Fig. 1.
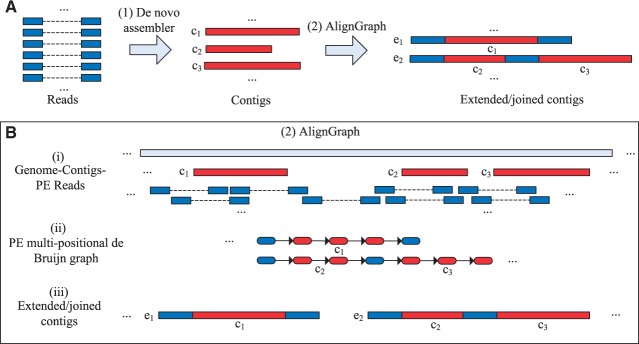
Overview of the AlignGraph algorithm. The outline on the top (A) shows AlignGraph in the context of common genome assembly workflows, and the one on the bottom (B) illustrates its three main processing steps. (A) In Step 1, the PE reads from a target genome are assembled by a de novo assembler into contigs (here and c3). Subsequently (Step 2), the contigs can be extended (blue) and joined by AlignGraph (e1 and e2). (B) The workflow of AlignGraph consists of three main steps. (i) The PE reads are aligned to the reference genome and to the contigs, and the contigs are also aligned to the reference genome. (ii) The PE multipositional de Bruijn graph is built from the alignment results, where the red and blue subpaths correspond to the aligned contigs and sequences from PE reads, respectively. (iii) The extended and/or joined contigs (here e1 and e2) are generated by traversing the graph
