Abstract
Protein prenylation is a post-translational modification required for proper cellular localization and activity of many important eukaryotic proteins. Farnesyltransferase inhibitors (FTIs) have been explored extensively for their antitumor activity. To assist in identifying potentially new and more useful markers for therapeutic applications, we developed a strategy that uses a combination of metabolic labeling and 2D DIGE (differential gel electrophoresis) to discover new prenylated proteins whose cellular levels are influenced by FTIs. In this approach, metabolic labeling of prenylated proteins was first carried out with an alkyne-modified isoprenoid analog, C15Alk, in the presence or absence of the FTI L-744,832. The resulting alkyne-tagged proteins were then labeled with Cy3-N3 and Cy5-N3 and subjected to 2D differential gel electrophoresis (DIGE). Multiple spots having altered levels of labeling in presence of the FTI were observed. Mass spectrometric analysis of some of the differentially labeled spots identified several known prenylated proteins, along with HisRS, PACN-3, GNAI-1 and GNAI-2, which are not known to be prenylated. In vitro farnesylation of a C-terminal peptide sequence derived from GNAI-1 and GNAI-2 produced a farnesylated product, suggesting GNAI-1 and GNAI-2 are potential novel farnesylated proteins. These results suggest that this new strategy could be useful for the identification of prenylated proteins whose level of post-translational modification has been modulated by the presence of an FTI. Additionally, this approach, which decreases sample complexity and thereby facilitates analysis, should be applicable to studies of other post-translational modifications as well.
Introduction
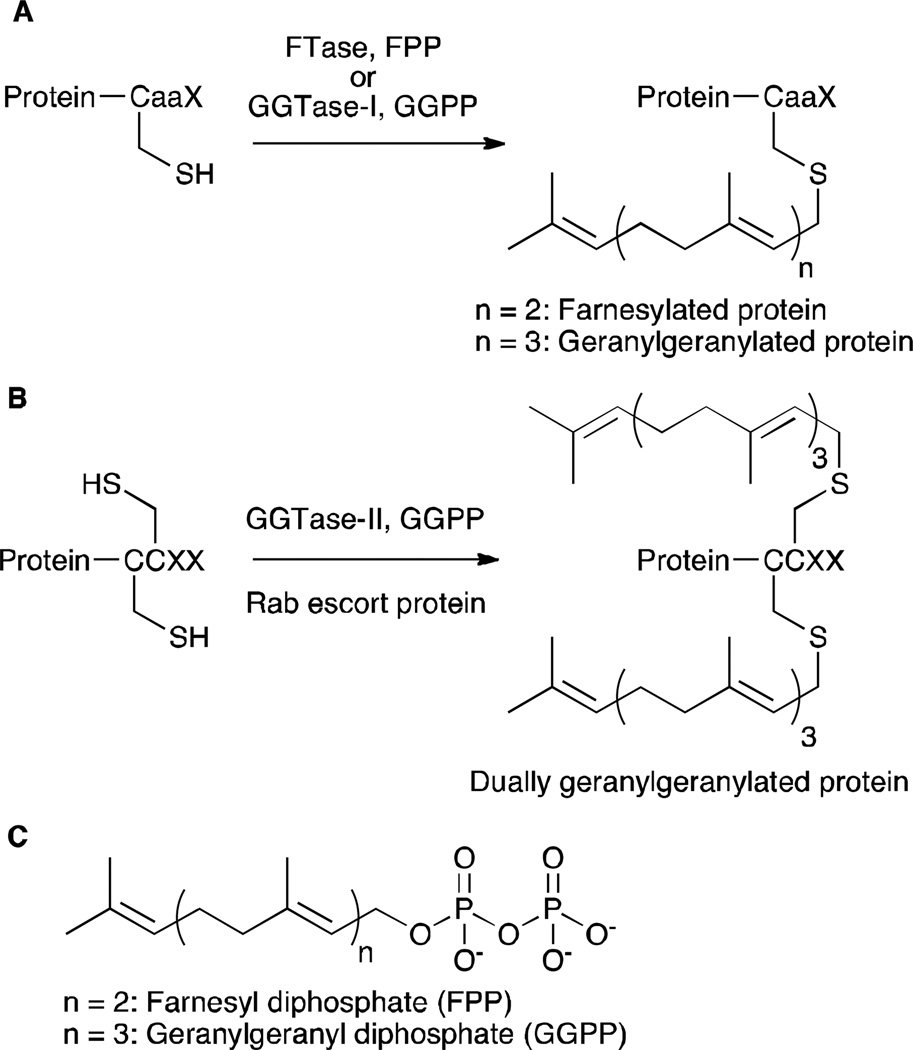
Protein prenylation is a post-translational modification, involving covalent attachment of either a farnesyl (15 carbon) or a geranylgeranyl (20 carbon) isoprenoid to Cys residues close to the C-termini of certain proteins. Protein farnesyltransferase (FTase) and protein geranylgeranyltransferase type-I (GGTase-I) catalyze transfer of the corresponding isoprenoid to the Cys residue in the C-terminal CaaX-box motif of proteins (Figure 1A), where “C” indicates Cys, “a” is generally an aliphatic amino acid and “X” is a variable residue that roughly determines which isoprenoid becomes attached.1 Geranylgeranyltransferase type-II (GGTase-II) catalyzes dual geranylgeranylation of Rab proteins possessing sequences with multiple Cys residues (such as CXC, CC), in the presence of Rab escort protein (REP) (Figure 1B).1 There is considerable interest in the process of protein prenylation since prenylated proteins play key roles in the progression of many diseases ranging from cancer and viral infections to aging-related disorders.2,3,4
Figure 1.
An overview of protein prenylation. (A) PFTase and GGTase-I catalyze covalent attachment of farnesyl and geranylgeranyl groups, respectively, to proteins with a CaaX motif. (B) GGTase-II catalyzes dual geranylgeranylation of Rab proteins (with CCXX or CXC motifs) in concert with Rab escort protein. (C) Structures of FPP and GGPP.
In an effort to detect and identify the prenylated proteome, several analogs of isoprenoid substrates, farnesyl diphosphate (FPP) and geranylgeranyl diphosphate (GGPP), have been synthesized and utilized. In chemical proteomic methods, prenylated proteins are first tagged with functionalized (alkyne,5,6,7 azide,8,9 biotin,10 or anilinogeraniol11) isoprenoid analogs by exploiting the promiscuous substrate specificity of prenyltransferase enzymes. Detection of tagged prenylated proteins is achieved via either a bioorthogonal reaction (click reaction, Staudinger ligation) or affinity methods (streptavidin or antibody against anilinogeranyl). Once tagged, the proteins can be identified either by mass spectrometry or western blotting using antibodies against known prenylated proteins. This method has led to the detection and successful identification of a number of farnesylated as well as singly and dually geranylgeranylated proteins.5–11 However, the number of prenylated proteins identified by these chemical proteomic methods is still much smaller compared to the predicted number of prenylated proteins (less than 100 prenylated proteins identified from several hundred predicted proteins)12.7
Several of the above mentioned studies describe efforts to characterize the effects of FTI treatment on the labeling of prenylated proteins with isoprenoid analogs, commonly employing 1D gel electrophoresis to visualize these effects.5,6,8,10 However, the limited separation of proteins by 1D electrophoresis restricts the use of such a method to monitor changes in prenylation of individual proteins. Nguyen et al. used multidimensional protein identification technology (MudPIT), a mass spectrometry based method, and a 15N-labeled internal standard to quantitate the in vivo effects of a GGTase-II inhibitor on prenylated proteins.10 Onono et al. characterized effects induced by two different FTIs on the farnesylated proteome using 2D electrophoresis and subsequent western blotting.11 While this latter method is useful for detecting overall changes in the farnesylated proteome using anilinogeraniol and antibodies specific for anilinogeranyl moiety, it requires antibodies against individual known farnesylated proteins to monitor changes in specific proteins. This makes it difficult to identify novel farnesylated proteins that might have altered levels of farnesylation in the presence of an FTI.
Based on the observation that alkyne-modified analogs manifest lower background labeling compared to azide-modified analogs,13 we previously synthesized alkyne-modified isoprenoids, including C15Alk (1), and utilized them for visualizing differences in prenylated proteins in several cell lines, and changes produced by FTI and an inhibitor of GGTase-I.5 In that work, alkyne-tagged prenylated proteins were also fractionated using 2D gels and several labeled proteins were identified via mass spectrometry. Charron et al. also used this C15Alk analog for proteomic profiling of prenylation, wherein they identified several previously uncharacterized prenylated proteins along with numerous known prenylated proteins.6,7 This C15Alk analog is of particular interest because it is accepted as a substrate by all three prenyltransferase enzymes.5,6 Therefore, it provides a good way to metabolically label and monitor all the prenylated proteins within a cell. This is important considering several FTIs also possess some inhibitory activity against GGTase-I and/or GGTase-II and hence can produce effects beyond inhibition of farnesylation.
Here, we demonstrate the use of a C15Alk in gel-based quantitative proteomic method that combines a metabolic labeling strategy with differential gel electrophoresis (DIGE) to quantitatively characterize the effects of an FTI on the labeling of prenylated proteins. Use of DIGE combines the advantages of greater separation of proteins on a 2D gel and the ability to run and visualize two different samples together on a single gel. Using this strategy and subsequent mass spectrometry analysis, we report the identification of several prenylated proteins suggested to be undergoing changes in prenylation level upon treatment of an FTI. In an effort to validate those results, synthetic peptides derived from the C-termini of GNAI-1 and GNAI-2 (proteins that have not previously been demonstrated to be prenylated) were prepared and used to confirm that they can be enzymatically prenylated by FTase.
Results
Overview of combining metabolic labeling with 2D-DIGE
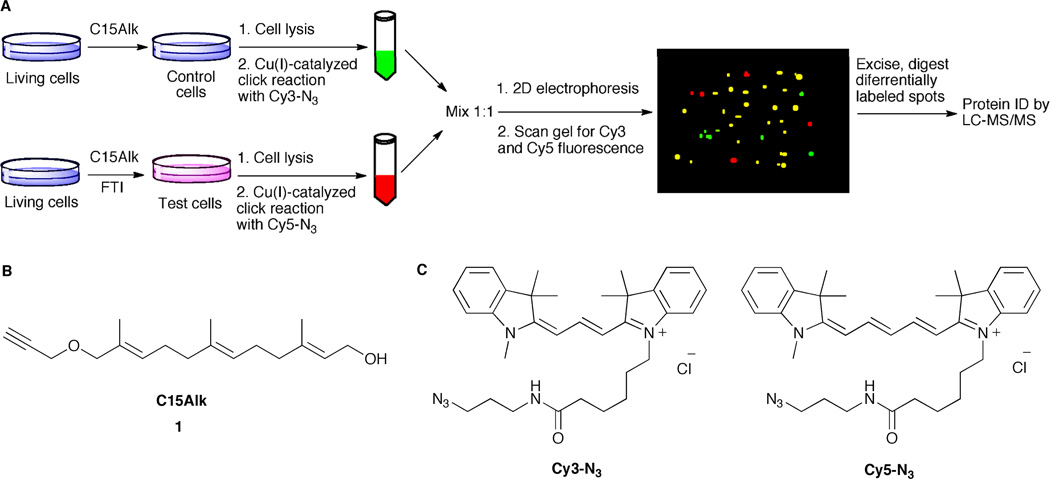
To extend the use of our C15Alk analog for quantitative characterization of effects of FTIs on the prenylated proteome, a workflow was developed and summarized in Figure 2A. First, the prenylated proteins are metabolically labeled with C15Alk either in the presence or absence of an FTI. Then the alkyne-modified proteins from the two different samples are individually reacted with Cy3-N3 or Cy5-N3 via a Cu(I)-catalyzed click reaction and mixed in equal amounts. Finally, the proteins are separated by 2D gel electrophoresis, and differences in the labeling of proteins in the two samples are characterized by comparing fluorescence intensities of labeled proteins in Cy3 and Cy5 channels of a scanning densitometer.
Figure 2.
An overview of the strategy of combining metabolic labeling with DIGE. (A) Schematic showing metabolic labeling and DIGE workflow; (B) Structure of C15Alk, 1; (C) Structures of Cy3-N3 and Cy5-N3.
While typically, Cy dyes used in DIGE are charge and mass-matched, size-matched azide-functionalized Cy dyes are not commercially available. There are two commercially available Cy3-N3 and Cy5-N3 dyes (Figure 2C), but there is a mass difference of 26 Da between them. Fortunately, Tsolakos et al. have reported that the mass difference of 26 Da between the two Cy dyes they used did not produce detectable changes in the migration of the labeled proteins.14 Therefore, we decided to use the commercially available dyes shown in Figure 2C in the DIGE experiments reported here. To verify this with the Cy3-N3 and Cy5-N3 dyes and to examine the utility of these dyes in the click reaction, HeLa cells were treated with 50 µM C15Alk (1) for 24 h in the presence of 25 µM lovastatin, to suppress endogenous production of FPP and GGPP and enhance C15Alk incorporation. Cells were lysed and the lysates were divided into two parts. These two different aliquots were reacted with either Cy3-N3 or Cy5-N3, respectively, under Cu(I)-catalyzed click reaction conditions. Equal amounts (based on protein concentration) of the Cy3- and Cy5-labeled proteins were mixed together and separated by 2D gel electrophoresis. Upon scanning the gel for Cy3 and Cy5 fluorescence, successful labeling was observed (Figure S1), which resembled our previously reported C15Alk labeling.5 Analysis (DeCyder software, DIA module) of the Cy3 and Cy5 images showed that more than 96% of the spots had equal intensities in Cy3 and Cy5 channels (Figure S1), indicating that the two azide-based dye reagents react with equal efficiency. Also, visual inspection of the gel images did not reveal any differences in the migration of the labeled spots in the overlay image of the Cy3 and Cy5 channels. Overall, these experimental results indicate that proteins labeled with Cy3-N3 and Cy5-N3 via the click reaction react with equal efficiency and co-migrate on 2D gels suggesting that this DIGE strategy should be useful for quantifying differences in prenylated proteins resulting from various drug treatments. It should be noted that in this and subsequent 2D-DIGE experiments, equal amounts of Cy3- and Cy5-labeled lysates were mixed together after estimating the lysate protein concentrations using standard protein assays. Minor differences in loading of the labeled samples as well as fluorescence characteristics of Cy3 and Cy5 are normalized in the Differential In-gel Analysis (DIA) module of the DeCyder software.
2D-DIGE visualizes effects of FTI treatment on C15Alk-labeled prenylome
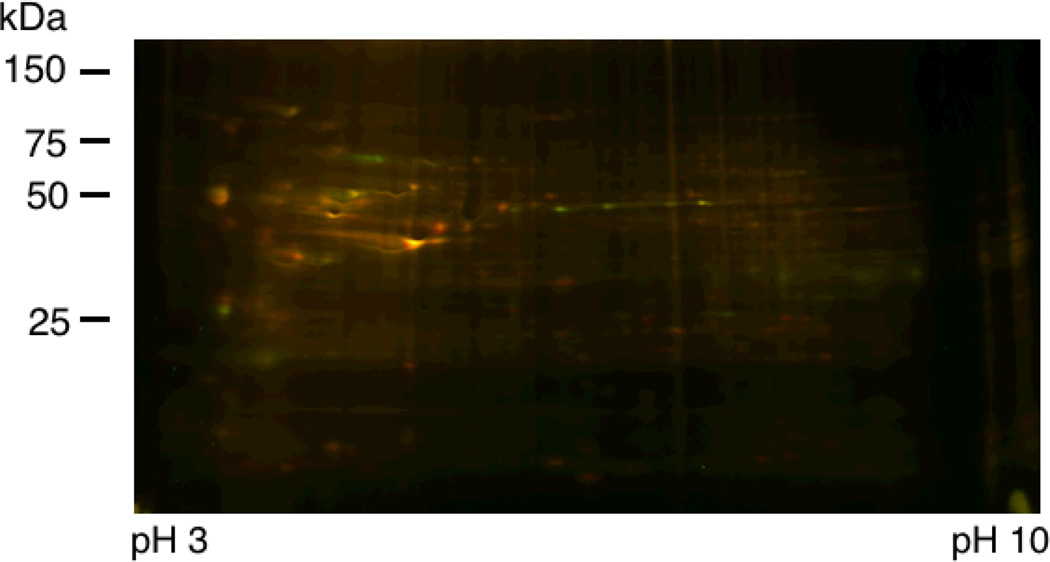
In order to assess the effects of FTI treatment on the labeling of prenylated proteins, HeLa cells were treated with 50 µM C15Alk in the presence or absence of 10 µM L-744,832, which is a commercially available potent FTase inhibitor.15 FTI treatment under these conditions did not cause any noticeable toxicity to HeLa cells, which is consistent with previously reported observations.16 Lysate from control cells (treated only with C15Alk) was reacted with Cy3-N3, and lysate of the FTI-treated cells was reacted with Cy5-N3. 2D-DIGE analysis of the mixed sample and subsequent DeCyder Differential In-gel Analysis detected 504 total spots of which 32 spots showed decreased labeling in the presence of the inhibitor while 23 spots showed an increase in labeling when the inhibitor was present (threshold of 2 model S.D). An overlay image of the fluorescence from the Cy3 and Cy5 channels is given in Figure 3 (see also Figure S2), while 3D views of spots showing differential labeling are shown in Figure 4 and Table S2. Two more replicates of this experiment were run using samples from new sets of cells and reversed labeling of samples with Cy3 and Cy5 dyes. DeCyder Biological Variation Analysis of these three replicates detected 394 spots in at least two of the three gels. 16 spots were detected to have decreased labeling and 17 spots with increased labeling in presence of FTI treatment, with T-test scores less than 0.05.
Figure 3.
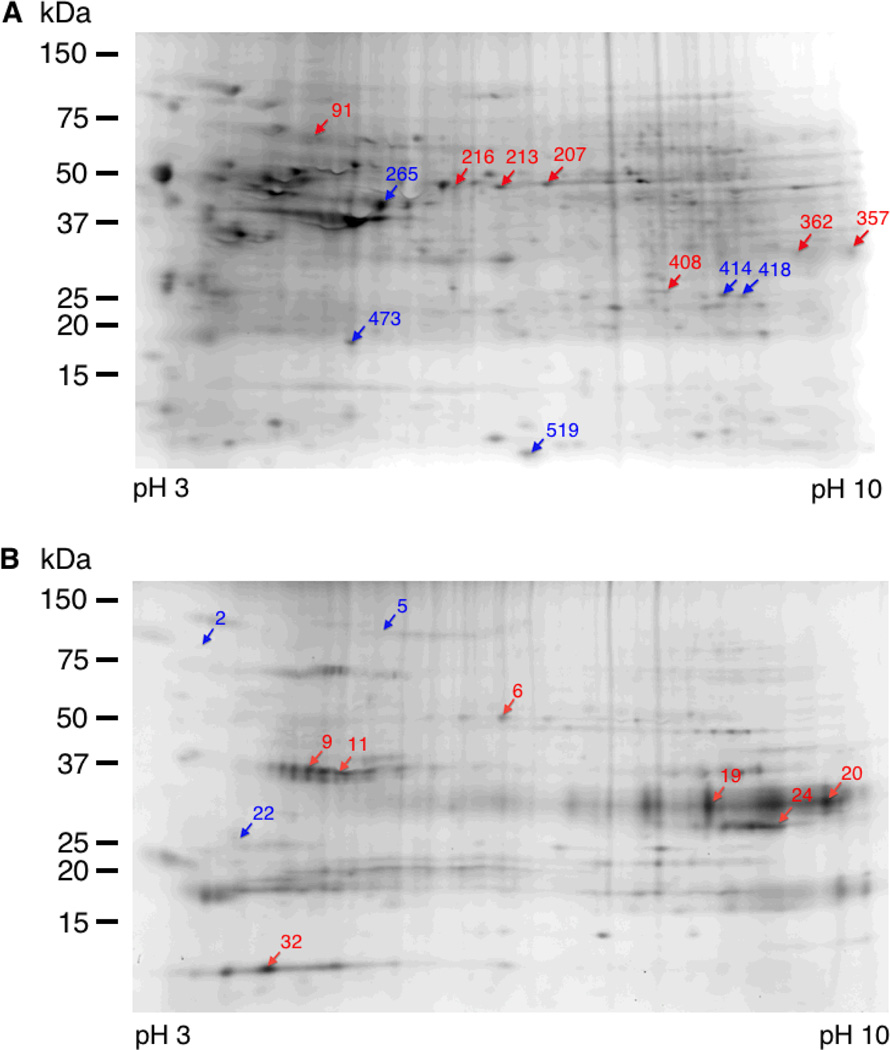
Merged 2D-DIGE image of proteins from HeLa cells treated with 50 µM C15Alk in absence (Green, reacted with Cy3-N3) or presence of 10 µM FTI (Red, reacted with Cy5-N3). Proteins were first resolved on a pH 3–10 NL IPG strip and then on a 10–20% polyacrylamide gradient gel.
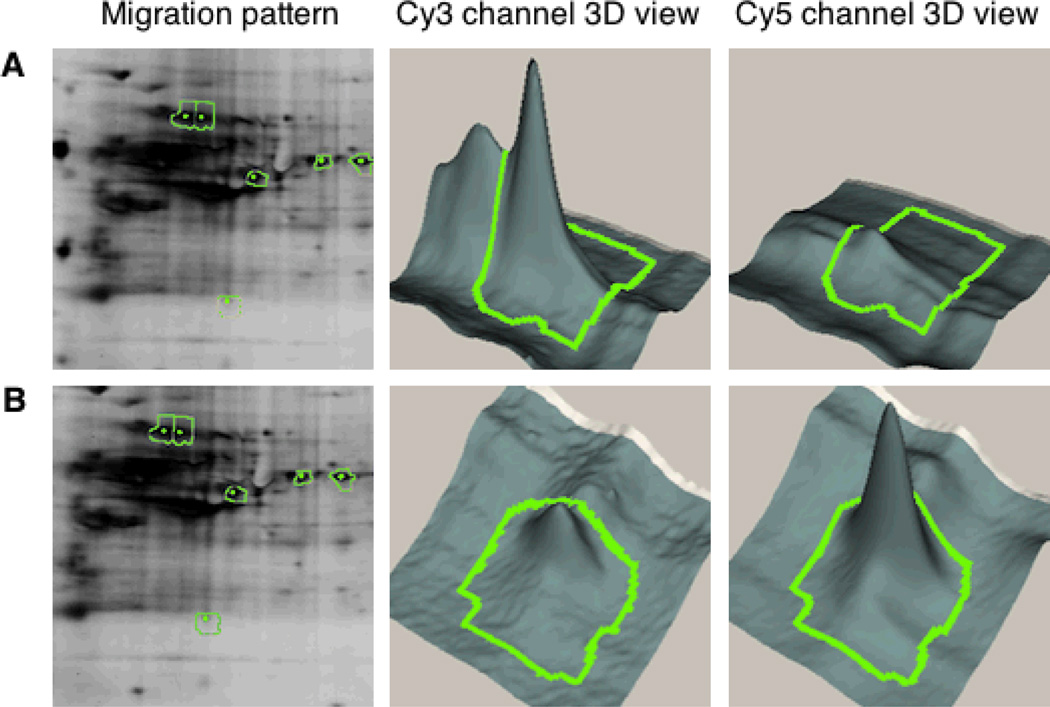
Figure 4.
Migration pattern and 3D view of representative spots from metabolic labeling and 2D DIGE analysis of prenylated proteins. Left panel: Enlarged view of migration pattern in 2D gel for spots 91 and 473 (green) along with adjacent spots selected for excision and protein identification (yellow). Middle and right panels: 3D view of specified spots in the Cy3 and Cy5 channels, where x and y coordinates indicate the position of the spot on the gel and the z axis (vertical dimension) shows the fluorescence signal intensity in that channel. (A) Spot 91 shows decrease in C15Alk labeling (Cy3 channel) upon FTI treatment (Cy5 channel); (B). Spot 473 shows an increase in C15Alk labeling (Cy3 channel) upon FTI treatment (Cy5 channel).
We selected the 12 most intense spots (7 with decreased labeling and 5 with increased labeling) from the first gel for excision (Figure 5A), tryptic digestion, and LC-MS/MS analysis. Most of these spots were detected in all the replicates of the gel and showed differential labeling with good T-test scores (Table S1). A total of 16 proteins containing potential prenylation motifs were identified from those spots and are listed in the Table 1; a complete list of all identified proteins is included in the supplementary information (Table S4). Most of the identified proteins (14) have been reported in previous studies,7–10 whereas, HisRS and PACN-3, were not previously known or computationally predicted to be prenylated.17 Interestingly, HisRS contains a LCIC sequence at the C-terminus, which could potentially be processed by GGTase-II. However, it is significantly bigger (57 kD) than Rab proteins (20–25 kDa) normally processed by GGTase-II. The C-terminus sequence of PACN-3 is CVGA, which is reported to be not a good substrate for FTase derived from yeast. In an in vitro screening of a peptide library based on CaaX sequences, Wang et al. observed that CVGA sequence was not an efficient substrate for yeast FTase.18 In vitro as well as in vivo studies of CaaX variants of a yeast pheromone a-factor performed by Trueblood et al. indicated that a-factor-CVGA had a significant decrease in farnesylation compared to its wt CVIA counterpart.19 Further studies are required to validate the prenylation status of HisRS and PACN-3 proteins. Efforts to identify the C-terminal peptides of these proteins containing the lipid modifications to serve as direct evidence of prenylation of these proteins were not successful which is in accordance with previous proteomic studies of protein prenylation. This could be due to the fact that the modified peptide is present in very small abundance and the fragmentation pattern of the modification on Cys is not precisely known. Additionally, many of the prenylated proteins have Lys and Arg residues in close proximity to the prenylated Cys, which results in very short C-terminal tryptic peptides that often escape detection.
Figure 5.
Spot maps showing protein spots excised for digestion and mass spectrometric protein identification. HeLa cells were treated with 50 µM 1 in presence or absence of 10 µM FTI. Blue and red arrows point to the spots whose intensities increased and decreased, respectively, in C15Alk labeling upon FTI treatment. (A) Gel image obtained from cell lysates subjected to click reaction and DIGE analysis without prior fractionation; (B) Gel image obtained from cell lysates that were first subjected to Triton X-114 fractionation followed by click reaction and DIGE analysis of the detergent-phase-derived samples.
Table 1.
List of identified proteins containing a possible C-terminal prenylation motif. The upper section of the table lists proteins identified from the whole cell lysate samples, and the bottom section of the table gives proteins identified from the Triton X-114 fractionated samples.
| Spot number |
Protein | Accession number |
Theoretical Mwa |
Theoretical plb |
Number of peptides |
% Coverage |
C15+FTV C15 ratio |
Protein identification probability % |
Peptides identifiedc |
C-terminus | Previously identified in pretomic studyd |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 91 | Lamin B1 | P20700 | 65961.72 | 5.11 | 75 | 83 | −2.37 | 100 | AEHDQLLLNVAK, ALYETELADAR |
CAIM | Yes |
| 91 | Lamin B2 | Q03252 | 67295.31 | 5.29 | 11 | 22 | −2.37 | 100 | EGELTVAQGR, SEVELAAALSDKR |
CYVM | Yes |
| 216 | HisRS | P12081 | 57,412.50 | 5.73 | 5 | 13 | −1.53 | 100 | ASAELIEEECAK, YDLTVPFAR |
LCIC | No |
| 216 | PACN-3 | Q9UKS6 | 48,486.70 | 5.83 | 3 | 10 | −1.53 | 100 | ADSAVSQEQLR, SPDEVTLTSIVPTR |
CVGA | No |
| 362 | Lamin A/C | P02545 | 74139.49 | 6.57 | 4 | 8 | −1.79 | 100 | QNGDDPLLTYR, SVGGSGGGSFGDNLVTR |
CSIM | Yes |
| 473 | Rab 10 | P61026 | 22540.93 | 8.58 | 2 | 11 | 1.63 | 100 | AFLTLAEDILR, IQIWDTAGQER |
SKCC | Yes |
| 473 | Rab 1A | P62820 | 22546.56 | 5.93 | 5 | 41 | 1.63 | 100 | FADDTYTESYISTIGVDFK, TITSSYYR |
GGCC | Yes |
| 473 | Rab 22A | Q9UL26 | 21855.06 | 8.32 | 3 | 29 | 1.63 | 100 | NAININELFIEISR, GSAAAIIVYDITKEETFSTLK |
RSCC | Yes |
| 473 | Rab 2A | P61019 | 23414.36 | 6.1 | 4 | 26 | 1.63 | 100 | GAAGALLVYDITR, LQIWDTAGQESFR |
GGCC | Yes |
| 473 | Rab 31 | Q13636 | 21568.78 | 6.59 | 4 | 33 | 1.63 | 100 | GSAAAVIVYDTITK, NAINIEELFQGISR |
RRCC | Yes |
| 473 | Rab 7A | P51149 | 23489.75 | 6.39 | 2 | 13 | 1.63 | 100 | DPENFPFVVLGNK, EAINVQAFQTIAR |
SCSC | Yes |
| 473 | Rap 1A | P62834 | 20647.7 | 6.39 | 8 | 55 | 1.63 | 100 | LVVLGSGGVGK, SALTVQFVQGIFVEK |
CLLL | Yes |
| 473 | Cdc42 | P60953 | 21,258.80 | 6.16 | 3 | 20 | 1.63 | 100 | TPFLLVGTQIDLR, NVFDEAILAALEPPEPK |
CVLL | Yes |
| 414 | Rab 5A | P20339 | 23658.68 | 8.32 | 3 | 17 | 2.39 | 100 | GVDLTEPTQPTR, QASPNIVIALSGNK |
CCSN | Yes |
| 414 | Rab 5B | P61020 | 23575.62 | 8.29 | 5 | 33 | 2.39 | 100 | SEPQNLGGAAGR, GVDLHEQSQQNK |
CCSN | Yes |
| 414 | Rab 5C | P51148 | 23482.56 | 8.64 | 4 | 26 | 2.39 | 100 | GVDLQENNPASR, GAQAAIVVYDITNTDTFAR |
CCSN | Yes |
| 11 | GNAI-1 | P63096 | 40229.89 | 5.7 | 4 | 14 | −10.5 | 100 | LKIDFGDSAR, IAQPNYIPTQQDVLR |
CGLF | No |
| 11 | GNAI-3 | P08754 | 40401 | 5.51 | 6 | 18 | −10.5 | 100 | LKIDFGEAAR, ISQSNYIPTQQDVLR |
CGLY | Yes |
| 11 | GNAI-2 | P04899 | 40319.71 | 5.34 | 7 | 25 | −10.5 | 100 | YDEAASYIQSK, IAQSDYIPTQQDVLR |
CGLF | No |
Theoretical mol wt. and theroretical pI are from ExPasy database.
When more than 2 peptides were matched in the MS analysis, 2 representative peptide sequences are listed in the “peptides identified column”.
Triton X-114 fractionation of prenylated and non-prenylated proteins prior to DIGE
Many prenylated proteins are present in low abundance.8 Therefore, it is desirable to enrich them to facilitate their detection via spectrometric methods. This can be achieved using Triton X-114 fractionation, which separates hydrophobic proteins from their more polar counterparts.20 Previously, it has been established that addition of an isoprenoid increases protein hydrophobicity causing prenylated proteins to partition into the detergent phase.21 Accordingly, HeLa cells were treated with C15Alk in the presence or absence of FTI, as described above. Cells were then lysed in Triton X-114 containing buffer and subjected to temperature dependent partitioning of the proteins into aqueous and detergent phases. Detergent phase-derived samples were used for click reactions with of Cy3-N3 and Cy5-N3 and subsequent separation of proteins via 2D gel electrophoresis.
Interestingly, the pattern of C15Alk-labeled and Triton X-114 fractionated protein (Figure S3) spots appeared different than the pattern obtained in the absence of fractionation (Figure S2), especially in 50–75 kDa region. DeCyder DIA analysis of Cy3 and Cy5 fluorescence images of the gel detected a total 208 spots, of which 45 spots showed decreases and 63 spots showed an increase in C15Alk labeling in presence of the FTI (threshold of 2 model S.D). We selected 10 spots (6 with decreased labeling and 4 with increased labeling) from this gel for excision (Figure 5B), tryptic digestion, and LC-MS/MS. This analysis identified 3 proteins containing potential prenylation motifs listed in Table 1; a complete list of all proteins identified within those spots is given in the supplementary information (Table S5). Interestingly, the proteins GNAI-1 and GNAI-2 were identified for the first time in this study. These proteins have the CaaX-box sequence CGLF at their C-termini, which is computationally not predicted to be prenylated.17 However, Hougland et al. reported that a short peptide based on this motif, Dansyl-TKCGLF, was accepted as a multiple turnover substrate by FTase.22 Therefore, we decided to further study prenylation of CGLF motif-containing peptides.
GNAI-1 and GNAI-2 are potential novel prenylated proteins
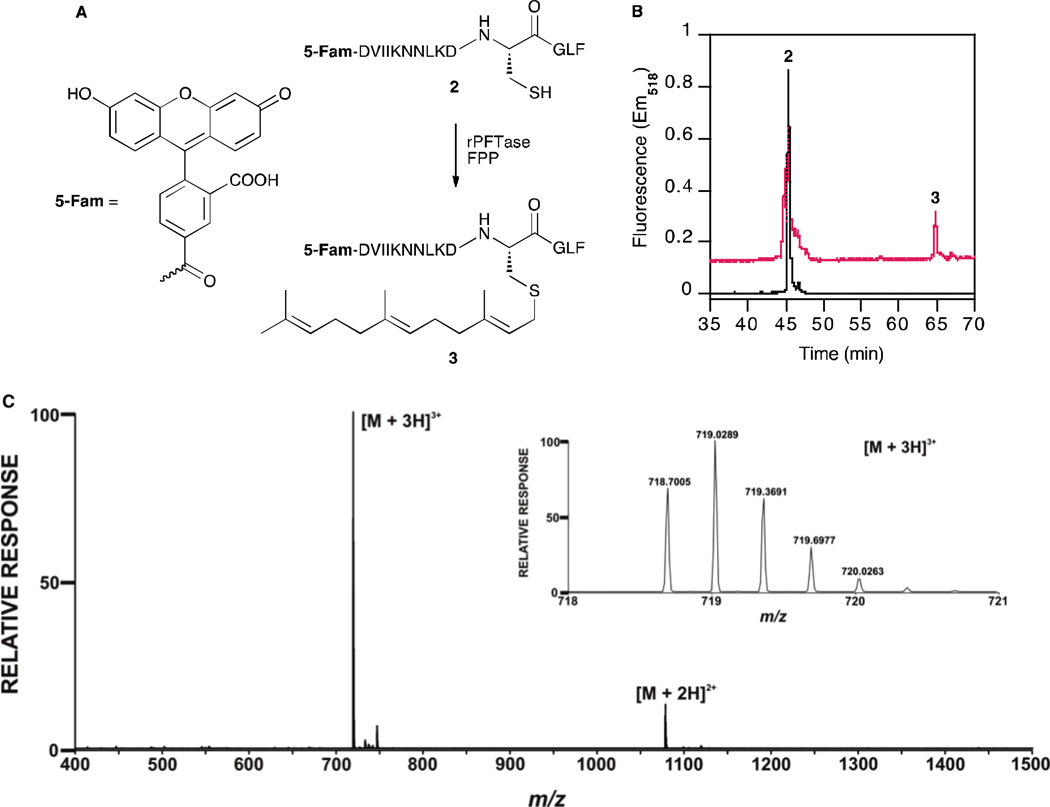
In order to investigate whether the CGLF motif of GNAI-1 and GNAI-2 proteins can be enzymatically farnesylated, we first synthesized a short peptide, dansyl-GCGLF (Figure S6), to determine the kinetic parameters for farnesylation. Using a previously established dansyl-based continuous fluorescence assay,23,24 a time-dependent increase in fluorescence was observed suggesting that the peptide is an enzyme substrate. Kinetic analysis performed by varying the concentration of the peptide 2 yielded kinetic constants kcat = 0.107 s−1 and Km = 7.93 µM (Figure S7). Amino acid residues upstream of the CaaX-box are known to affect binding and reactivity in FTase-catalyzed reactions.25 Therefore, we next synthesized a peptide containing 14 residues from the C-terminus of GNAI-1 and GNAI-2, 2 (Figure 6A). Having 10 residues upstream of the CaaX box, this peptide is a better mimic of GNAI-1 and GNAI-2, compared to the shorter peptide dansyl-GCGLF. Peptide 2 was subjected to in-vitro mammalian FTase-catalyzed farnesylation with FPP and the crude reaction mixture was analyzed by HPLC and LC-MS to probe for the presence of the farnesylated product. The presence of a new peak in the HPLC chromatogram at 65 min (Figure 6B) (with higher retention time in reverse-phase HPLC), indicated the formation of the farnesylated peptide 3.
Figure 6.
Analysis of the farnesylation of a C-terminal peptide derived from GNAI-2. (A) Scheme for rPFTase-catalyzed farnesylation of the peptide, 2; (B) HPLC analysis of peptide 2 before (black) and after farnesylation (red) with rPFTase shows 40% farnesylation of peptide when 2.5 µM peptide 2 was reacted with 10 µM FPP and 100 nM PFTase overnight. (C) Mass spectrum clearly shows the presence of [M+2H]2+ (calculated m/z = 1077.55, observed m/z = 1077.54) and [M+3H]3+ (calculated m/z = 718.70, observed m/z = 718.70) peaks for the farnesylated peptide 3. Inset shows enlarged view of [M+3H]3+ peak.
Discussion
In this study, we describe a strategy for combining metabolic labeling with 2D-DIGE to quantitatively characterize the effects of inhibitors of prenyltransferases on prenylation levels of various proteins. To accomplish this, an alkyne-modified isoprenoid, C15Alk, was employed for metabolic labeling of prenylated proteins in HeLa cells, either in the presence or absence of an FTI. The resulting alkyne-tagged proteins present in these two samples were then conjugated to Cy3-azide and Cy5-azide dyes via Cu(I)-catalyzed click reaction. After mixing equal amounts of the two samples, the proteins were separated via 2D gel electrophoresis and visualized by scanning the gel for Cy3 and Cy5 fluorescence. Fluorescence intensities of a given protein spot in the Cy3 and Cy5 channels correspond to the extent of labeling of the protein with C15Alk in the presence and absence of the FTI. Imaging analysis of the 2D gels using DeCyder software allowed us to identify global changes in the prenylation levels of many proteins that occur in HeLa cells upon treatment with the FTI.
Excision of a subset of the protein spots showing differential labeling in the presence of FTI and subsequent mass spectrometric analysis of these spots identified several prenylated proteins including potentially novel prenylated proteins HisRS, PACN-3, GNAI-1 and GNAI-2 (Table 1). We found that FTI treatment decreased labeling of Lamin A/C, Lamin B1 and Lamin B2, which is in accordance with previous findings. Lamin proteins, components of nuclear lamina, are known FTase substrates and are required for nuclear envelope assembly.26 Currently, FTIs are being investigated as possible therapeutic agents against Hutchinson-Giford progeria syndrome (HGPS) by inhibiting farnesylation of a mutant form of prelamin A, a precursor to mature lamin A.26 FTIs are also known to inhibit farnesylation of lamin B1 and lamin B2; however, long-term inhibition of farnesylation of these proteins can have adverse effects of nuclear function.27 We observed an increase in labeling of several Rab proteins when HeLa cells were treated with the FTI. This is in accordance with the findings by Si et al., that treatment with FTI increased Rab geranylgeranylation in several tissues,28 and is consistent with the notion that inhibition of farnesylation leads to a larger pool of FPP that can be converted to GGPP resulting in increased geranylgeranylation. FTI treatment was also observed to decrease the labeling of HisRS and PACN-3. These proteins have not been previously reported or predicted to be prenylated, and more analysis is required to establish their prenylation status. We also observed a decrease in labeling of guanine nucleotide-binding proteins GNAI-1, GNAI-2 and GNAI-3 in the presence of FTI. In our previous proteomic analysis of prenylated proteins, we had observed labeling of GNAI-3 with a shorter alkyne-modified FPP analog.5 In this study, we report GNAI-1 and GNAI-2 as potentially novel substrates of FTase. We show here that FTase catalyzes in vitro farnesylation of a 14 amino acid peptide derived from the C-termini of these proteins. Several protein lacking canonical farnesylation motifs were also identified from some of the protein spots (reported in the supplementary information). One possibility is that these proteins migrate very close to the labeled prenylated proteins on a 2D gel under the conditions used. Alternatively, they could be unanticipated isoprenoid-modified proteins, as observed by Charron et al.7
Through the use of 2D-DIGE, we were able to achieve better separation of proteins compared with our earlier work that utilized 1D electrophoresis for detecting labeled prenylated proteins. To further increase the separation, prior pH fractionation and subsequent narrow pH range 2D gel electrophoresis methods could be utilized, as described by Chan et al.9 The method of DIGE uses mass and charge-matched Cy3- and Cy5- dyes to label two different samples to be compared. This enables multiplexing of the samples on a single gel, eliminating gel-to-gel variations for easy matching of the protein spots from the two samples. In this study, we used Cy3-N3 and Cy5-N3, which differ in mass by 26 Da. But this difference causes less than 0.3% difference in Cy3- and Cy5-labeled versions of a protein of a molecular weight of 10 kDa, and is therefore not detectable by gel electrophoresis, consistent with previous reports by Tsolakos et al.14 Dontsova and co-workers recently reported the synthesis of mass- and charge-matched Cy3- and Cy5-azide dyes, which can be useful for similar analysis, especially for low molecular weight proteins.29
The experiments described here set the stage for future work aimed at identifying improved markers for protein prenylation that may be useful for drug development. In this regard, the alkyne-modified isoprenoid analog used here, C15Alk, has the advantage of being useful for monitoring not only farnesylated but also singly and dually geranylgeranylated proteins, as it is accepted as a substrate by all three prenyltransferase enzymes.5,6 Overall, the ability to selectively metabolically label prenylated proteins decreases the overall complexity of the samples for analysis and can potentially facilitate the detection of low abundance prenylated proteins that may be relevant top disease.4 Combining this with 2D DIGE increases resolution and allows facile quantitation of differences due to drug treatment. Such information should be useful for the development of therapeutic strategies based on prenylation inhibition. Moreover, this method could also be applied to studying other post-translation modifications, including S-palmitoylation,30 N-myristoylation,30 and glycosylation,31 which can be studied by similar alkyne-modified reporters.
Conclusion
In conclusion, we report here a method of combining metabolic labeling of proteins with 2D-DIGE for quantitative detection of the effects of treatment with FTIs on prenylation levels. This was achieved by using an alkyne-modified isoprenoid analog to tag prenylated proteins, followed by labeling with Cy3-N3 and Cy5-N3 dyes via Cu(I)-catalyzed click reaction, and DIGE analysis of labeled proteins. Subsequent mass spectrometry based analysis was used for identification of some of the proteins having altered levels of prenylation, and hence, potential therapeutically significant targets of FTIs. This method could readily be expanded for rapid and comprehensive comparison of the effects of various prenyltransferase inhibitors on prenylation levels of proteins and elucidate differences in the mechanisms of their action.
Materials and methods
Chemicals and reagents
HeLa cells were generously provided by Dr. Audrey Minden (Department of Chemical Biology, Rutgers University). C15Alk was synthesized as previously described.32 Cy3-N3 and Cy5-N3 were purchased from Lumiprobe. Dulbecco’s Modified Eagle Medium (DMEM), fetal bovine serum (FBS), Sypro Ruby protein stain were from Life Technologies. Benzonase (250 units/µL), phenylmethylsulfonyl fluoride (PMSF), protease inhibitor cocktail, Tris(2-carboxyethyl)phosphine (TCEP), tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl] amine (TBTA) were purchased from Sigma Aldrich. Detergent Compatible Protein Assay (DC assay), Bradford assay and Criterion™ Tris–HCl gels were obtained from Bio-Rad. ProteoExtract Protein Precipitation kit and FTI L-744,832 were from Calbiochem. Immobiline™ DryStrips, ampholyte buffer, DeStreak reagent and Deep Purple protein stain solution were purchased from GE Healthcare.
Cell culture and metabolic labeling
HeLa cells were grown in DMEM supplemented with 10% FBS. For metabolic labeling, cell media were supplemented with 50 µM of C15Alk (at 50–70% cell confluence) and cells were grown for 24 h. Where appropriate, 10 µM of FTI was added along with C15Alk into the cell media. After the treatment period, cells were washed twice with ice-cold PBS, suspended in PBS by scraping and pelleted by centrifuging at 1000 × g for 10 min. 200–300 µL of lysis buffer [300 µL PBS containing 0.2% or 1% SDS, 0.64 µL of 1 mM PMSF in ethanol, 0.26 µL of benzonase (250 units/µL) and 5 µL of protease inhibitor cocktail] was added to the pellets and followed by 10 min incubation on ice. Cells were then lysed using a Sonic Dismembrator (Fisher Scientific) at 4 W for 6–10 pulses of 2–10 sec after 20 sec rest periods. The protein concentration in the cell lysate was measured by the DC assay according to the manufacturer’s instructions.
Triton X-114 fractionation
Triton X-114 fractionation was performed as previously reported.20,21,33 Briefly, cells from 100 mm culture plates were lysed in 300–400 µL of 1% Triton X-114 in TBS [50 mM Tris, 150 mM NaCl, pH 7.5] by incubating for 1 h at 4 °C with rotation. Lysates were centrifuged for 10 min at 10,000 × g at 4 °C to remove insoluble debris. An aliquot (~50 µL) of this lysate was stored as total cell lysate. The remaining lysate was subjected to temperature-dependent phase separation by 5 min incubation at 37 °C followed by 10 min centrifugation at 10,000 × g at RT. After removal of the aqueous phase (top layer), TBS was added to the detergent phase to reduce the concentration of Triton X-114 to 1%. Cu(I)-catalyzed click reactions of the detergent phase with Cy3- and Cy5-azides were performed as describe below, without estimating the protein concentration. Subsequently, proteins were precipitated by adding 1 volume of 100% trichloroacetic acid (TCA) and 8 volume chilled acetone, and incubating at −20 °C for 1 h. Precipitated proteins were pelleted by centrifugation, and pellets were washed three times with chilled acetone. After resuspending the pellets in IEF rehydration buffer, the protein concentration was estimated using Bradford assay per manufacturer’s instructions.
Click reactions
To 50–150 µg (1 mg/mL) of HeLa lysate proteins were added Cy3-N3 or Cy5-N3 (final concentration 25 µM), TCEP (final concentration of 1 mM) and TBTA (final concentration of 100 µM). After vortexing, CuSO4 (final concentration of 1 mM) was added and reactions were incubated at RT for 1–2 h, with rotation. Equal quantities of Cy3- and Cy5-labeled proteins were mixed together and immediately precipitated using a ProteoExtract Protein Precipitation kit to remove excess click reaction reagents.
2D gel electrophoresis
Protein pellets were resuspended in 250 µL of IEF rehydration buffer [7.0 M urea, 2.0 M thiourea, 2.0% CHAPS, and 1.0% n-dodecyl-β-D-maltoside, 12 µM DeStreak reagent, 0.5% v/v ampholyte buffer and 12 mM dithiothreitol (DTT)]. Samples (80 µg protein for Triton X-114-fractionated samples, 125–150 µg protein for other samples) were rehydrated into 13 cm pH 3–10 NL Immobiline™ DryStrips overnight at 30 V for 6 h, 60 V for 7 h, then resolved in an Ettan™ IPGphor™ IEF apparatus per manufacturer's protocol. IEF strips were equilibrated in SDS equilibration buffer (50 mM Tris, pH 8.8, 8.0 M urea, 30% glycerol (v/v), 4.0% SDS, 1.0% DTT) for 0.5 h and resolved on 10–20% polyacrylamide Criterion™ Tris•HCl gels. Cy3- and Cy5-labeled protein spots were visualized with a Typhoon 8610 scanner (GE Healthcare) or a Typhoon FLA 7000 scanner (GE Healthcare). To obtain total protein stain images, gels were stained with either Deep Purple protein stain or Sypro Ruby protein stain and visualized with a Typhoon 8610 scanner or a FX Molecular Imager (Bio-Rad), respectively.
DeCyder analysis
Gel images were cropped with Image Quant v5.2 software (Molecular Dynamics) and imported into DeCyder v5.02 (GE Healthcare) software. Spot detection and quantification of spot intensities (for the gels used for spot excision) was performed using the Differential In-gel Analysis (DIA) module of the DeCyder software, using the double detection algorithm (Cy3 and Cy5 images). The threshold for differential protein labeling to be considered significant was 2 standard deviations. To align and compare spots from three replicates of the FTI-treated samples (without Triton X-114 fractionation), the Biological Variation Analysis (BVA) module was used. The channel pertaining to the C15Alk-treated sample was duplicated and imported as a Cy2 image for normalizing in the BVA analysis.
Spot excision and in-gel tryptic digestion
Gel images were imported into a Genomic Solutions® Investigator ProPicTM instrument (Genomic Solutions, Ann Arbor, MI, USA) for robotic excision. Spots of interest were excised and digested as described by Anderson et al.34
MS analysis of protein spots and database search
Lyophilized tryptic peptides were desalted with C18 resin according to the “Stage Tip” procedure.35 Aliquots of ~0.25 µg of total peptide were dissolved in 5.5 µl of loading solvent (98:2:0.01, H2O:CH3CN:HCO2H) and analyzed using a Velos Orbitrap instrument (Thermo Fisher Scientific) as described previously.36 The .RAW data files were converted to MzXML format using msconvert (Proteowizard) and then to .MGF files using TINT raw-to-mgf converter. Sequest (version 27) searches were performed against the UniProt Homo sapiens (taxon 9606; March 7, 2013 version) database with canonical and isoform sequences (175436 proteins), to which a contaminant database (thegpm.org/crap/index) was appended. Search parameters included variable modifications of methionine oxidation and cysteine iodoacetamide; partial trypsin cleavage (1 missed cleavage); Peptide tolerance 100 ppm, fragment tolerance 0.80 Da; and False Discovery Rate analysis against reversed database.
Synthesis of 5-FAM-DVIIKNNLKDCGLF (2) and Ds-GCGLF (4)
Peptides 2 and 4 (0.05 mmol scale) were synthesized using standard Fmoc solid phase peptide synthesis using an automated peptide synthesizer, following manufacturer’s instructions. In brief, Fmoc-Phe-Wang resin (1 eq) was deprotected using piperidine and amino acids (4 eq) were coupled using HCTU (4 eq) and DIEA (1 eq) in DMF solvent. For peptide 4, 4 eq of Fmoc-dansyl-Gly was used. For peptide 2, the Fmoc group of the precursor peptide (lacking the 5-FAM group) was deprotected in the automated peptide synthesizer and 5-Carboxyfluorescein (5-Fam) was coupled to it while still on the resin in the following manner: A solution of 5-FAM-hydroxysuccinimidyl ester (1.1 eq) in 2 mL DMF was added to the peptide on resin. DIEA (2.2 eq) was added and reaction was allowed to proceed overnight on a rotisserie wrapped in aluminum foil. After overnight reaction, resin was treated with 20% piperidine in DMF for 30 min. After washing the resin three times with DMF and three times with CH2Cl2, the peptide was cleaved off the resin by treatment with Reagent K for 30 min, precipitated in the presence of Et2O and pelleted by centrifugation. Pellets of crude peptides 2 and 4 were then dissolved in CH3OH, and analyzed by HPLC, and purified by preparative HPLC (Buffer A: 0.1% aq TFA, Buffer B: 0.1% TFA in CH3CN). Purified peptides were analyzed by HPLC and ESI-MS (Figure S4, Figure S5).
Enzymatic farnesylation of peptide 2
Enzymatic reactions contained 50 mM Tris, pH 7.4, 25 mM MgCl2, 25 µM ZnCl2, 12.5 mM DTT, 2.5 µM of 2, 10 µM FPP and 100 nM rat FTase. First, the peptide was incubated with DTT at room temperature for 1 h, in the dark. The reaction was initiated by adding FTase and incubated overnight at room temperature. 100 µL of crude reaction mixture was analyzed by HPLC. For LC-MS analysis, 5 mL of reaction mixture was lyophilized and then resuspended in 250 µL of 20% CH3CN and 20% MeOH in water. 5 µL of it was analyzed using a Waters Acquity UPLC/Synapt G2 QTOF mass spectrometer.
Continuous fluorescence assay
Assays were performed in 96 well format as described by DeGraw et al.37 Briefly, the assay solution contained 0.2–25 µM dansyl-GCGLF, and 10 µM FPP in 50 mM Tris, pH 7.5, 5 mM DTT, 5 mM MgCl2, 50 µM ZnCl2, and 0.2 % (w/v) n-octyl-β-D-glucopyranoside at 25 °C. Peptides were incubated in this buffer for 30 min and the initial fluorescence reading of the dansyl group was recorded (λex = 340 nm, λem = 505 nm) on a DTX880 Multi-mode plate reader (Beckman Coulter). Purified rPFTase was first diluted in a buffer containing BSA (50 mM Tris, pH 7.5, 1 mM DTT, 5 mM MgCl2, 50 µM ZnCl2, 20 µM KCl and 1 mg/mL BSA) and then added to the peptide solution 100 nM concentration to initiate the farnesylation reaction. Fluorescence of dansyl group was measured as a function of time. The total fluorescence change observed upon reaction completion was divided by the initial concentration of the peptide substrate in a given reaction to yield a conversion from fluorescence units to product concentration (Ampconv). The linear initial rate, V, in fluorescence intensity per minute, was then converted to a velocity (µM product produced per minute) by dividing V with Ampconv.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants GM058842 and GM084152, and a Department of Chemistry Excellence in Graduate Studies fellowship (University of Minnesota). A part of this work was carried out using instruments in the University of Minnesota Center for Mass Spectrometry and Proteomics, and facilities provided by the Minnesota Supercomputing Institute (University of Minnesota). We thank Dr. LeeAnn Higgins at the University of Minnesota Center for Mass Spectrometry and Proteomics, and Dr. Joseph J. Dalluge Department of Chemistry Mass Spectrometry Laboratory (University of Minnesota) for their assistance and technical guidance.
References
- 1.Zhang FL, Casey PJ. Annu. Rev. Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 2.Ochocki JD, Distefano MD. MedChemComm. 2013;4:476–492. doi: 10.1039/C2MD20299A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berndt N, Hamilton AD, Sebti SM. Nat. Rev. Cancer. 2011;11:775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z, Meray RK, Grammatopoulos TN, Fredenburg RA, Cookson MR, Liu Y, Logan T, Lansbury PT., Jr. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4635–4640. doi: 10.1073/pnas.0806474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeGraw AJ, Palsuledesai C, Ochocki JD, Dozier JK, Lenevich S, Rashidian M, Distefano MD. Chem. Biol. Drug Des. 2010;76:460–471. doi: 10.1111/j.1747-0285.2010.01037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charron G, Tsou LK, Maguire W, Yount JS, Hang HC. Mol. BioSyst. 2011;7:67–73. doi: 10.1039/c0mb00183j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charron G, Li MMH, MacDonald MR, Hang HC. Proc. Natl. Acad. Sci. U.S.A. 2013;110:11085–11090. doi: 10.1073/pnas.1302564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kho Y, Kim SC, Jiang C, Barma D, Kwon SW, Cheng J, Jaunbergs J, Weinbaum C, Tamanoi F, Falck J, Zhao Y. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12479–12484. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan LN, Hart C, Guo L, Nyberg T, Davies BSJ, Fong LG, Young SG, Agnew BJ, Tamanoi F. Electrophoresis. 2009;30:3598–3606. doi: 10.1002/elps.200900259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen UTT, Guo Z, Delon C, Wu Y, Deraeve C, Fraenzel B, Bon RS, Blankenfeldt W, Goody RS, Waldmann H, Wolters D, Alexandrov K. Nat. Chem. Biol. 2009;5:227–235. doi: 10.1038/nchembio.149. [DOI] [PubMed] [Google Scholar]
- 11.Onono FO, Morgan MA, Spielmann HP, Andres DA, Subramanian T, Ganser A, Reuter CWM. Mol. Cell. Proteomics. 2010;9:742–751. doi: 10.1074/mcp.M900597-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan LN, Tamanoi F. Enzymes, Vol 29: Protein Prenylation, Pt A. 2011;29:195–206. [Google Scholar]
- 13.Speers AE, Cravatt BF. Chem. Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Tsolakos N, Techanukul T, Wallington A, Zhao Y, Jones C, Nagy J, Wheeler JX. Proteomics. 2009;9:1727–1730. doi: 10.1002/pmic.200800563. [DOI] [PubMed] [Google Scholar]
- 15.Nørgaard P, Law B, Joseph H, Page DL, Shyr Y, Mays D, Pietenpol JA, Kohl NE, Oliff A, Coffey RJ, Poulsen HS, Moses HL. Clin. Cancer Res. 1999;5:35–42. [PubMed] [Google Scholar]
- 16.Węsierska-Gądek J, Kramer MP, Schmid G. J. Cell. Biochem. 2008;104:189–201. doi: 10.1002/jcb.21612. [DOI] [PubMed] [Google Scholar]
- 17.Maurer-Stroh S, Eisenhaber F. Genome Biol. 2005;6 doi: 10.1186/gb-2005-6-6-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y-C, Distefano MD. Chem. Comm. 2012;48:8228–8230. doi: 10.1039/c2cc31713c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trueblood CE, Boyartchuk VL, Picologlou EA, Rozema D, Poulter CD, Rine J. Mol. Cell. Biol. 2000;20:4381–4392. doi: 10.1128/mcb.20.12.4381-4392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coxon FP, Ebetino FH, Mules EH, Seabra MC, McKenna CE, Rogers MJ. Bone. 2005;37:349–358. doi: 10.1016/j.bone.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 21.Reinicke AT, Hutchinson JL, Magee AI, Mastroeni P, Trowsdale J, Kelly AP. J. Biol. Chem. 2005;280:14620–14627. doi: 10.1074/jbc.M500076200. [DOI] [PubMed] [Google Scholar]
- 22.Hougland JL, Hicks KA, Hartman HL, Kelly RA, Watt TJ, Fierke CA. J. Mol. Biol. 2010;395:176–190. doi: 10.1016/j.jmb.2009.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cassidy PB, Dolence JM, Poulter CD. Methods Enzymol. 1995;250:30–43. doi: 10.1016/0076-6879(95)50060-x. [DOI] [PubMed] [Google Scholar]
- 24.DeGraw AJ, Hast MA, Xu J, Mullen D, Beese LS, Barany G, Distefano MD. Chem. Biol. Drug Des. 2008;72:171–181. doi: 10.1111/j.1747-0285.2008.00698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hicks KA, Hartman HL, Fierke CA. Biochem. 2005;44:15325–15333. doi: 10.1021/bi050951v. [DOI] [PubMed] [Google Scholar]
- 26.Bishop WR, Doll R, Kirschmeier P. Enzymes, Vol 29: Protein Prenylation, Pt A. 2011;29:275–303. [Google Scholar]
- 27.Adam SA, Butin-Israeli V, Cleland MM, Shimi T, Goldman RD. Nucleus. 2013;4:142–150. doi: 10.4161/nucl.24089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Si XN, Zeng Q, Ng CH, Hong WJ, Pallen CJ. J. Biol. Chem. 2001;276:32875–32882. doi: 10.1074/jbc.M010400200. [DOI] [PubMed] [Google Scholar]
- 29.Osterman IA, Ustinov AV, Evdokimov DV, Korshun VA, Sergiev PV, Serebryakova MV, Demina IA, Galyamina MA, Govorun VM, Dontsova OA. Proteomics. 2013;13:17–21. doi: 10.1002/pmic.201200393. [DOI] [PubMed] [Google Scholar]
- 30.Hannoush RN, Sun J. Nat. Chem. Biol. 2010;6:498–506. doi: 10.1038/nchembio.388. [DOI] [PubMed] [Google Scholar]
- 31.Agard NJ, Bertozzi CR. Acc. Chem. Res. 2009;42:788–797. doi: 10.1021/ar800267j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hosokawa A, Wollack JW, Zhang Z, Chen L, Barany G, Distefano MD. Int. J. Pept. Res. Ther. 2007;13:345–354. [Google Scholar]
- 33.Berg TJ, Gastonguay AJ, Lorimer EL, Kuhnmuench JR, Li R, Fields AP, Williams CL. J. Biol. Chem. 2010;285:35255–35266. doi: 10.1074/jbc.M110.129916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen JD, Boylan KLM, Xue FS, Anderson LB, Witthuhn BA, Markowski TW, Higgins L, Skubitz APN. Electrophoresis. 2010;31:599–610. doi: 10.1002/elps.200900441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rappsilber J, Ishihama Y, Mann M. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 36.Lin-Moshier Y, Sebastian PJ, Higgins L, Sampson ND, Hewitt JE, Marchant JS. J. Biol. Chem. 2013;288:355–367. doi: 10.1074/jbc.M112.405761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeGraw AJ, Keiser MJ, Ochocki JD, Shoichet BK, Distefano MD. J. Med. Chem. 2010;53:2464–2471. doi: 10.1021/jm901613f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.