Abstract
Summary: Development of effective tools such as oligo-microarrays and next-generation sequencing methods for monitoring gene expression on a large scale has resulted in the discovery of gene signatures with prognostic/predictive value in various malignant neoplastic diseases. However, with the exponential growth of gene expression databases, biologists are faced with the challenge of extracting useful information from these repositories. Here, we present a software package, BioPlat (Biomarkers Platform), which allows biologists to identify novel prognostic and predictive cancer biomarkers based on the data mining of gene expression signatures and gene expression profiling databases. BioPlat has been designed as an easy-to-use and flexible desktop software application, which provides a set of analytical tools related to data extraction, preprocessing, filtering, gene expression signature calculation, in silico validation, feature selection and annotation that leverage the integration and reuse of gene expression signatures in the context of follow-up data.
Availability and implementation: BioPlat is a platform-independent software implemented in Java and supported on GNU/Linux and MS Windows, which is freely available for download at http://www.cancergenomics.net.
Contact: mcabba@gmail.com
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Human cancer transcriptomes have been extensively profiled over the past decade, allowing the identification of different cancer molecular subtypes and the development of prognostic and predictive gene expression signatures. Gene expression signatures have been curated from the literature and collected into publicly available databases such as MSigDB 3.0 and GeneSigDB 4.0, with >10 000 gene signatures related to human diseases (Culhane et al., 2012; Liberzon et al., 2011). On the other hand, databases such as GEO (Gene Expression Omnibus) and AE (ArrayExpress) are the primary repositories for the raw data from functional genomic studies (Barrett et al., 2013; Rustici et al., 2013). A central focus on translational cancer research is that patient diagnosis and prognosis can be improved by stratification of patients based on functional genomics data beside relevant follow-up data. Therefore, we developed BioPlat (Biomarkers Platform), a user-friendly bioinformatics resource, which provides a set of analytical tools for the in silico identification of novel prognostic and predictive cancer biomarkers. It encompasses all the stages of data mining and analysis of the vast number of gene expression signatures and profiling data, including data extraction, preprocessing, filtering, in silico validation and feature selection.
Currently, there exists a myriad of applications and software focused on the integration and analysis of oncogenomics and clinicopathological data (e.g. Oncomine, ITTACA, PAPAyA, IntOGene, Cancer Genomics Browser, MeV, GenePattern). However, the salient feature that makes BioPlat unique is that it allows the direct and easy integration of gene expression signatures and gene expression profiling data to further perform survival analysis. Moreover, BioPlat implements features that are not included in other biomarker discovery tools. These features include statistic methods to measure the performance of prediction models and also algorithms to perform feature selection. Although several of these statistical tests and algorithms are mostly available in R packages, our software provides a unified framework that facilitates the use of these components.
2 METHODS
2.1 System implementation
BioPlat is a desktop client application implemented in Java and based on Rich Client Platform and Standard Widget Toolkit. It uses object-oriented programming, a local H2 in-memory database and Hibernate to perform the object-relational mapping. The Java-embedded database contains all the required annotation data for mapping probes and gene identifiers (Entrez ID and Ensembl ID) to gene symbols and related information. Statistical and data mining analyses are performed with the R statistical package and Bioconductor (Gentleman et al., 2004). Briefly, we use several R/Bioconductor packages (e.g. frma, inSilicoDb, fpc, affy, genefu, limma, survival, survcomp) and functions (e.g. dist, hclust, cutree) for data retrieval, preprocessing, management and analysis. The integration between Java and R was achieved using an R bridge developed by us (named R4J) based on Rserve. BioPlat was designed using extension points for allowing users with programming knowledge to incorporate new behavior and algorithms easily.
2.2 Features and algorithms implementation
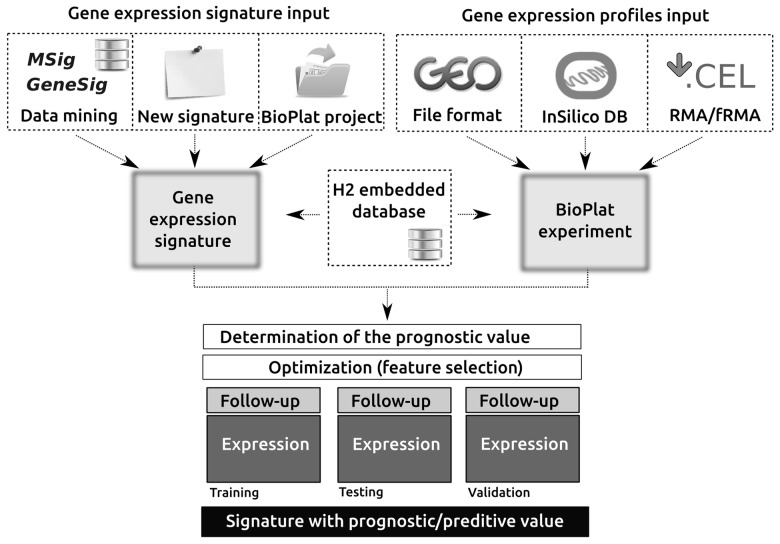
BioPlat consists of an integrated set of tools that allow the access and analysis of data deposited in gene expression signatures and gene expression profiling repositories (Fig. 1 and Supplementary Data S1). Gene expression signatures can be queried and filtered by tumor localization, tissue types or signature identifiers directly from the two main curated repositories: MSigDB (Liberzon et al., 2011) and GeneSigDB (Culhane et al., 2012). BioPlat allows the creation/edition of gene lists by using any of the identifiers included in the embedded database (e.g. Gene name, Entrez, Ensembl and Probe IDs). In addition, BioPlat integrates information and tools provided by other well-known online resources such as DAVID (Database for Annotation, Visualization and Integrated Discovery), STRING (Search Tool for the Retrieval of Interacting Genes/Proteins), Enrichr, Expression Atlas, RNA-seq Atlas, Gene Cards and others.
Fig. 1.
Diagrammatic workflow of BioPlat. The first two steps in BioPlat are the gene signature creation and the import of experiments containing follow-up and gene expression profiling data. BioPlat provides several processes for the gene list generation such as access and data mining of gene signature databases. Gene expression data, provided by experiments incorporated in the platform, are used to cluster the samples of the experiments based on K-means method. This clustering will let the platform validate the statistical significance level of the biomarker using the follow-up data of the same experiment in terms of C-index, Log-rank test and Kaplan–Meier survival analysis
Gene expression profiles (BioPlat experiments) are used for the validation and optimization steps of gene signatures previously defined. A BioPlat experiment contains follow-up data and gene expression profiles of samples. BioPlat experiments can be programmatically queried and imported from InSilico database (Taminau et al., 2011), local GEO-formatted files or preprocessing Affymetrix CEL files with RMA/fRMA (frozen robust multiarray analysis) algorithms. Although the platform is prepared for incorporating experiment mining processes as a new way to propose candidate gene lists, the main use of the experiments in BioPlat is to estimate the prognostic/predictive value of a gene expression signature. In the validation process, given a pair of gene signature experiments, the results shown are the annotated data matrix with the patient’s cluster, the concordance index (C-index), the Log-Rank test P-value and the Kaplan–Meier curves.
Because of the large number of features usually present in the gene expression signature analyzed, signature optimization is a key step that was considered in the development of the software. We focus on selecting compact feature subsets while maximizing prediction accuracy for biomarker discovery to reduce complexity. BioPlat provides two optimization processes—‘blind search’ and particle swarm optimization (PSO)—to search a better candidate gene signature in the solution space of the signature to be optimized. The implementation of the PSO algorithm in BioPlat was based on the PSO binary version previously described (Kennedy and Eberhart, 1997). Briefly, PSO is a machine learning metaheuristic whose aim is to reduce the solution space for finding the optimum gene signature without going over the whole space based on a metric of quality associated with the outcome (e.g. C-index). Considering that the metric of performance is a critical point for the algorithms and that new metrics are being worked on, the comparison strategy was designed in the platform to be easily replaced without affecting the core of the algorithm. Moreover, to avoid the overfitting of the found solution on the training experiment used for running the heuristic logics, any optimizer allows configuring not only a training experiment but also a testing experiment and a validation one.
3 USAGE EXAMPLE
We wanted to provide a systematic analysis of breast cancer gene expression signatures, in an effort to identify breast cancer biomarkers with prognostic value. The integration of 655 breast cancer signatures obtained from MSig and GeneSig databases, using the Metasignature wizard, revealed a set of 140 genes that were the most commonly deregulated transcripts among breast cancer studies (Supplementary Data S2A). K-means clustering of the metasignature in three independent publicly available datasets (GSE25066 n = 508, NKI dataset n = 295 and a compiled dataset of 737 carcinomas derived from GSE2034, GSE3494 and GSE11121) identified two main clusters of breast carcinomas that differed in their relapse-free survival (Supplementary Data S2B).
Feature selection analysis of the 140 gene metasignature in the GSE25066 study (training dataset) using PSO-based process allows reducing the candidate gene signature to 60 genes (Supplementary Data S2C and S3) with improved prognostic performances as reflected by having the highest C-index and the lowest nominal log-rank P-values for relapse-free survival. Interestingly, cluster 1 was highly associated with luminal A/B breast cancer intrinsic subtypes, whereas cluster 2 was associated with basal-like breast carcinomas. Gene enrichment analysis revealed two functional modules significantly affected: one related with the response to steroid hormone (upmodulated in cluster 1) and another related with the cell cycle signaling pathway (upmodulated in cluster 2) (Supplementary Data S2A–E). Finally, we compared the prognostic performance of the 60-gene signature with the PAM50 and genomic grade Index signatures on the GSE25066 dataset. This analysis demonstrated that the 60-gene signature outperformed the genomic grade Index, having similar prognostic value that the PAM50 signature in predicting the relapse-free survival of patients with early-stage breast cancers (Supplementary Data S4).
4 CONCLUSION
BioPlat facilitates the integration, analysis, validation and feature selection of gene signatures derived from different databases in the context of follow-up data obtained from publicly available gene expression profiling repositories.
Funding: National Agency of Scientific and Technological Promotion (PICT-0275) and The National Cancer Institute of Argentina.
Conflict of Interest: none declared.
REFERENCES
- Barrett T, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullhane AC, et al. GeneSigDB: a manually curated database and resource for analysis of gene expression signatures. Nucleic Acid Res. 2012;40:D1060–D1066. doi: 10.1093/nar/gkr901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy J, Eberhart R. Particle swarm optimization. IEEE Neural Netw. Proc. 1995;4:1942–1948. [Google Scholar]
- Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustici G, et al. ArrayExpress update—trends in database growth and links to data analysis tools. Nucleic Acids Res. 2013;41:D987–D990. doi: 10.1093/nar/gks1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taminau J, et al. InSilicoDb: an R/Bioconductor package for accessing human Affymetrix expert-curated datasets from GEO. Bioinformatics. 2011;27:3204–3205. doi: 10.1093/bioinformatics/btr529. [DOI] [PubMed] [Google Scholar]