Abstract
Ovarian cancer is a silent killer as most patients have non-specific symptoms and usually present in advanced stage of the disease. It occurs due to certain genetic alterations and mutations namely founder mutations, 187delAG and 5385insC in BRCA1 and 6174delT in BRCA2 which are associated with specific family histories. These highly penetrant susceptibility genes responsible for approximately half of families containing 2 or more ovarian cancer cases account for less than 40% of the familial excess malignancy risk. The remaining risk may be due to single nucleotide polymorphisms (SNPs) which are single base change in a DNA sequence with usual alternatives of two possible nucleotides at a given position. Preliminary study involving 30 women with histologically proven epithelial ovarian cancer was conducted and their detailed genetic analysis was carried out. Regions of founder mutations on BRCA1 and BRCA2 were amplified and sequenced using primers designed based on 200 bp upstream and downstream regions of the mutation sites. Five sequence variants in BRCA1 were identified of which three novel sequence variants were found in 23 patients while in BRCA2, one novel sequence variant was found. The three founder mutations 187delAG, 5385insC in BRCA1 and 6174delT in BRCA2 were not seen in any of the subjects.
Keywords: Epithelial ovarian cancer, BRCA1, BRCA2, single nucleotide polymorphism
Introduction
Ovarian cancer is a major public health problem being the number one killer amongst gynecological malignancies with more than two thirds of patients presenting with late stage disease [1,2]. Globally it is the fourth most common cause of cancer related death among women [3]. In India, the age adjusted rate (AAR) of ovarian cancer is 5.3/100,000 women [4]. Ovarian cancer seems to be the result of a multistep process due to accumulation of genetic alterations, which in women with familiarity for ovarian tumors, could be inherited. The hereditary cancers are associated with a dominant autosomal genetic predisposition with high penetrance conferred primarily by BRCA1 and BRCA2 genes [5].
The cumulative life time risk of developing epithelial ovarian cancer associated with mutations of BRCA1 And BRCA2 ranges from 40%-50% and 20%-30% respectively [5]. BRCA1 mutations are four times more common than BRCA2 mutations [3]. BRCA1 and BRCA2 participates to a large multiprotein complex that acts as a sensor for DNA damage [6-8]. Though these highly penetrant susceptibility genes are responsible for approximately half of families containing 2 or more ovarian cases, these account for less than 40% of the familial excess risk of ovarian cancer [9,10]. An increased-risk family history is seen in two percent of adult women in the general population [11]. The familial risks also seem to be due to combinations of multiple alleles conferring moderate to low penetrance susceptibility which are caused by single nucleotide polymorphisms (SNPs) in proto-ontogenesis [12,13]. SNPs are genetically stable, hence, they serve as excellent biological markers. On an average, frequency of SNPs in the human population is more than 1%. SNPs found within a coding sequence are more likely to alter the biological function of a protein.
We intend, by means of this study, to assess the genetic alterations that persist in BRCA1/BRCA2 genes that are likely to increase the carriers’ risk of developing epithelial ovarian cancer. An important aspect of the study was also to evaluate the differences in symptomatology between ovarian cancer cases with above mentioned mutations and those with somatic alterations.
Material and method
Sample collection
This pilot study was conducted in the Department of Obstetrics and Gynaecology, Pathology and Biochemistry at Guru Teg Bahadur Hospital, Delhi. The study group included 30 patients with histologically proven epithelial ovarian cancer. After carefully coding the patients, their paraffin block samples were studied for BRCA1 and BRCA2 founder mutations.
Patients up to 60 years of age with histopathologically proven epithelial ovarian cancer who had no prior chemotherapy or radiotherapy were included in the study. Women with other types of ovarian cancer like germ cell or sex cord tumors, those with recurrent ovarian cancer and epithelial ovarian cancer patients who had received prior chemotherapy or radiotherapy were excluded from the study.
DNA isolation
DNA was isolated from paraffin blocks of patients using QIAGEN genomic DNA extraction kit. DNA was checked for its quality and quantified using both Agarose Gel Electrophoresis as well as PicoGreen quantification to study polymorphisms.
PCR amplification of founder mutations
Founder mutations namely 187delAG, 5385insC and 6174delT of BRCA1 and BRCA2 genes were validated in our study group. The regions of interest were amplified and directly sequenced to detect the mutations. A 340 bp region covering 187delAG in exon 2 and a 271 bp region covering 5382insC in exon 20 of BRCA1 was selected for amplification. A 270 bp region in exon 11 covering 6174delT in BRCA2 was also amplified. Primers designed from the upstream and downstream regions of the mutation sites were as follows: 187delAG_F_int: TTCGTATTCTGAGAGGCTGCT; 187delAG_R_int: ACGCCTCTCAGGTTCCG; 5385insC_F_exn: TGCAGATGCTGAGTTTGTGT; 5385insC_R_int: AGCTTATCTGAACAAAGTGATATT; 6174delT_F_exn: ATATGTCTGGATTGGAGAAAGTT; 6174delT_R_exn: AGCTGGTCTGAATGTTCGTT.
PCRs were set up in a 25 μl volumes, using 10-20 ng template DNA, 1 × PCR Buffer, 1.5-2.5 mM of MgCl2, 2 mM dNTPs, 5-10 pmoles of each Forward and Reverse primers and 1 unit Taq Polymerase (Promega). Initial denaturation of the DNA was done at 95° for 5 min followed by 35 cycles of denaturation at 95° for 60 sec, annealing at 55° for 30 sec and extension at 72° for 1 min followed by final extension at 72° for 10 min. An aliquot of 2 ul of each PCR product was used for Agarose gel electrophoresis analysis. The above conditions were the same for all the three regions. Samples that did not amplify in these conditions underwent troubleshooting by altering the number of cycles.
Sequencing
Sequencing performed using Big dye terminator (BDTv3.1) chemistry (Applied Biosystems). Full scale sequencing reactions set up in 20 μl volume using 20-50 ng of purified PCR products and 5-10 picomoles of sequencing primers with 5% DMSO as an additive. Initial denaturation was done at 96°C for 1 min followed by 30 cycles of denaturation at 96°C for 10 sec, annealing at 50°C for 5 sec and extension at 60° for 5 min. These sequenced products were purified using Sodium acetate and ethanol precipitation method and sequenced on 3730 DNA Sequencer. Sequencing data of samples were viewed using Sequence Scanner (v1.0) software (Applied Biosystems).
Sequence analysis
The sequence output from the instrument was taken for sequence analysis in several softwares and the sequences were studied for any variation. Briefly, the forward and the reverse sequences were trimmed and they were individually analyzed to ensure no disparity in the forward and reverse sequences. All the sequences were then aligned with the corresponding region from the actual sequence downloaded from NCBI. Alignment was done using Bioedit software. After the alignment, the sequences were studied for identifying mutations and single nucleotide polymorphisms. For this purpose, softwares like Mutation Surveyor, Seqscape and FinchTV were used for analysis and confirmation and generation of mutation report.
Results
The founder mutations namely 187delAG and 5382insC in BRCA1 and 6174delT in BRCA2 genes were evaluated in patients (n=30) of EOC. Age at diagnosis, clinical presentation, stage, tumor markers, histopathology and the outcome were also studied. We observed that 16.5% (n=5) patients were nulliparous and were diagnosed at an average age of 29±9.46 years which was lower than the overall average of 46.27±10.66 years. It was also observed that delayed pregnancy, low parity, infertility and use of ovulation induction agents were risk factors for ovarian cancer. On histopathological examination all women (n=30) were found to have papillary serous adenocarcinoma.
All samples were amplified for sequence variants both in exon as well as exon-intron boundaries for the three founder mutations and any other mutations within a region of approx. 200 bps upstream and downstream to these sites in BRCA1 and BRCA2. The sequences were aligned after trimming with the Forward and Reverse sequences to highlight the regions of sequence showing variations. After aligning, the samples that showed variations were analyzed further and mutation report generated.
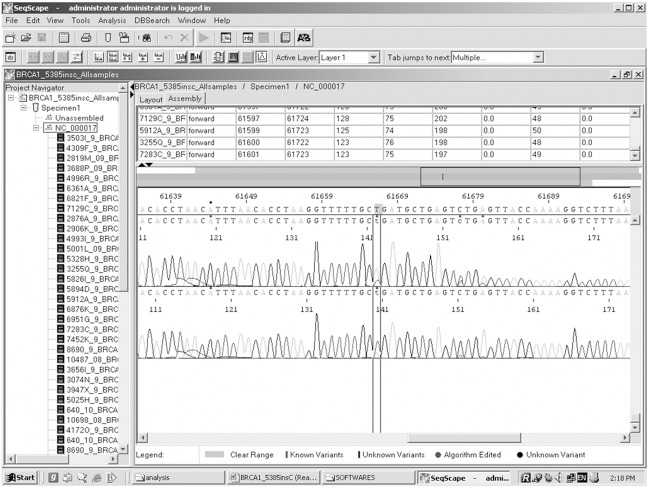
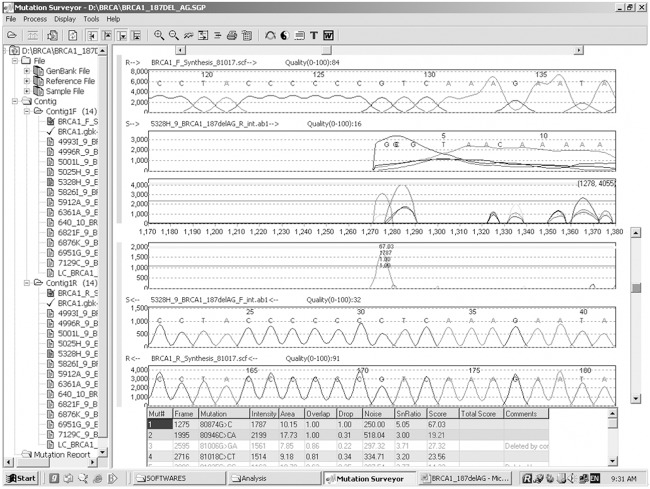
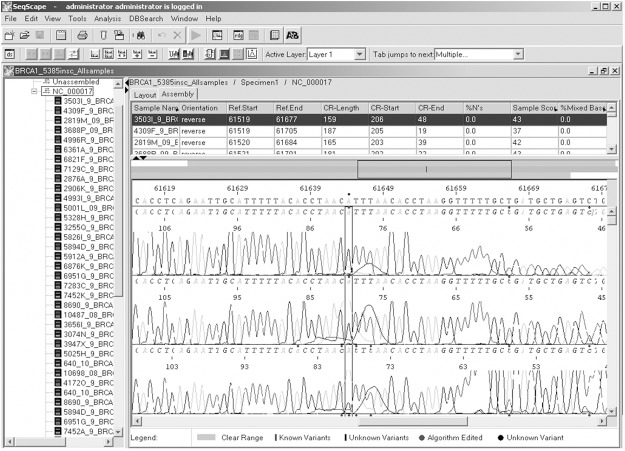
Genetic analysis of BRCA1 revealed five sequence variants. A novel sequence variant G>A at nucleotide position 38530776 was seen in one patient (Figure 1); T>A at nucleotide position 3869359, was observed in 14 patients (Figure 2) and another novel sequence variation (C>T) at nucleotide position 38469369 was identified in eight patients (Figure 3). A reported SNP, C>G at nucleotide position 38530676 was seen in 19 patients (Figure 4). Reported SNP, G>A at nucleotide position 38469338 was identified in 23 patients (Figure 5). However, both the founder mutations, namely 187delAG and 5385insC were not found in BRCA1 gene. In BRCA2, one novel sequence variant A>T at nucleotide position 31818421 was seen in 1 patient (Figure 6) while the founder mutation 6174delT was not observed in BRCA2 gene.
Figure 1.
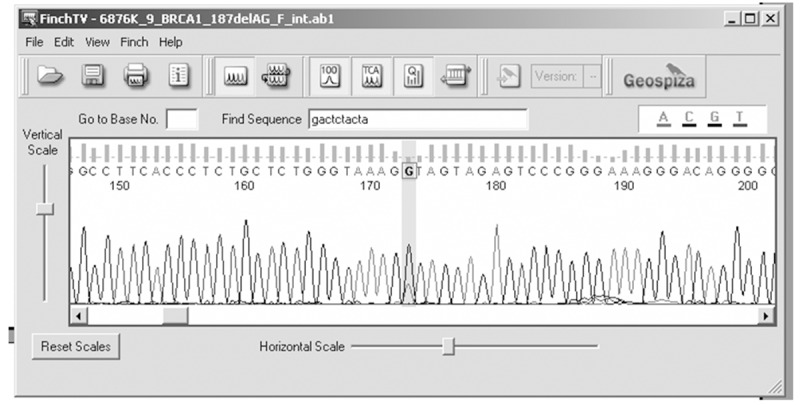
Electropherogram showing G>A at nucleotide position 38530776.
Figure 2.
Identification of sequence variant T>A at nucleotide position 3869359 using Seqscape.
Figure 3.
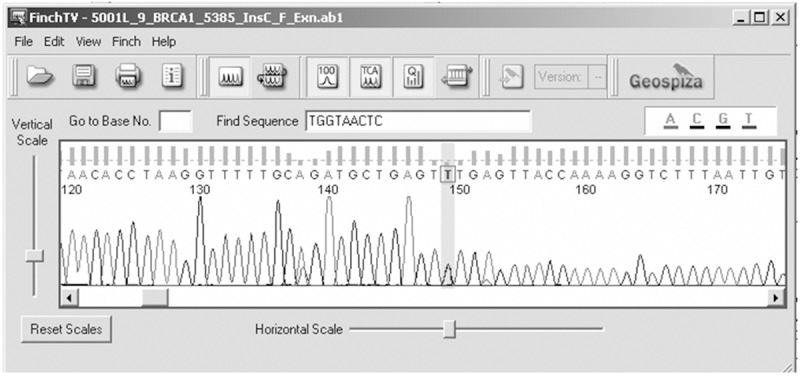
Electropherogram showing (C>T) at nucleotide position 38469369.
Figure 4.
SNP identification at nucleotide position 38530676 using Mutation Surveryor.
Figure 5.
Identification of SNP, G>A at nucleotide position 38469338 using Seqscape.
Figure 6.
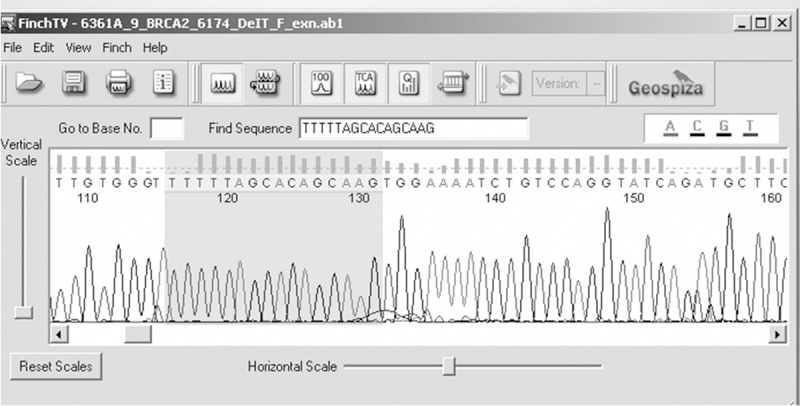
Electropherogram showing A>T at nucleotide position 31818421.
All the 30 patients were followed-up for a mean period of 6 months with a range of 3-9 months. Their response to chemotherapy, trend of CA-125 levels and quality of life were assessed. Twenty of them were tumor free by either CT or PET scan and hence showed a complete response by WHO and RECIST criteria, while 3 had some remnant deposits on either CT or PET scan showing a partial response by the same criteria.
Discussion
Epidemiologic studies indicate that hereditary cancers constitute 5-10% [6] and may be up to 14% [2,14,15] of all epithelial ovarian cancers. These cancers are autosomal dominant conferred primarily by mutations in BRCA1 and BRCA2 genes [16]. BRCA1 and 2 mutations account for >90% of hereditary cancers. It is estimated that in general population approximately 1 in 300 to 1 in 800 individuals carry a mutation in BRCA1 or BRCA2 genes. Estimates of the frequency of BRCA1 and BRCA2 germ line mutations in ovarian cancer have ranged between 2-12% and 2-6% for BRCA1 and BRCA2 mutations respectively [17].
The present study analyzes mutations namely 187delAG and 5382insC in BRCA1 and 6174delT in BRCA2 genes in 30 patients of histologically proven epithelial ovarian cancer. The profile we considered in the study were age at diagnosis, clinical presentation, stage, tumor markers, histopathology and the outcome.
The average age at diagnosis for sporadic tumors was 62.3 years and that of familial cases was 49-54.3 years as reported by Zweemer et al. However, in our study we observed that the average age at diagnosis was 46.27 (SD±10.66) years with range of 19-60 years which was also lower than the average age at diagnosis of 57 years for sporadic cases, 53 years for BRCA1, and 58 years for BRCA2 carriers as reported by Pal et al [2]. Thus the average age at presentation was lower in our study vis-à-vis the above studies but comparable to a published study of five years between 2005 and 2009, from our hospital where the mean age at diagnosis of epithelial ovarian cancers (n=147) was 45.8±12.4 [18]. This is an interesting finding which needs to be explored further as epithelial ovarian cancers are mostly seen in the 6th decade of life in Western studies [19].
Previous studies indicated that having at least one child is protective with risk reduction to 0.3-0.4 and the risk reduces as the age at last pregnancy increases [20,21] while delayed pregnancy, low parity, infertility and use of ovulation induction agents increase the risk of developing ovarian cancer. In our study we observed that the 16.5% (n=5) of nulliparous patients were diagnosed at a lower average age of 29 (SD±9.46) years as compared to the overall average of 46.27 (SD±10.66) years. Of the five nulliparous patients, 60% (n=3) were in stage 3 of the disease and one of them also had a history of use of ovulation induction agents.
In this study, 340 bp region covering 187delAG on exon 2 and a 271 bp region covering 5382insC on exon 20 of BRCA1 as well as 270 bp region covering 6174delT on exon 11 of BRCA2 were amplified and evaluated. The three founder mutations namely 187delAG and 5382insC in BRCA1 and 6174delT in BRCA2 gene, implicated in familial ovarian cancer and observed to have an autosomal pattern of inheritance, were not found in any of the 30 patients. However, these mutations were observed in other previous studies [22,23] and we have also found few common reported as well as 4 novel mutations. A total of five sequence variants, including two reported single nucleotide polymorphisms and three novel sequence variants, were seen in BRCA1 gene while only one novel sequence variant was observed in the BRCA2 gene. A reported SNP, C>G at position 38530676 was seen in 63.3% (n=19) patients and another SNP, G>A at position 38469338 was seen in 76.7% (n=23) of patients in BRCA1 gene. A sequence variant, G>A was identified at position 38530776 in 3.33% (n=1) patient while another sequence variant was, T>A at position 3869359 was seen in 46.67% (n=14) patients. A novel sequence variant C>T in the same gene was identified at position 38469369 in 26.7% (n=8) patients in BRCA1 gene. However, only one novel sequence A>T at 31818421 was seen in 3.3% (n=1) patient in BRCA2 gene. Thus, the mutation frequency of BRCA1 was 76.7% and BRCA2 was 3.3% in our study as compared to the mutation frequency of 24.6% and 3.28% in BRCA1 and BRCA2 respectively in the south Indian population as identified by Vaidyanathan et al [22]. The mutation frequency in BRCA2 was comparable between the two studies unlike the mutation frequency in BRCA1 which was much higher in our study.
In our study there were four patients who had four out of five mutations in BRCA1 gene, including one reported SNP on exon 2 and one SNP as well as two novel sequence variants on exon 20. Their mean age at diagnosis was 52.75 years as compared to the overall average of 46.27 years and the mean CA-125 level at presentation was 2418.6 U/ml as compared to the average of 2039.75 U/ml. 50% (n=2) of these four patients presented in stage I and the other 50% (n=2) in stage III. Grade 3 picture was seen in 50% of them while one each had grade 1 and grade 2 picture. One out of these four patients was not found to be tumor free after surgery and chemotherapy and was also observed to have a poor quality of life. A case-control study involving larger number of patients is needed to see if these sequence variants have a role in disease causation, surgical outcome and response to chemotherapy.
In the Indian scenario as observed by Saxena et al breast cancer patients (n=20), 18 sequence variants in total (9 in BRCA1 and 9 in BRCA2) were identified along with rare sequence variants in 15 out of 105 (14.2%) early onset cases without family history and 4 out of 34 (11.7%) breast cancer cases with family history [23]. This study was carried out on the entire gene and sequencing of all exons was done as compared to our study which was confined to exons 2 and 20 in BRCA1 and exon 11 in BRCA2 and identified 5 sequence variations in BRCA1 and 1 in BRCA2 gene. The prevalence of these genes in the Indian population appears to be low compared to other Asian countries but is similar to that reported from Shanghai [24]. However, the role of BRCA1 and 2 mutations and SNPs can be better established by a larger case-control study involving sequencing of the entire gene.
In conclusion this pilot study identified 5 sequence variants (2 reported SNPs and 3 novel variants) in BRCA1 gene and 1 novel sequence variant in BRCA2 gene. The three founder mutations, namely 187delAG and 5382insC in BRCA1 and 6174delT in BRCA2 genes were not seen in any of the patients. It is recommended that a case control study including larger number of patients be designed to establish the role of BRCA1 and BRCA2 gene mutations in epithelial ovarian cancer causation.
Disclosure of conflict of interest
None.
References
- 1.Meinhold-Heerlein I, Bauerschlag D, Zhou Y, Sapinoso LM, Ching K, Frierson H Jr, Bräutigam K, Sehouli J, Stickeler E, Könsgen D, Hilpert F, von Kaisenberg CS, Pfisterer J, Bauknecht T, Jonat W, Arnold N, Hampton GM. An integrated clinical genomic approach identifies a candidate multi-analyte blood test for serous ovarian cancer. Clin Cancer Res. 2007;13:458–66. doi: 10.1158/1078-0432.CCR-06-0691. [DOI] [PubMed] [Google Scholar]
- 2.Pal T, Permuth-Wey J, Kapoor R, Cantor A, Sutphen R. Improved survival in BRCA2 carriers with ovarian cancer. Fam Cancer. 2007;6:113–9. doi: 10.1007/s10689-006-9112-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd J, Sonoda Y, Federici MG, Bogomolniy F, Rhei E, Maresco DL, Saigo PE, Almadrones LA, Barakat RR, Brown CL, Chi DS, Curtin JP, Poynor EA, Hoskins WJ. Clinicopathologic features of BRCA-linked and sporadic ovarian cancer. JAMA. 2000;283:2260–5. doi: 10.1001/jama.283.17.2260. [DOI] [PubMed] [Google Scholar]
- 4.National Cancer Registry Programme. New Delhi: ICMR; 2002. Five year consolidated report of the hospital based carcinoma registries 1994-98. [Google Scholar]
- 5.Russo A, Calò V, Bruno L, Rizzo S, Bazan V, Di Fede G. Hereditary ovarian cancer. Crit Rev Oncol Hematol. 2009;69:28–44. doi: 10.1016/j.critrevonc.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Tagliaferri P, Ventura M, Baudi F, Cucinotto I, Arbitrio M, Di Martino MT, Tassone P. BRCA1/2 genetic background-based therapeutic tailoring of human ovarian cancer: hope or reality? J Ovarian Res. 2009;2:14. doi: 10.1186/1757-2215-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–39. [PMC free article] [PubMed] [Google Scholar]
- 8.Drew Y, Calvert H. The potential of PARP inhibitors in genetic breast and ovarian cancers. Ann N Y Acad Sci. 2008;1138:136–45. doi: 10.1196/annals.1414.020. [DOI] [PubMed] [Google Scholar]
- 9.Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, Sobol H, Teare MD, Struewing J, Arason A, Scherneck S, Peto J, Rebbeck TR, Tonin P, Neuhausen S, Barkardottir R, Eyfjord J, Lynch H, Ponder BA, Gayther SA, Zelada-Hedman M, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–89. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antoniou AC, Easton DF. Risk prediction models for familial breast cancer. Future Oncol. 2006;2:257–74. doi: 10.2217/14796694.2.2.257. [DOI] [PubMed] [Google Scholar]
- 11.U.S. Preventive Services Task F. Genetic risk assessment and BRCA mutation testing for breast and ovarian cancer susceptibility: recommendation statement. Ann Intern Med. 2005;143:355–61. doi: 10.7326/0003-4819-143-5-200509060-00011. [DOI] [PubMed] [Google Scholar]
- 12.Frank TS, Deffenbaugh AM, Reid JE, Hulick M, Ward BE, Lingenfelter B, Gumpper KL, Scholl T, Tavtigian SV, Pruss DR, Critchfield GC. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J. Clin. Oncol. 2002;20:1480–90. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 13.Pharoah PD, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BA. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31:33–6. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- 14.Narod S, Ford D, Devilee P, Barkardottir RB, Eyfjord J, Lenoir G, Serova O, Easton D, Goldgar D. Genetic heterogenicity of breast ovarian cancer revisited. Breast cancer linkage consortium. Am J Hum Genet. 1995;57:957–8. [PMC free article] [PubMed] [Google Scholar]
- 15.Frank TS, Manley SA, Olopade OI, Cummings S, Garber JE, Bernhardt B, Antman K, Russo D, Wood ME, Mullineau L, Isaacs C, Peshkin B, Buys S, Venne V, Rowley PT, Loader S, Offit K, Robson M, Hampel H, Brener D, Winer EP, Clark S, Weber B, Strong LC, Thomas A, et al. Sequence analysis of BRCA1 and BRCA2: correlation of mutations with family history and ovarian cancer risk. J. Clin. Oncol. 1998;16:2417–25. doi: 10.1200/JCO.1998.16.7.2417. [DOI] [PubMed] [Google Scholar]
- 16.Boyd J, Sonoda Y, Federici MG, Bogomolniy F, Rhei E, Maresco DL, Saigo PE, Almadrones LA, Barakat RR, Brown CL, Chi DS, Curtin JP, Poynor EA, Hoskins WJ. Clinicopathologic features of BRCA-linked and sporadic ovarian cancer. JAMA. 2000;283:2260–5. doi: 10.1001/jama.283.17.2260. [DOI] [PubMed] [Google Scholar]
- 17.Pal T, Permuth-Wey J, Betts JA, Krischer JP, Fiorica J, Arango H, LaPolla J, Hoffman M, Martino MA, Wakeley K, Wilbanks G, Nicosia S, Cantor A, Sutphen R. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer. 2005;104:2807–16. doi: 10.1002/cncr.21536. [DOI] [PubMed] [Google Scholar]
- 18.Sinha S, et al. Gynaecological malignancy: 5 year analysis from a tertiary hospital (Abstract) Scientific Proceedings, XVIII Annual Conference of Association of Gynaecological Oncologists of India. 2009:113. [Google Scholar]
- 19.Berek JS, et al. Ovarian and fallopian tube cancer. In: Berek JS, editor. Berek & Novak’s Gynecology. Philadelphia: Williams & Wilkins; 2007. pp. 1457–1548. [Google Scholar]
- 20.Negri E, Franceschi S, Tzonou A, Booth M, La Vecchia C, Parazzini F, Beral V, Boyle P, Trichopoulos D. Pooled analysis of 3 European case-control studies: I. Reproductive factors and risk of epithelial ovarian cancer. Int J Cancer. 1991;49:50–6. doi: 10.1002/ijc.2910490110. [DOI] [PubMed] [Google Scholar]
- 21.Modan B, Hartge P, Hirsh-Yechezkel G, Chetrit A, Lubin F, Beller U, Ben-Baruch G, Fishman A, Menczer J, Struewing JP, Tucker MA, Wacholder S National Israel Ovarian Cancer Study Group. Parity, oral contraceptives, and the risk of ovarian cancer among carriers and noncarriers of a BRCA1 or BRCA2 mutation. N Engl J Med. 2001;345:235–40. doi: 10.1056/NEJM200107263450401. [DOI] [PubMed] [Google Scholar]
- 22.Vaidyanathan K, Lakhotia S, Ravishankar HM, Tabassum U, Mukherjee G, Somasundaram K. BRCA1 and BRCA2 germline mutation analysis among Indian women from south India: identification of four novel mutations and high-frequency occurrence of 185delAG mutation. J Biosci. 2009;34:415–22. doi: 10.1007/s12038-009-0048-9. [DOI] [PubMed] [Google Scholar]
- 23.Saxena S, Chakraborty A, Kaushal M, Kotwal S, Bhatanager D, Mohil RS, Chintamani C, Aggarwal AK, Sharma VK, Sharma PC, Lenoir G, Goldgar DE, Szabo CI. Contribution of germline BRCA1 and BRCA2 sequence alterations to breast cancer in Northern India. BMC Med Genet. 2006;7:75. doi: 10.1186/1471-2350-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suter NM, Ray RM, Hu YW, Lin MG, Porter P, Gao DL, Zaucha RE, Iwasaki LM, Sabacan LP, Langlois MC, Thomas DB, Ostrander EA. BRCA1 and BRCA2 mutations in women from Shanghai China. Cancer Epidemiol Biomarkers Prev. 2004;13:181–9. doi: 10.1158/1055-9965.epi-03-0196. [DOI] [PubMed] [Google Scholar]