is used, such that specifying a tissue for a particular training example can take the form [0 1 0 0 0] to denote the second tissue out of five possible types. We have two such inputs totaling 10 variables that specify tissue types. The reduced feature set and tissue variables become input into the second hidden layer. The weights connected to this and the final hidden layer of the DNN are then trained together in a supervised manner, with targets being PSI and ΔPSI. After training these final two layers, weights from all layers of the DNN were fine-tuned by backpropagation.
Each training example consists of 1393 genomic features and two tissue types as input. The targets consist of (i) PSI for each of the two tissues and (ii) ΔPSI between the two tissues. Given a particular exon and five possible tissue types, we constructed 5 × 5 = 25 training examples. This construction has redundancy in that we generate examples where both tissues are the same in the input to teach the model that it should predict no change for ΔPSI given identical tissue indices. Also, if the tissues are swapped in the input, a previously increased inclusion label should become decreased inclusion. The same rationale extends to the LMH code. Generating these additional examples is one method to incorporate this knowledge without explicitly specifying it in the model architecture.
We applied a threshold to exclude examples from training if the total number RNA-Seq junction reads is below 10. This removed 45.8% of the total number of training examples. We further define exons as having large tissue variability if ΔPSI ≥ ±0.15 for at least one tissue pair profiled. These exons make up 28.6% of the total number of remaining exons that have more than 10 junction reads. Additional information about the read coverage of the dataset can be found in Section 1 of the Supplementary Material.
To promote the neural network to better discover the meaning of the inputs representing tissue types, we biased the distribution of training examples in the minibatches. We first selected all events which exhibit large tissue variability, and constructed minibatches based only on these events. At each training epoch, we further sampled (without replacement) training cases from the larger pool of events with low tissue variability, of size equal to one fifth of the minibatch size. The purpose is to have a consistent backpropagation signal that updates the weights connected to the tissue inputs and bias learning towards the event with large tissue variability early on before overfitting occurs. As training progresses, the splicing pattern of the events with low tissue variability is also learned. This arrangement effectively gives the events with large tissue variability greater importance (i.e. more weight) during optimization. A side effect is that it also places more importance to the medium category of the LMH code during training, since they tend to be present more often in exons with tissue-specific splicing patterns.
Both the LMH and DNI codes are trained together. Because each of these two tasks might be learning at different rates, we allowed their learning rates to differ. This is to prevent one task from overfitting too soon and negatively affecting the performance of another task before the complete model is fully trained (Silver and Mercer, 1996). This is implemented by having different learning rates for the weights between the connections of the last hidden layer and the softmax functions for each task.
The performance of the model was assessed using the area under the Receiver-Operating Characteristic curve (AUC) metric. To evaluate the PSI predictions for the LMH code, we used the 1 versus all formulation. This produces three AUCs (AUCLow, AUCMed, AUCHigh), one for each class. For ΔPSI predictions, since the no change class is much more abundant, we find that the multi-class 1 versus all formulation tends to overestimate the tissue-specificity performance of the model due to class skew (Fawcett, 2006). Furthermore, the model can predict, based on the genomic features alone, that there is tissue-specific splicing for a given exon (which is biologically meaningful), but not necessarily how different tissues change the splicing pattern. We therefore provide two metrics to evaluate the DNI code. The first is to compute the AUCDvI based on the decrease versus increase class between two tissues. The second is to compute AUCChange by comparing no change versus the other two classes.
To train and test the DNN, data was split into approximately five equal folds at random for cross validation. Each fold contains a unique set of exons that are not found in any of the other folds. Three of the folds were used for training, one used for validation and one held out for testing. We trained for a fixed number of epochs and selected the hyperparameters that give the optimal AUC performance on the validation data. The model was then retrained using these selected hyperparameters with both the training and validation data. Five models were trained this way from the different folds of data. Predictions from all five models on their corresponding test set were used to evaluate the code's performance. To estimate the confidence intervals, the data were randomly partitioned five times, and the above training procedure was repeated.
The DNN weights were initialized with small random values sampled from a zero-mean Gaussian distribution. Learning was performed with stochastic gradient descent with momentum and dropout, where minibatches were constructed as described above. A small L1 weight penalty was included in the cost function (4) (Tibshirani, 1994). The model's weights were updated after each minibatch. The learning rate ε was decreased with epochs e, and also included a momentum term μ that starts out at 0.5, increasing to 0.99, and then stays fixed. The momentum term accelerates learning, and stabilizes learning near the end of training when the momentum is high by distributing gradient information over many updates. The weights of the model parameters θ were updated as follows:
| (5) |
We used a dropout rate of 50% for all layers except for the input layer (the autoencoder), where we did not use dropout, as it empirically decreased the model's predictive performance. Training was carried out for 1500 epochs for both the pretraining with the autoencoder and supervised learning.
The performance of a DNN depends on a good set of hyperparameters. Instead of doing a grid search over the hyperparameter space, we used a Bayesian framework called spearmint to automatically select the model's hyperparameters (Snoek et al., 2012). The method uses a Gaussian Process to search for a joint setting of hyperparameters that optimizes an algorithm's performance on validation data. It uses the performance measures from previous experiments to decide which hyperparameters to try next, taking into account the trade-off between exploration and exploitation. This method eliminates many of the human judgments involved with hyperparameter optimization and reduces the time required to find such hyperparameters. The algorithm requires only the search range of hyperparameter values to be specified, as well as how long to run the optimization for. We used the expected improvement criterion in the optimization, as it does not require its own tuning parameter, unlike other methods in the framework. We score each experiment by the sum of the AUCs from both the LMH and DNI codes, requiring the set of hyperparameters to perform well on both tasks. Detailed information on the selected hyperparameters and search procedure are described in Section 2 of the Supplementary Material.
The DNN was implemented in Python, making use of Gnumpy for GPU-accelerated computation (Tieleman, 2010). The GPU used was a Nvidia GTX Titan. For the configuration with the optimal hyperparameters, the GPU provided ∼15-fold speedup over our original CPU implementation. This was crucial as otherwise hyperparameter optimization would not have been practical.
We compared the splicing code's performance trained with the DNN with the BNN, as well as an MLR classifier as a baseline. The MLR was trained by removing the hidden layer while keeping the training methodology identical to the neural networks. Because the predictions of the BNN consist only of the PSI prediction for each tissue separately at the output (Xiong et al., 2011), for the BNN to make tissue difference predictions in the same manner as the DNI code, we used a MLR on the predicted outputs for each tissue pair. For a fair comparison, we similarly trained a MLR on the LMH outputs of the DNN to make DNI predictions, and report that result separately. In both cases, the inputs to the MLR are the LMH predictions for two tissues as well as their logarithm. Schematic of the BNN and MLR architecture can be found in Supplementary Figures S3 and S4.
3 RESULTS
We present three sets of results that compare the test performance of the BNN, DNN and MLR for splicing pattern prediction. The first is the PSI prediction from the LMH code tested on all exons. The second is the PSI prediction evaluated only on targets where there are large variations across tissues for a given exon. These are events where ΔPSI ≥ ±0.15 for at least one pair of tissues, to evaluate the tissue specificity of the model. The third result shows how well the code can classify ΔPSI between the five tissue types. Hyperparameter tuning was used in all methods. The averaged predictions from all partitions and folds are used to evaluate the model's performance on their corresponding test dataset. Similar to training, we tested on exons and tissues that have at least 10 junction reads.
For the LMH code, as the same prediction target can be generated by different input configurations, and there are two LMH outputs, we compute the predictions for all input combinations containing the particular tissue and average them into a single prediction for testing. To assess the stability of the LMH predictions, we calculated the percentage of instances in which there is a prediction from one tissue input configuration that does not agree with another tissue input configuration in terms of class membership, for all exons and tissues. Of all predictions, 91.0% agreed with each other, 4.2% have predictions that are in adjacent classes (i.e. low and medium, or medium and high), and 4.8% otherwise. Of those predictions that agreed with each other, 85.9% correspond to the correct class label on test data, 51.2% for the predictions with adjacent classes and 53.8% for the remaining predictions. This information can be used to assess the confidence of the predicted class labels. Note that predictions spanning adjacent classes may be indicative that the PSI value is somewhere between the two classes, and the above analysis using hard class labels can underestimate the confidence of the model.
3.1 Performance comparison
Table 1a reports AUCLMH_All for PSI predictions from the LMH code on all tissues and exons. The performance of the DNN in the low and high categories are comparable with the BNN, but excels at the medium category, with especially large gains in brain, heart and kidney. Because a large portion of the exons exhibit low tissue variability (Section 1 of Supplementary Material), evaluating the performance of the model on all exons may mask the performance gain of the DNN. This assumes that exons with high tissue variability are more difficult to predict, where a computational model must learn how AS interprets genomic features differently in different cellular environments. To more carefully see the tissue specificity of the different methods, Table 1b reports AUCLMH_TV evaluated on the subset of events that exhibit large tissue variability. Here, the DNN significantly outperforms the BNN in all categories and tissues. The improvement in tissue specificity is evident from the large gains in the medium category, where exons are more likely to have large tissue variability. In both comparisons, the MLR performed poorly compared with both the BNN and DNN.
Table 1.
Comparison of the LMH code's AUC performance on different methods
| (a) AUCLMH_All | ||||
|---|---|---|---|---|
| Tissue | Method | Low | Medium | High |
| Brain | MLR | 81.3 ± 0.1 | 72.4 ± 0.3 | 81.5 ± 0.1 |
| BNN | 89.2 ± 0.4 | 75.2 ± 0.3 | 88.0 ± 0.4 | |
| DNN | 89.3 ± 0.5 | 79.4 ± 0.9 | 88.3 ± 0.6 | |
| Heart | MLR | 84.6 ± 0.1 | 73.1 ± 0.3 | 83.6 ± 0.1 |
| BNN | 91.1 ± 0.3 | 74.7 ± 0.3 | 89.5 ± 0.2 | |
| DNN | 90.7 ± 0.6 | 79.7 ± 1.2 | 89.4 ± 1.1 | |
| Kidney | MLR | 86.7 ± 0.1 | 75.6 ± 0.2 | 86.3 ± 0.1 |
| BNN | 92.5 ± 0.4 | 78.3 ± 0.4 | 91.6 ± 0.4 | |
| DNN | 91.9 ± 0.6 | 82.6 ± 1.1 | 91.2 ± 0.9 | |
| Liver | MLR | 86.5 ± 0.2 | 75.6 ± 0.2 | 86.5 ± 0.1 |
| BNN | 92.7 ± 0.3 | 77.9 ± 0.6 | 92.3 ± 0.5 | |
| DNN | 92.2 ± 0.5 | 80.5 ± 1.0 | 91.1 ± 0.8 | |
| Testis | MLR | 85.6 ± 0.1 | 72.3 ± 0.4 | 85.2 ± 0.1 |
| BNN | 91.1 ± 0.3 | 75.5 ± 0.6 | 90.4 ± 0.3 | |
| DNN | 90.7 ± 0.6 | 76.6 ± 0.7 | 89.7 ± 0.7 | |
| (b) AUCLMH_TV | ||||
|---|---|---|---|---|
| Tissue | Method | Low | Medium | High |
| Brain | MLR | 71.1 ± 0.2 | 58.8 ± 0.2 | 70.8 ± 0.1 |
| BNN | 77.9 ± 0.5 | 61.1 ± 0.5 | 76.5 ± 0.7 | |
| DNN | 82.8 ± 1.0 | 69.5 ± 1.1 | 81.1 ± 0.4 | |
| Heart | MLR | 73.9 ± 0.3 | 58.6 ± 0.4 | 72.7 ± 0.1 |
| BNN | 78.1 ± 0.3 | 58.9 ± 0.3 | 75.7 ± 0.3 | |
| DNN | 82.0 ± 1.1 | 67.4 ± 1.3 | 79.7 ± 1.2 | |
| Kidney | MLR | 79.7 ± 0.3 | 64.3 ± 0.2 | 79.4 ± 0.2 |
| BNN | 83.9 ± 0.5 | 66.4 ± 0.5 | 83.3 ± 0.6 | |
| DNN | 86.2 ± 0.6 | 73.2 ± 1.3 | 85.3 ± 1.2 | |
| Liver | MLR | 80.1 ± 0.5 | 63.7 ± 0.3 | 79.4 ± 0.3 |
| BNN | 84.9 ± 0.7 | 65.4 ± 0.7 | 84.4 ± 0.7 | |
| DNN | 87.7 ± 0.6 | 69.4 ± 1.2 | 84.8 ± 0.8 | |
| Testis | MLR | 77.3 ± 0.2 | 60.8 ± 0.3 | 77.0 ± 0.1 |
| BNN | 81.1 ± 0.5 | 63.9 ± 0.9 | 81.0 ± 0.5 | |
| DNN | 84.6 ± 1.1 | 67.8 ± 0.9 | 83.5 ± 0.9 | |
Notes: ± indicates 1 standard deviation; top performances are shown in bold.
Next, we look at how well the different methods can predict ΔPSI between two tissues, where it must determine the direction of change. This is shown in Table 2. As described above, ΔPSI predictions for the BNN were made by training a MLR classifier on the LMH outputs (BNN-MLR). To make the comparison fair, we included the performance of the DNN in making ΔPSI predictions by also using a MLR classifier (DNN-MLR) on the LMH outputs. Finally, we evaluated the ΔPSI predictions directly from the DNI code, as well as the MLR baseline method, where the inputs include the tissue types.
Table 2.
Comparison of the DNI code's performance in terms of the AUC for decrease versus increase (AUCDvI) and change versus no change (AUCChange)
| (a) AUCDvI |
(b) AUCChange |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Brain versus Heart | Brain versus Kidney | Brain versus Liver | Brain versus Testis | Heart versus Kidney | Heart versus Liver | Heart versus Testis | Kidney versus Liver | Kidney versus Testis | Liver versus Testis | Change versus No change |
| MLR | 50.3 ± 0.2 | 48.8 ± 0.8 | 48.3 ± 1.1 | 51.2 ± 0.5 | 50.0 ± 1.5 | 47.8 ± 1.7 | 51.1 ± 0.5 | 49.4 ± 0.8 | 51.9 ± 0.5 | 51.3 ± 0.6 | 74.7 ± 0.1 |
| BNN-MLR | 65.3 ± 0.3 | 73.7 ± 0.2 | 69.1 ± 0.4 | 72.9 ± 0.5 | 72.6 ± 0.3 | 66.7 ± 0.4 | 68.3 ± 0.7 | 54.7 ± 0.6 | 65.0 ± 0.8 | 65.0 ± 0.9 | 76.6 ± 0.8 |
| DNN-MLR | 77.9 ± 0.1 | 83.0 ± 0.1 | 81.6 ± 0.1 | 82.3 ± 0.2 | 82.4 ± 0.1 | 81.3 ± 0.1 | 82.4 ± 0.1 | 76.8 ± 0.5 | 79.9 ± 0.2 | 79.1 ± 0.1 | 79.9 ± 0.8 |
| DNN | 79.4 ± 0.7 | 83.3 ± 0.8 | 82.5 ± 0.6 | 82.9 ± 0.7 | 86.1 ± 1.0 | 85.1 ± 1.1 | 84.8 ± 0.8 | 76.2 ± 1.0 | 82.5 ± 1.0 | 81.8 ± 1.3 | 86.5 ± 1.0 |
Note: ± indicates 1 standard deviation; top performances are shown in bold.
Table 2a shows the AUCDvI for classifying decrease versus increase inclusion for all pairs of tissue. Both the DNN-MLR and DNN outperform the BNN-MLR by a good margin. Comparing the DNN with DNN-MLR, the DNN shows some gain in differentiating brain and heart AS patterns from other tissues. The performance of differentiating the remaining tissues (kidney, liver and testis) with each other is similar between the DNN and DNN-MLR. We note that the similarity between the DNN and DNN-MLR in terms of performance can be due to the use of soft labels for training. Using MLR directly on the genomic features and tissue types performs rather poorly, where predictions are no better than random.
The models are further evaluated on predicting whether there is a difference in splicing patterns for all tissues, without specifying the direction. AUCChange is computed on all exons and tissue pairs. This is shown in Table 2b. The results indicate that this is a less demanding task, as the models can potentially use just the genomic features to determine whether an exon will have tissue variability. The difference in performance between all methods is less compared with AUCDvI. However, as the evaluation is over all pairs of tissues, the DNN, which has access to the tissue types in the input, does significantly better. Although this is also true for the MLR, it still performed worst overall. This suggests that in the proposed architecture where tissue types are given as an input, the MLR lacks the capacity to learn a representation that can jointly use tissue types and genomic features to make predictions that are tissue-specific. Both results from Table 2 show that there is an advantage to learning a DNI code rather than just learning the LMH code.
To test whether the predictions generalize to RNA-Seq data from a different experiment, we selected data for two mouse tissues, namely the brain and the heart, from (Barbosa-Morais et al., 2012), and analyzed how our model, which is trained with data from (Brawand et al., 2011), performs. Table 3 shows the set of evaluations on the DNN identical to that of Tables 1 and 2, tested on this RNA-Seq data. For the brain, there is an ∼1–4% decrease in AUCLMH_All and ∼4–5% decrease for AUCLMH_TV. For the heart, the model's performance on both dataset is equivalent to within 1 standard deviation for both AUCLMH_All and AUCLMH_TV. A decrease in performance of ∼7% is observed in AUCDvI for brain versus heart. There is an increase in AUCChange but that is owing to only two tissues being evaluated as opposed to five, where the AUC would be pulled down by the other tissues with lower performances if they were present.
Table 3.
Performance of the DNN evaluated on a different RNA-Seq experiment
| (a) AUCLMH_All | |||
|---|---|---|---|
| Tissue | Low | Medium | High |
| Brain | 88.1 ± 0.5 | 76.1 ± 1.0 | 87.0 ± 0.6 |
| Heart | 90.7 ± 0.5 | 78.4 ± 1.3 | 89.0 ± 1.0 |
| (b) AUCLMH_TV | |||
|---|---|---|---|
| Tissue | Low | Medium | High |
| Brain | 79.1 ± 0.9 | 66.1 ± 1.0 | 77.6 ± 0.8 |
| Heart | 82.6 ± 1.0 | 65.3 ± 1.2 | 78.8 ± 1.1 |
| (c) AUCLMH_TV |
(d) AUCChange |
||
|---|---|---|---|
| Method | Brain versus Heart | Method | Change versus No Change |
| DNN-MLR | 72.9 ± 0.1 | DNN-MLR | 81.7 ± 1.0 |
| DNN | 74.2 ± 1.5 | DNN | 91.9 ± 0.7 |
Overall, the decrease in performance is not unexpected, owing to differences in PSI estimates from variations in the experimental setup. To see how PSI differed, we computed the expected PSI values for brain and heart in all exons from both sets of experiments, and evaluated their Pearson correlation. For the brain, the correlation is 0.945, and for the heart, it is 0.974. This can explain why there is a larger decrease in performance for brain, which is a particularly heterogeneous tissue, and hence can vary more between experiments depending on how the samples were prepared. We note that the performance of the DNN on this dataset is still better than the BNN's predictions on the original dataset. Viewed as a whole, the results indicate that our model can indeed be useful for splicing pattern predictions for PSI estimates computed from other datasets. It also shows that our RNA-Seq processing pipeline is consistent.
We believe there are several reasons why the proposed DNN has improved predictive performance in terms of tissue-specificity compared with the previous BNN splicing code. One of the main novelties is the use of tissue types as an input feature, which stringently required the model's hidden representations be in a form that can be well-modulated by information specifying the different tissue types for splicing pattern prediction. This requirement would be relaxed if each tissue was trained separately. Furthermore, this hidden representation is described by thousands of hidden units and multiple layers of non-linearity. In contrast, the BNN only has 30 hidden units to represent the variables that can be used by the model to modulate splicing based on the cellular condition, which may not be sufficient. Another difference is that for the DNI code, a training objective was specifically designed to learn to predict ΔPSI, which is absent from the BNN. However, the performance gain of the DNN-MLR over BNN-MLR shows that this is only part of the improvement.
In addition, we performed hyperparameter search to optimize the DNN, where we gained considerable improvements over our original hand-tuned models, at ∼4.5% for the DNI code and ∼3.5% for the LMH code. Interestingly, the final set of hyperparameters found opt for a much larger (∼4×) number of hidden units than our initial attempt (with matching hyperparameters). Manually trying to find these hyperparameters would have been difficult, where a user may settle for a suboptimal set of hyperparameters owing to the substantial effort and time required for training a model with millions of parameters.
Another performance boost came from the use of dropout, which contributed ∼1–6% improvement in the LMH code for different tissues, and ∼2–7% in the DNI code, compared with without. The performance difference would likely be larger if hyperparameter optimization were not performed on the model that did not use dropout. We note also that even with dropout, a small L1 weight penalty was found to be beneficial, which may explain our model's tolerance for a large number of hidden units with sparse weights.
One additional difference compared with previous work is that training was biased toward the tissue-specific events (by construction of the minibatches), thereby promoting the model to learn a good representation about cellular context. We were able to get some small performance gains (within 2 standard deviations) of ∼1–2% in AUCLMH_TV and AUCDVI using this methodology compared with training with all events treated equally. More importantly, biasing the training examples encourages the model to learn about the tissues as input, which has a significantly different meaning compared with the genomic features and make up only a small number of the input dimension. We find that without this learning bias, the model more frequently settles to a bad local minimum, or does not learn to use the tissues as input at all. Together, all these changes allowed us to train a model that significantly improves on previous work.
With regards to training the two tasks jointly, we found that with hyperparameter tuning, the performance of the model when each task was trained separately compared with being trained together was not statistically different. This is likely because both tasks are too similar for any transfer learning to take place, as evident by the similarity in performance in the DNI code between the DNN and DNN-MLR models. Nevertheless, we find that training both codes together stabilizes learning, specifically, training becomes more tolerant to a larger range of hyperparameters leading to reduced variance between models.
3.2 Model and feature analysis
A major contribution to the success of splicing pattern predictions that generalize well comes from the richness of our feature set. For example, we observed a significant decrease in the performance of the splicing code if the reduced feature vector dimension is too small by either principle component analysis or an autoencoder with small number of hidden units. We found that the performance of both the LMH code and the DNI code drops by up to 4% when the reduced dimension is at 150 (down from 1393). This suggests a sufficiently large number of hidden variables denoting genomic features are required to interact with tissue inputs to achieve good performance.
It can be useful to see how the genomic features are used by the DNN to perform splicing pattern predictions. We analyzed our model in two different ways.
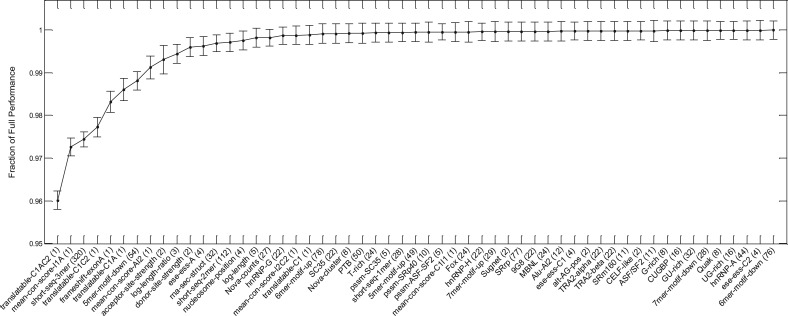
In the first method, to see which feature types are important to the model, we substituted genomic features to their median across all exons and looked at how the predictive performance changed. We divided the full feature set into 55 groups based on what they represent. The grouping, along with additional descriptions, can be found in Section 4 of the Supplementary Material. Here, the performance measure is defined as the sum of the three classes from AUCLMH_All. The decrease in test performance (as a fraction of that obtained with the full feature set) when each group of features is substituted by their median is shown in Figure 2. Feature groups that cause large decrease in performance are presumably important to the splicing code. The standard deviation is computed from the five trained models with random partitions of the data as described above. The order of the feature group toward the right of the plot should not be used to determine their order of importance owing to the small difference they make to the model relative to their standard deviations. It is interesting to see how small the decrease in AUC is when each feature group is effectively removed. Many features contain redundant information and therefore can compensate for missing features from other groups. For example, some of the motifs for splicing factors are represented in features representing n-mer counts. The most influential features describe the translatability of the exon, conservation scores and whether the alternative exon introduces a frame shift. The feature groups corresponding to counts of 3-mers and 5-mers are also important.
Fig. 2.
Plot of the change in AUCLMH_All by substituting the values in each feature groups by their median. Feature groups that are more important to the predictive performance of the model have lower values. The groups are sorted by the mean over multiple partitions and folds, with the standard deviations shown. The number of features for each feature group are indicated in brackets
To examine how each individual feature affects the DNN's predictions, we adapted the method from (Simonyan et al., 2014). Briefly, examples from the dataset are given as input to the trained model and forward propagated through the neural network. At the output, the target is modified to a different value, for example, in classification, by changing the class label. The error signal is then backpropagated to the inputs. The resulting signal describes how much each input feature needs to change to make the modified prediction, as well as the direction. The computation is extremely quick, as it only requires a single forward and backward pass through the DNN, and all examples can be calculated in parallel. We used this procedure on exons with low tissue variability, and modified the low PSI targets to high, and the high PSI targets to low. Table 4 lists the top 25 features with the largest backpropagated signal magnitude (which indicate that these features need to change the least to affect the prediction the most, and are hence important; note also that all of our features are normalized). The table also indicates general trends in the direction of change for each feature over the dataset. If more than 5% of the examples do not follow the general direction of change, it is indicated by both an up and down arrow. Some of the splicing rules inferred by the model can be seen. For example, presence of splicing silencers inhibits the splicing of the alternative exon leading to higher inclusion, a shorter alternative exon is more likely to be spliced out, and the strength and position of acceptor and donor sites can lead to different splicing patterns.
Table 4.
The top 25 features (unordered) of the splicing code that describes low and high percent inclusion
| Feature description | Low | High |
|---|---|---|
| Strength of the I1 acceptor site | ↓ | ↑ |
| Strength of the I2 donor site | ↓ | ↑ |
| Strength of the I1 donor site | ↑ | ↓ |
| Mean conservation score of first 100 bases in 3′ end of I1 | ↑↓ | ↑↓ |
| Mean conservation score of first 100 bases in 5′ end of I2 | ↑↓ | ↑↓ |
| Counts of Burge's exonic splicing silencer in A | ↓ | ↑ |
| Counts of Chasin's exonic splicing silencer in A | ↓ | ↑ |
| Log base 10 length of exon A | ↓ | ↑ |
| Log base 10 length ratio between A and I2 | ↓ | ↑ |
| Whether exon A introduces frame shift | ↑↓ | ↑↓ |
| Predicted nucleosome positioning in 3′ end of A | ↑↓ | ↑↓ |
| Frequency of AGG in exon A | ↑ | ↓ |
| Frequency of CAA in exon A | ↓ | ↑ |
| Frequency of CGA in exon A | ↓ | ↑ |
| Frequency of TAG in exon A | ↑ | ↓ |
| Frequency of TCG in exon A | ↓ | ↑ |
| Frequency of TTA in exon A | ↑ | ↓ |
| Translatability of C1-A | ↓ | ↑ |
| Translatability of C1-A–C2 | ↓ | ↑ |
| Translatability of C1–C2 | ↑ | ↓ |
| Counts of Yeo's ‘GTAAC’ motif cluster in 5′ end of I2 | ↓ | ↑ |
| Counts of Yeo's ‘TGAGT’ motif cluster in 5′ end of I2 | ↓ | ↑ |
| Counts of Yeo's ‘GTAGG’ motif cluster in 5′ end of I2 | ↓ | ↑ |
| Counts of Yeo's ‘GTGAG’ motif cluster in 5′ end of I2 | ↓ | ↑ |
| Counts of Yeo's ‘GTAAG’ motif cluster in 5′ end of I2 | ↓ | ↑ |
Note: The direction of the arrows indicate that a feature's value should in general be increased (↑) or decreased (↓) to change the PSI predictions to low or high. Feature details can be found in Section 4 of the Supplementary Material.
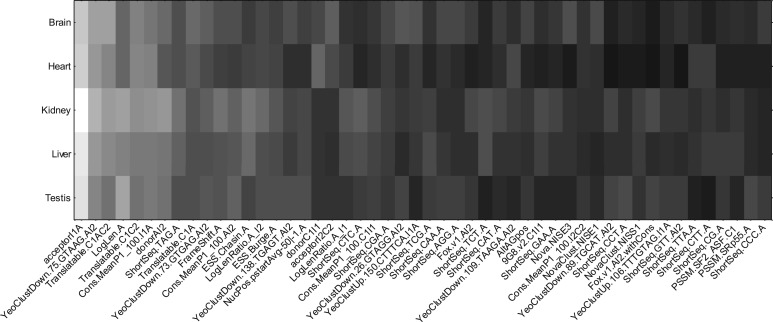
Next, we wanted to see how features are used in a tissue-specific manner. Using the set of exons with high tissue variability, we computed the backpropagation signal to the inputs with the output targets changed in the same manner as above, for each tissue separately. Figure 3 shows the sum of the magnitudes of the gradient, normalized by the number of examples in each tissue for the top 50 features. We can observe that the sensitivity of each feature to the model's predictions differs between tissues. The profile for kidney and liver tend to be more similar with each other than others, which associates well with the model's weaker performance in differentiating these two tissues. This figure also provides a view of how genomic features are differentially used by the DNN, modulated by the input tissue types. In both Table 4 and Figure 3, the backpropagation signals were computed on examples from the test set, for all five partitions and folds.
Fig. 3.
Magnitude of the backpropagated signal to the input of the top 50 features computed when the targets are changed from low to high, and high to low. White indicates that the magnitude of the signal is large, meaning that small perturbations to this input can cause large changes to the model's predictions. The features are approximately sorted left to right by the magnitude
4 CONCLUSIONS
In this work, we introduced a computational model that extends the previous splicing code with new prediction targets and improved tissue-specificity, using a learning algorithm that scales well with the volume of data and the number of hidden variables. The approach is based on DNNs, which can be trained rapidly with the aid of GPUs, thereby allowing the models to have a large set of parameters and deal with complex relationships present in the data. We demonstrate that deep architectures can be beneficial even with a sparse biological dataset. We further described how the input features can be analyzed in terms of the predictions of the model to gain some insights into the inferred tissue-regulated splicing code.
Our architecture can easily be extended to the case of more data from different sources. For example, using the same architecture, we may be able to learn a hidden representation that spans additional tissue types as well as multiple species. Through transfer learning, training such model with multiple related targets might be beneficial particularly if the number of training examples in certain species or tissues is small.
ACKNOWLEDGEMENT
The authors thank Andrew Delong for helpful discussion and assistance with GPU computing.
Funding: B.J.F. received funding from the Canadian Institutes for Health Research, the Natural Sciences and Engineering Research Council of Canada, and a John C Polanyi Award. M.K.K.L. was supported by the Natural Science and Engineering Research Council Canada Graduate Scholarship.
Conflict of Interest: None declared.
REFERENCES
- Ahn S, et al. Proceedings of the 29th International Conference on Machine Learning. 2012. Bayesian posterior sampling via stochastic gradient fisher scoring; pp. 1591–1598. [Google Scholar]
- Barash Y, et al. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- Barash Y, et al. AVISPA: a web tool for the prediction and analysis of alternative splicing. Genome Biol. 2013;14:R114. doi: 10.1186/gb-2013-14-10-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Bengio Y. Learning deep architectures for AI. Foundations and Trends in Machine Learning. 2009;2:1–127. [Google Scholar]
- Bengio Y, et al. Representation learning: a review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013;35:1798–1828. doi: 10.1109/TPAMI.2013.50. [DOI] [PubMed] [Google Scholar]
- Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- Caruana R, Niculescu-Mizil A. Proceedings of the 23rd International Conference on Machine Learning - ICML’06. 2006. An empirical comparison of supervised learning algorithms; pp. 161–168. [Google Scholar]
- Eickholt J, Cheng J. Predicting protein residue-residue contacts using deep networks and boosting. Bioinformatics. 2012;28:3066–3072. doi: 10.1093/bioinformatics/bts598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhan D, et al. Why does unsupervised pre-training help deep learning? J. Mach. Learn. Res. 2010;11:625–660. [Google Scholar]
- Fawcett T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006;27:861–874. [Google Scholar]
- Glorot X, et al. Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics. 2011. Deep sparse rectifier neural networks. [Google Scholar]
- Hinton G, Srivastava N. Improving neural networks by preventing co-adaptation of feature detectors. arXiv Tech. Rep. 2012:1207.0580. [Google Scholar]
- Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313:504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- Kakaradov B, et al. Challenges in estimating percent inclusion of alternatively spliced junctions from RNA-seq data. BMC Bioinformatics. 2012;13(Suppl 6):S11. doi: 10.1186/1471-2105-13-S6-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, et al. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lena P, et al. Deep architectures for protein contact map prediction. Bioinformatics. 2012;28:2449–2457. doi: 10.1093/bioinformatics/bts475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Silver D, Mercer R. The parallel transfer of task knowledge using dynamic learning rates based on a measure of relatedness. Connect. Sci. Spec. Issue Transf. Inductive Syst. 1996;8:277–294. [Google Scholar]
- Simonyan K, et al. Workshop at International Conference on Learning Representations. 2014. Deep inside convolutional networks: visualising image classification models and saliency maps. [Google Scholar]
- Snoek J, et al. Proceedings of Neural Information and Processing Systems. 2012. Practical Bayesian optimization of machine learning algorithms. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. B. 1994;73:273–282. [Google Scholar]
- Tieleman T. 2010. Gnumpy: an easy way to use GPU boards in Python. Technical Report. University of Toronto, Toronto. [Google Scholar]
- Touw WG, et al. Data mining in the Life Sciences with Random Forest: a walk in the park or lost in the jungle? Brief. Bioinform. 2013;14:315–326. doi: 10.1093/bib/bbs034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, et al. Bayesian prediction of tissue-regulated splicing using RNA sequence and cellular context. Bioinformatics. 2011;27:2554–2562. doi: 10.1093/bioinformatics/btr444. [DOI] [PubMed] [Google Scholar]