Abstract
Bone-marrow (BM)-derived cells have been shown to be capable of aiding skin regeneration in vivo by differentiating into keratinocytes. However, the conditions under which this occurs are not fully understood. Characterizing innate mechanisms of skin regeneration by stem cells in vivo is important for the area of stem cell biology. In this study, we investigate the use of novel in vivo imaging technology for characterizing the contribution of BM-derived cells to regeneration of the epidermis in mouse skin in vivo. In vivo imaging provides the ability to non-invasively observe the spatial positions and morphology of the BM-derived cells. Using a GFP BM-transplanted mouse model and in vivo multimodal microscopy, BM-derived cells can be observed in the skin. Our in vivo imaging method was used to search for the presence and identify the 3D spatial distribution of BM-derived cells in the epidermis of the skin under normal conditions, following wound healing, and after syngeneic skin grafting. We did not observe any evidence of BM-derived keratinocytes under these conditions, but we did observe BM-derived dendritic cells in the skin grafts. In vivo multimodal imaging has great potential for characterizing the conditions under which BM-derived cells contribute to skin regeneration.
Multimodal in vivo images of a skin graft on a GFP bone-marrow-transplanted recipient mouse seven days after grafting. Modalities include (A) optical coherence tomography (OCT), (B) microvascular phase-variance OCT, (C) second harmonic generation (SHG), and (D) two-photon-excited fluorescence (TPEF) microscopy.
Keywords: in vivo microscopy, multimodal, skin regeneration, bone marrow cells
1. Introduction
Bone-marrow (BM)-derived cells found in skin are known to be primarily involved in immune processes, such as immune surveillance and inflammation. However, several recent studies have demonstrated that BM-derived cells can also contribute to skin regeneration during skin repair processes such as wound healing [1, 2]. One example of this is the differentiation of BM-derived mesenchymal stem cells into keratinocytes for regenerating the epidermis. Several studies have reported finding BM-derived keratinocytes under normal conditions and following wound healing [3–7]. Others studies have reported very little, if any, contribution of BM-derived cells to epidermal regeneration [8]. A recent study detected few BM-derived keratinocytes following wound healing, but significantly higher numbers following skin grafting [9]. The conditions under which BM-derived stem cells are recruited to the skin and give rise to keratinocytes remain to be fully characterized. Understanding these innate mechanisms of skin regeneration by stem cells is crucial for translating stem cell therapies to clinical applications.
A large proportion of the studies attempting to find evidence of BM-derived keratinocytes following skin repair processes have made use of a GFP BM-transplanted mouse model. In this model, BM transplants are performed on wild-type mice using marrow from GFP-expressing donors. In these mice, all nucleated BM cells and cells derived from the BM express GFP. Searching for evidence of GFP-expressing keratinocytes is typically achieved using histological analysis of the skin following various skin repair processes [3–7]. While these methods have been used successfully, they are limited by the fact that they require the skin to be removed, fixed, sectioned, and stained. This greatly limits the ability to study the temporal dynamics of the BM-derived cells in the skin, an important factor for better understanding how these cells are recruited to the skin. In addition, processing of the tissue can introduce artifacts, such as distorting cellular morphology or inactivating GFP, which may impact the ability to detect BM-derived keratinocytes [10]. Furthermore, processing and analyzing wide areas of skin can be prohibitively time-consuming. As the presence of BM-derived keratinocytes appears to be a rare event [8], improved methods for detecting them would greatly aid studies attempting to characterize this effect.
One promising method for studying BM-derived cells and their contribution to skin regeneration is in vivo optical microscopy. Non-invasive visualization of the skin using in vivo imaging can offer several benefits. Most importantly, skin can be observed while keeping the three-dimensional spatial organization and the cellular morphology intact. In addition, the non-invasive nature enables repeated imaging of the same skin sites, allowing the temporal dynamics of the tissue to be directly observed. Among the different in vivo optical imaging techniques available, multiphoton microscopy (MPM) is particularly well-suited for in vivo study of BM-derived cells found in skin. MPM can make use of different contrast mechanisms to visualize different tissue components, such as two-photon-excited fluorescence (TPEF) [11] for visualizing GFP expressing cells and second-harmonic-generation (SHG) [12] for imaging collagen fibers, a major constituent in the dermis of skin. The combination of these contrast mechanisms is ideally suited for detecting the presence and location of BM-derived, GFP-expressing cells in reference to the collagen-rich dermal layer of skin in GFP BM-transplanted mice. While the BM-derived cells are visualized based on TPEF, the co-registered SHG signal can discriminate between the dermis and epidermis layers of the skin.
An additional imaging modality that is useful for in vivo skin imaging is optical coherence tomography (OCT) [13, 14]. OCT noninvasively visualizes tissue structure based on its optical scattering properties. In addition to structural information, various methods using OCT have been developed for visualizing the vasculature based on measuring time-varying changes in the optical scattering signal [15, 16]. This combined structural and vascular contrast using OCT can be useful for assessing the state of the skin during skin repair processes. This capability is beneficial for studying the BM-derived cell contribution to skin regeneration by providing a more comprehensive view of the functional skin microenvironment. Despite the unique capabilities and advantages provided by in vivo optical microscopy, this technology has not yet been utilized for studying the contribution of BM-derived stem cells to skin regeneration.
In this study, we made use of in vivo multimodal optical imaging techniques to identify BM-derived cells in the epidermis of the skin in GFP BM-transplanted mice. In vivo imaging of the skin was performed under various conditions, including skin wounding and skin grafting. Multimodal MPM imaging allowed the presence of BM-derived cells in the epidermis to be detected. No evidence of BM-derived keratinocytes was observed during normal conditions and wound healing. While epidermal BM-derived cells were detected under the condition of skin grafting, the morphology and spatial organization of these cells suggest that they are not keratinocytes but instead dendritic skin cells.
2. Experimental methods and materials
2.1 Multimodal microscope
The multimodal microscope used in this study was a custom-built integrated multi-photon microscope (MPM) and optical coherence tomography (OCT) system. The schematic of the microscope is shown in Figure 1. The system used a dual-spectrum laser source based on a tunable titanium-sapphire laser (Mai Tai HP, Spectra Physics) which has been previously reported [17]. The spectral-domain OCT system was based on a free-space interferometer and a custom-built spectrometer in the detection arm. TPEF and SHG signals were detected by a pair of photo-multiplier tubes (PMTs) using appropriate emission filters.
Figure 1.
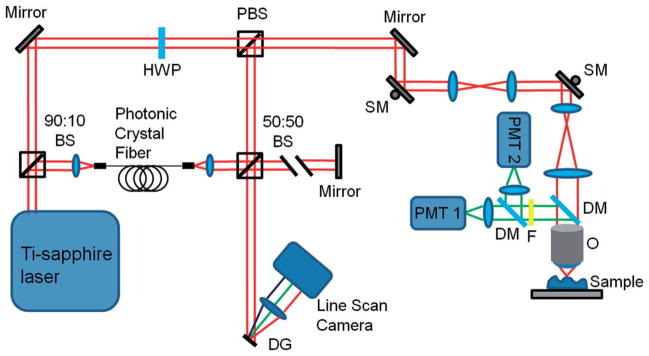
Schematic of the multimodal microscope. Abbreviations: BS, beam splitter; DG, diffraction grating; DM, dichroic mirror; F, low-pass filter; HWP, half-wave plate; O, objective; PBS, polarizing beam splitter; PMT, photo-multiplier tube; SM, scanning mirror.
TPEF/SHG and OCT datasets were acquired sequentially. The axial and transverse resolutions for TPEF and SHG were 0.8 μm and 0.5 μm, respectively. Single images consist of 256 by 256 pixels and the field-of-view was typically 380 × 380 μm2. A motorized stage enabled the acquisition of wide-area mosaics covering up to 3 × 3 mm2. This field-of-view was chosen to reasonably match the resolution of the system and to hold acquired mosaics to a reasonable file size for efficient processing. For OCT, the axial and transverse resolutions were 5 μm and 25 μm, respectively.
2.2 Bone marrow transplant
All animal procedures were performed under protocols approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Illinois at Urbana-Champaign. Bone marrow (BM) transplants were performed in 6–10 week-old female wild-type C57BL/6 mice, which were used as recipients. Bone-marrow donors were male transgenic mice with global GFP expression (C57BL/6-Tg (CAG-EGFP) 10sb/J) [18]. Donor mice were sacrificed by CO2 inhalation and BM was harvested from their hind limbs (femur and tibia). The hind limb bones were crushed with a mortar and pestle and filtered with a 40 μm filter. Following lysing of red blood cells with an ACK lysing buffer, cells were counted and diluted to a concentration of approximately 7 × 106 cells/ml and kept on ice prior to transplantation. Recipient mice were treated with radiation from a cobalt-60 source (2 doses of 6 GY, administered 4 hours apart). Donor BM cells were transplanted by tail vein injection (150 μl, 106 cells).
2.3 Wounding and grafting
Full-thickness excisional wounds were made in the ear skin of BM-transplanted mice at least 8 weeks after BM transplantation. Under the guidance of a stereo microscope, a 500 μm square portion of the skin was removed using a scalpel. The wound was imaged periodically to monitor the progress of the wound healing.
Skin grafts were performed using wild-type C57BL/6 mice as donors. Donor mice were sacrificed by CO2 inhalation and ear skin was harvested. The ear skin was dissected apart, and skin from the dorsal ear was used for grafting. Recipient BM-transplanted mice (at least 8 weeks after BM transplantation) were prepared by carefully removing a region of skin from the back while the animals were anesthetized with isoflurane gas. The skin graft was placed on the recipient mouse and immediately covered with petrolatum gauze and wrapped in a bandage. Following seven days of recovery, the bandages were removed.
2.4 In vivo multimodal imaging
For in vivo multimodal imaging, mice were anesthetized with isoflurane gas and placed on a heating pad. The skin site to be imaged was pressed against a coverslip mounted on the motorized stage of the multimodal microscope and held in place by gently clamping the skin. Glycerol was first directly applied to the skin surface as an index-matching agent. Once mounted, MPM and OCT imaging of the skin was performed through the coverslip.
3. Results and discussion
To investigate the potential for in vivo multimodal microscopy to characterize BM-derived cells in the skin, OCT and MPM imaging was performed on ear skin in vivo in GFP BM-transplanted mice (Figure 2). MPM imaging was performed using both SHG and TPEF contrast mechanisms to visualize the collagen network in the dermis and the BM-derived GFP cells, respectively. Overlaying these two different types of image data allowed the spatial position of the BM-derived cells to be non-invasively determined, as seen in Figure 2A. The dermal layer of the skin can be distinguished due to the presence of SHG signal from collagen. It is apparent from these images that none of the GFP cells reside above the dermis layer, in the epidermis, a result that is consistent with previous findings [8].
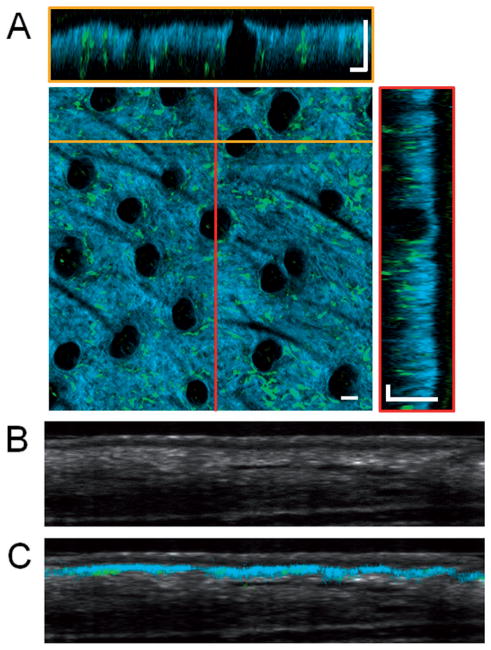
Figure 2.
Volumetric in vivo imaging of GFP bone-marrow-transplanted mouse ear skin. (A) Overlaid SHG (blue) and TPEF images (green) visualize the collagen network in the dermis and the bone-marrow-derived GFP cells, respectively. En face and cross-sectional slices of the volumetric data set allow the spatial locations of bone-marrow-derived cells to be visualized. Two different cross-sectional slices can be seen at locations corresponding to the yellow and red lines. En face images were obtained at a depth of approximately 40 μm below the skin surface. In this intact skin sample, no GFP cells are seen in the epidermis. Dark circular features seen in the en face section are hair follicles. Cross-sectional OCT images of the skin (B) without and (C) with the overlaid TPEF and SHG image data showing the location of the SHG and GFP signal within the skin. Scale bars are 50 μm.
While the SHG signal is limited to the dermal layer of the skin, OCT provides a more general view of the structure of the skin. The cross-sectional OCT image in Figure 2B demonstrates the ability to distinguish different layers of the skin, including the epidermis, dermis, and subcutaneous layers. Overlaying the co-registered data (Figure 2C) clearly demonstrates the boundary between the dermis and epidermis. SHG signal is only visible from the superficial layer of the dermis, as the penetration depth of MPM imaging is significantly lower than OCT.
The multimodal information provided by OCT and MPM can be used to non-invasively visualize the skin during skin repair processes. OCT and MPM imaging was performed 12 weeks following a full thickness excisional wound in the ear of a GFP BM-transplanted mouse (Figure 3). The extent of the original wound is indicated by the region within the yellow line. The structural OCT image (Figure 3A) demonstrates the formation of new tissue while the microvascular OCT image (Figure 3B) demonstrates the growth of new blood vessels within a portion of the wound site. The SHG image (Figure 3C) shows that new collagen has formed while the TPEF image shows the presence of BM-derived GFP cells. This multimodal information allows fully-healed regions to be distinguished from regions that have yet to heal completely (area indicated by red arrow). This can potentially provide important information about the microenvironment needed for BM-derived cells to contribute to epidermal regeneration.
Figure 3.
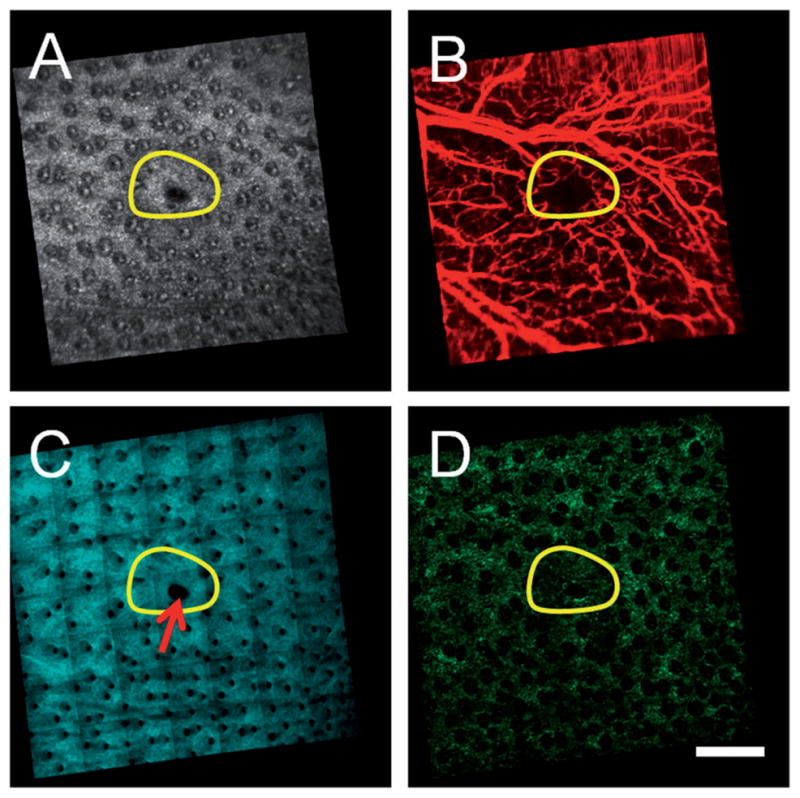
Multimodal in vivo en face images of ear skin on a GFP bone-marrow-transplanted mouse taken twelve weeks following an excisional wound. (A) Structural OCT, (B) microvascular OCT, (C) SHG and (D) TPEF images demonstrate structural repair, synthesis of new collagen and revascularization in the wound site (initial size of the wound indicated by the yellow circular region). A region of the wound site that failed to fully heal is indicated by the red arrow. Images acquired from a depth of approximately 50 μm below the skin surface. Scale bar is 500 μm.
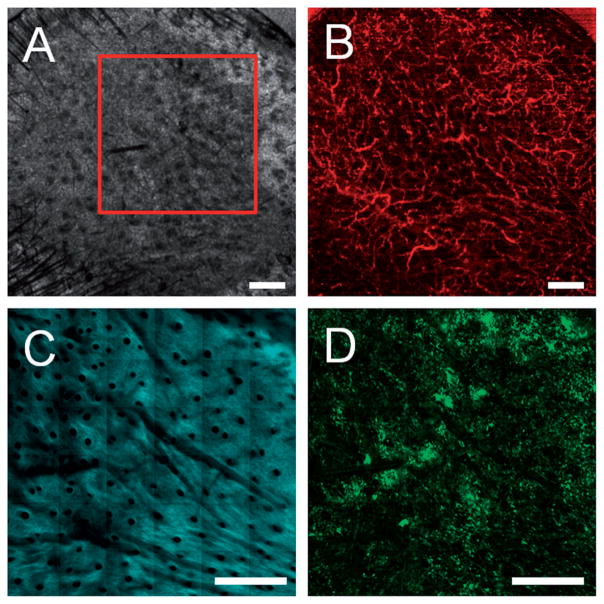
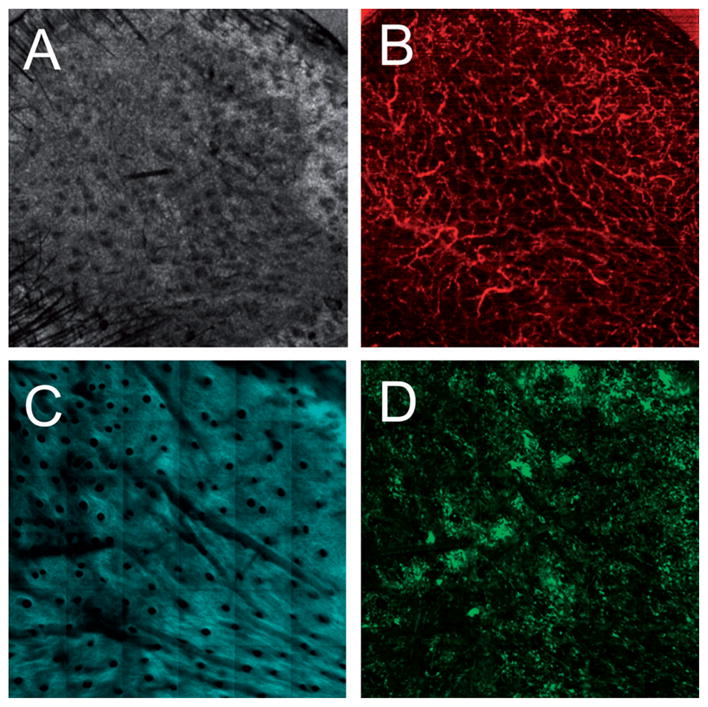
To investigate the contribution of BM-derived cells to epidermal regeneration during skin grafting, skin grafts from strain-matched inbred wild-type mice were grafted on a GFP BM-transplanted mouse. The skin graft was imaged with OCT and MPM imaging seven days following the skin graft procedure (Figure 4). The OCT microvascular image (Figure 4B) demonstrates the successful revascularization of the graft while the TPEF image (Figure 4D) shows a large influx of GFP cells into the graft.
Figure 4.
Multimodal in vivo images of a skin graft on a GFP bone-marrow-transplanted recipient mouse seven days after grafting. (A) Structural OCT, (B) microvascular OCT, (C) SHG and (D) TPEF images of the graft verify successful engraftment indicated by the revascularization of the graft and a large influx of bone-marrow-derived cells. The red box in (A) indicates the approximate region of overlap between the OCT and SHG/TPEF images. Images acquired from a depth of approximately 50 μm below the skin surface. Scale bars are 500 μm.
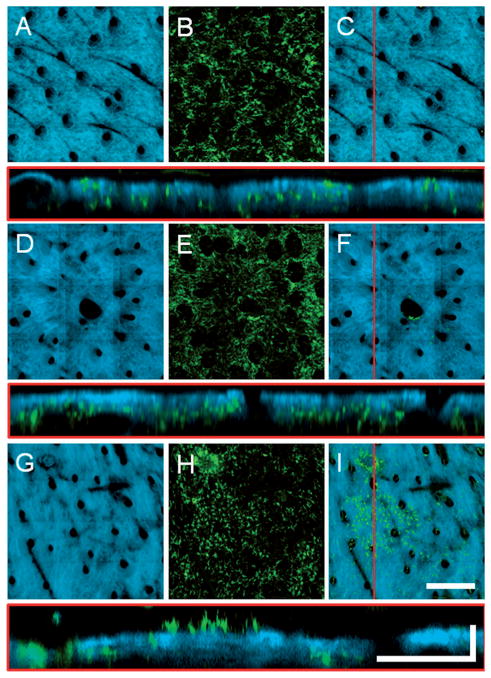
To search for evidence of BM-derived epidermal cells in skin, the volumetric MPM data sets can be represented to highlight epidermal GFP cells. In the multimodal overlays, the SHG can be assigned a high opacity to mask GFP cells that have co-registered SHG signal (i.e. cells that are in the dermis). Subsequent projection along the axial dimension of the volume results in a 2D image of the skin with epidermal GFP cells visible and dermal GFP cells hidden. In Figure 5 the SHG, GFP and multimodal overlays are shown for normal, wounded and grafted skin. It is apparent from these images that both normal and wounded skin do not contain epidermal GFP cells while the grafted skin has a significant number of epidermal GFP cells.
Figure 5.
Axial projections of in vivo volumetric SHG and TPEF images of (A–C) normal, (D–F) wounded, and (H–I) grafted skin in GFP bone-marrow-transplanted mice. (A, D, G) SHG projections visualize the collagen network while (B, E, H) TPEF projections visualize the bone-marrow-derived cells present throughout the depth of the skin. (C, F, I) Overlaid projections with high SHG opacity allowing the GFP cells in the epidermis to be visualized while masking the GFP cells in the dermis (see text for more detail). Cross-sectional images shown below each image set are from the location of the red line in (C, F, I). The presence of a focal cluster of epidermal GFP cells is apparent in the skin graft (I). En face images (A, B, D, E, G, H) are axial projections of volumetric data sets with a depth range of approximately 150 μm into the skin. Scale bar for en face images is 300 μm while the vertical and horizontal scale bars for cross sectional images are 50 μm and 200 μm, respectively.
Many of the epidermal cells in the grafted skin are found in a distinct cluster. The cells in this cluster form a monolayer with relatively uniform spacing between cells. In addition, these cells have a distinct morphology which consists of a main cell body and many tube-like extensions. All of these parameters are characteristic of a class of cell known as dendritic cells. Dendritic cells are antigen presenting cells that reside in the skin and play an important role in adaptive immunity. The in vivo MPM images of the graft epidermal cells were compared to images from a transgenic CD11C-YFP mouse (Figure 6). In this strain, the expression of YFP is limited to the dendritic cells of the immune system. It is apparent from Figure 6 that the spatial organization and morphology of the dendritic cells in the CD11C mouse are very similar to the cells found in the epidermis of the grafted skin, thus providing further evidence that the vast majority of observed BM-derived cells in the graft are clearly dendritic cells and not keratinocytes.
Figure 6.
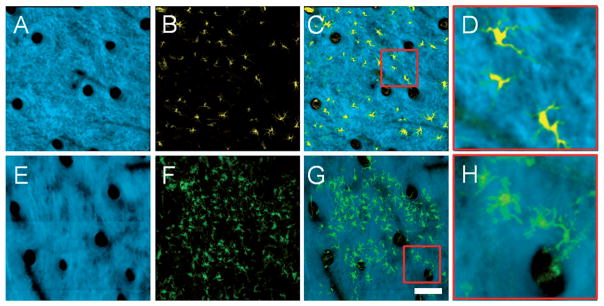
Comparison of in vivo volumetric SHG and TPEF axial projections from (A–D) YFP-CD11C mouse skin and (E–H) grafted skin on a GFP BM-transplanted mouse 12 days after grafting. (A, E) SHG, (B, F) TPEF and (C, G) overlaid projections with high SHG opacity allows the morphology of the epidermal cells in the two different mice to be compared. (D, H) High magnification images corresponding to the red boxes in (C) and (G), respectively, demonstrate that these cells have similar morphology, indicating that the epidermal cells in the graft are dendritic cells. Images are axial projections of volumetric data sets with a depth range of approximately 150 μm into the skin. Scale bar is 100 μm, and applies to (A–C, E–G).
No direct evidence of BM-derived keratinocytes was observed under the conditions tested. However, it is regularly reported that the presence of keratinocytes in the process of skin repair can be a very rare occurrence [8]. The sizeable presence of BM-derived dendritic cells in grafted skin was not expected. A recent study did not report detecting the presence of BM-derived dendritic cells during either wounding or skin grafting [9]. In contrast to this, a different study did report detecting Langerhans cells, a class of epidermal dendritic cells, during wound healing [8]. While dendritic cells are known to be regularly replaced from BM-derived progenitors, Langerhans cells are renewed from progenitors located in the skin under normal conditions [19]. However, replacement of Langerhans cells from the BM has been observed to occur under conditions of strong inflammation [20, 21]. The mechanisms leading to Langerhans cells being replaced from the BM is a topic of interest in immunology and could also benefit from the application of advanced multimodal in vivo imaging technology.
4. Conclusions
In this study, in vivo 3D multimodal optical imaging was used to search for evidence of epidermal regeneration by BM-derived cells during various skin repair processes. In vivo MPM imaging was demonstrated for visualizing GFP BM-derived cells in the skin, as well as for detecting collagen in the dermis layer. Together these images can clearly discriminate between GFP cells in the dermis versus the epidermis. OCT structural and microvascular imaging was demonstrated for assessing the state of skin following wounding and grafting. Based on the in vivo MPM images, no evidence of BM-derived keratinocytes was observed under normal conditions, wound healing, and skin grafting. However, it is possible that BM-derived keratinocytes may have been present at another time point which was not imaged. This is the subject of an ongoing longitudinal study to examine the presence of BM-derived cells in skin repair processes [22]. The conditions under which BM-derived cells can contribute to epidermal regeneration remain to be fully characterized. Under the test conditions of this study, the lack of evidence for BM-derived keratinocytes, as well as the considerable presence of BM-derived dendritic cells following wounding and skin grafting, highlights the variability and complexity of the role of BM-derived cells in skin regeneration. This study demonstrates the potential for in vivo multimodal imaging technology to contribute to better understanding the general mechanisms by which skin regeneration is influenced by BM-derived cells.
Supplementary Material
Acknowledgments
We thank Darold Spillman for his logistical and information-technology support toward this project. This project was supported in part by a grant from the U.S. National Science Foundation (CBET 10–33906, S.A.B.). Additional information can be found at: http://biophotonics.illinois.edu.
Footnotes
Author biographies Please see Supporting Information online.
References
- 1.Wu YJ, Zhao RCH, Tredget EE. Stem Cells. 2010;28:905–915. doi: 10.1002/stem.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu YJ, Wang JF, Scott PG, Tredget EE. Wound Repair Regen. 2007;15:S18–S26. doi: 10.1111/j.1524-475X.2007.00221.x. [DOI] [PubMed] [Google Scholar]
- 3.Fathke C, Wilson L, Hutter J, Kapoor V, Smith A, Hocking A, Isik F. Stem Cells. 2004;22:812–822. doi: 10.1634/stemcells.22-5-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng WM, Han Q, Liao LM, Li CH, Ge W, Zhao ZG, You SG, Deng HY, Murad F, Zhao RCH. Tissue Eng. 2005;11:110–119. doi: 10.1089/ten.2005.11.110. [DOI] [PubMed] [Google Scholar]
- 5.Brittan M, Braun KM, Reynolds LE, Conti FJ, Reynolds AR, Poulsom R, Alison MR, Wright NA, Hodivala-Dilke KM. J Pathol. 2005;205:1–13. doi: 10.1002/path.1682. [DOI] [PubMed] [Google Scholar]
- 6.Badiavas EV, Abedi M, Butmarc J, Falanga V, Quesenberry P. J Cell Physiol. 2003;196:245–250. doi: 10.1002/jcp.10260. [DOI] [PubMed] [Google Scholar]
- 7.Borue X, Lee S, Grove J, Herzog EL, Harris R, Diflo T, Glusac E, Hyman K, Theise ND, Krause DS. Am J Pathol. 2004;165:1767–1772. doi: 10.1016/S0002-9440(10)63431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan Q, Yee CL, Ohyama M, Tock C, Zhang G, Darling TN, Vogel JC. Experimental Hematology. 2006;34:672–679. doi: 10.1016/j.exphem.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Tamai K, Yamazaki T, Chino T, Ishii M, Otsuru S, Kikuchi Y, Iinuma S, Saga K, Nimura K, Shimbo T, Umegaki N, Katayama I, Miyazaki J, Takeda J, McGrath JA, Uitto J, Kaneda Y. Proc Natl Acad Sci USA. 2011;108:6609–6614. doi: 10.1073/pnas.1016753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter I, Fleischmann M, Klein D, Muller M, Salmons B, Gunzburg WH, Renner M, Gelbmann W. The Histochemical Journal. 2000;32:99–103. doi: 10.1023/a:1004014211408. [DOI] [PubMed] [Google Scholar]
- 11.Denk W, Strickler J, Webb W. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 12.Campagnola PJ, Loew LM. Nat Biotechnol. 2003;21:1356–1360. doi: 10.1038/nbt894. [DOI] [PubMed] [Google Scholar]
- 13.Huang D, Swanson EA, Lin CP, Schuman JS, Stinson WG, Chang W, Hee MR, Flotte T, Gregory K, Puliafito CA, Fujimoto JG. Science. 1991;254:1178–1181. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gambichler T, Moussa G, Sand M, Sand D, Altmeyer P, Hoffmann K. J Dermatol Sci. 2005;40:85–94. doi: 10.1016/j.jdermsci.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Zhao YH, Chen ZP, Saxer C, Xiang SH, de Boer JF, Nelson JS. Opt Lett. 2000;25:114–116. doi: 10.1364/ol.25.000114. [DOI] [PubMed] [Google Scholar]
- 16.Makita S, Hong Y, Yamanari M, Yatagai T, Yasuno Y. Opt Express. 2006;14:7821–7840. doi: 10.1364/oe.14.007821. [DOI] [PubMed] [Google Scholar]
- 17.Graf BW, Jiang Z, Tu H, Boppart SA. Journal of Biomedical Optics. 2009;14:034019. doi: 10.1117/1.3147422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 19.Merad M, Manz MG, Karsunky H, Wagers A, Peters W, Charo I, Weissman IL, Cyster JG, Engleman EG. Nat Immunol. 2002;3:1135–1141. doi: 10.1038/ni852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merad M, Ginhoux F, Collin M. Nat Rev Immunol. 2008;8:935–947. doi: 10.1038/nri2455. [DOI] [PubMed] [Google Scholar]
- 21.Ginhoux F, Tacke F, Angeli V, Bogunovic M, Loubeau M, Dai XM, Stanley ER, Randolph GJ, Merad M. Nat Immunol. 2006;7:265–273. doi: 10.1038/ni1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graf BW, Valero MC, Chaney EJ, Marjanovic M, Boppart MD, Boppart SA. Proc SPIE. 2011;7902:790206. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.