Abstract
Glioblastoma (GBM) stem cells (GSC) are a subpopulation of tumor cells that display stem-like characteristics (stemness) and play unique roles in tumor propagation, therapeutic resistance and tumor recurrence. Therapeutic targets in GSC are a focus of increasing interest to improve GBM therapy. Here we report that the hyaluronan-mediated motility receptor (HMMR) is highly expressed in GBM tumors where it supports the self-renewal and tumorigenic potential of GSC. HMMR silencing impairs GSC self-renewal and inhibits the expression of GSC markers and regulators. Furthermore, HMMR silencing suppresses GSC-derived tumor growth and extends the survival of mice bearing GSC xenografts. Conversely, HMMR overexpression promotes GSC self-renewal and intracranial tumor propagation. In human GBM tumor specimens, HMMR expression is correlated positively with the expression of stemness-associated markers and regulators. Our findings identify HMMR as a candidate therapeutic target to GSC as a GBM treatment strategy.
Keywords: Cancer stem cell, brain tumor, oncogene, self-renewal, RNA interference
Introduction
Gliobastoma multiforme (GBM) is the most common and lethal primary brain tumor with a median survival time of approximately 14 months (1, 2). GBM consists of morphological and functionally heterogeneous populations of cells (3, 4). Only a minority of the GBM cells has the capacity to initiate and sustain a hierarchical and heterogeneous cancer cell population when injected into immune-compromised mice (5). These tumor-initiating GBM cells, alternatively called GBM stem cells (GSCs), display stem-like characteristics (stemness) including extensive self-renewal, multi-lineage differentiation potential and propagation of tumors that recapitulate the tissue architecture and cellular hierarchy of the parental tumor (6-9). During the past decade, considerable evidence has demonstrated that GSCs interact with tumor microenvironment to promote tumor angiogenesis, immune evasion, therapeutic resistance and tumor recurrence (10-13). It is becoming increasingly important to understand the stemness-supporting signaling in GSCs and develop novel therapeutic strategies for efficiently inhibiting brain tumor stemness and improving patient survival.
GBM-associated oncogenic pathways, such as EGF, HGF/Met, PDGF, Notch, Sonic Hedgehog, hypoxia-induced factor and VEGF, contribute to the malignant characteristics of GBM, including uncontrolled proliferation, invasion and angiogenesis (2, 14-17). The activation of these oncogenic pathways has also been shown to support the hierarchy of self-renewing tumor-initiating stem cells in GBM (18-21). Evaluating their therapeutic efficacy in GSC models is valuable to determine their potential application in not only blocking tumor cell proliferation but also inhibiting tumor stemness and preventing tumor recurrence.
Hyaluronan-mediated motility receptor (HMMR, also known as receptor for hyaluronate-mediated motility (RHAMM)) is an oncogene that is hyper-expressed and plays essential roles during the neoplastic progression of human leukemias and solid tumors (22-26). High levels of HMMR in breast cancer are associated with poor disease outcome (27). HMMR has been identified as a novel breast cancer susceptibility gene. Homozygous variation in the HMMR locus associates with higher risk of breast cancer (28). In human gliomas, HMMR expression is virtually ubiquitous in glioma tumor specimens. GBM expresses more HMMR than do lower grade lesions. Glioma cell lines also have higher level of HMMR than that in normal human astrocytes (29).
HMMR is a multifunctional oncogenic protein, the overexpression of which is transforming and essential for maintaining H-ras-mediated transformation (30). HMMR and CD44 are two ubiquitous receptors for hyaluronan, which is a prominent component of the microenvironment in most malignant tumors. CD44 has been identified as a cancer stem cell marker and CD44 directly regulates cancer stem cells in a variety of cancers including glioblastoma (31, 32). Extracellular HMMR forms a complex with CD44 that upon binding to hyaluronan activates intracellular signaling pathways, such as extracellular signal-regulated kinase (ERK), that regulate tumor cell survival, proliferation and invasion (33). Intracellular HMMR associates with microtubules, interacts with the mitotic spindle and contributes to tumor progression by promoting genomic instability (34). Currently, critical evidence is still lacking regarding the function of HMMR in the context of tumor-initiating stem cells in GBM and other cancers.
Here we used human-GBM-derived neurosphere cultures to examine the function of HMMR in tumor-initiating GSCs. HMMR was found as an novel regulator of GBM stemness. HMMR silencing in GSCs caused loss of self-renewal and blocked GSC-initiated xenograft growth. HMMR overexpression promoted GSC self-renewal and in vivo tumor propagation. In human GBM tumors, HMMR expression was found to positively correlate with the expression of stemness-associated markers and regulators. Overall, our results identify HMMR as a novel therapeutic target for inhibiting GBM stemness and tumor propagation.
Materials and Methods
Reagents
All reagents were purchased from Sigma-Aldrich unless otherwise stated.
GBM tumor specimens
HMMR IHC on human GBM tissue array (GL806b, US Biomax) was performed using anti-HMMR antibody (Origene) and Vectastain Elite ABC kit (Vector Laboratories). We analyzed three random fields per tumor tissue to generate an average value per individual GBM specimens following our established protocol (35). HMMR expression value was determined by calculating the percent area of antibody staining using ImageJ software (http://rsb.info.nih.gov/ij/). The average IgG control values were determined in adjacent serial sections and subtracted from the raw HMMR expression value. Human GBM tumors were collected at Johns Hopkins Hospital. All human materials were obtained and used in compliance with the Johns Hopkins Medicine Institutional Review Boards.
Cell culture
Human GBM neurosphere lines, 0913 (GBM1A) and 0627 (GBM1B), were originally established by Vescovi and colleagues (36). Cells were cultured in serum-free medium supplemented with EGF/FGF and incubated in 5% CO2/95% air condition at 37°C. The primary GBM neurospheres (JH273 and JH551) were established from GBM tumors at Johns Hopkins University using the same methods and culture conditions as described by Galli et al (36), and have been validated by us (37, 38). Primary neurospheres were used at less than 10 passages.
Lentiviral transduction
The sequences for HMMR shRNA lentiviral vectors (TRCN0000061553, TRCN0000061555, Thermo Scientific) are listed in Supplemental Table 1. Human HMMR cDNA was cloned into pTRIPZ vector (Thermo Scientific) with AgeI and MluI. The GFP and RFP genes were cloned into the pLEX-MCS vector (Thermo Scientific). Trans-Lentiviral Packaging System (Thermo Scientific) was used for lentivirus packaging. Cells were infected by lentivirus (MOI = 5) for 24h with TransDux Virus Infection solution (System Biosciences). Stable GBM neurosphere lines were established by puromycin selection (1μg/ml).
Neurosphere formation and soft agar clonogenic assays
Viable cells (2 × 104/well) were cultured in 6 well plates. After 6 days, neurospheres were fixed in medium with 1% agarose and counted (>100μm in diameter, 3 random fields per well) after Wright staining (1%) using computer-assisted morphometry (MCID).
Flow cytometric assay
Unfixed cells were stained with CD133/2(293C3)-PE antibody (Miltenyi Biotec) following manufacturer's protocol.
Western blot
Total cellular proteins were extracted with RIPA buffer (Sigma-Aldrich) containing protease and phosphatase inhibitors (Calbiochem). SDS-PAGE was performed with 50μg total cellular proteins using 4–12% gradient Tris-glycine gels (LI-COR Biosciences). Western blot was performed using Quantitative Western Blot System using secondary antibodies were labeled with IRDye infrared dyes (LI-COR Biosciences). The primary antibodies were: anti-HMMR (Origene), anti-SOX2, anti-BMI1, anti-pERK1/2, anti-pMEK1/2, anti-MEK1/2 (Cell Signaling), anti-OLIG2 (Santa Cruz), anti-ERK1 (BD), and anti-β-actin (Sigma).
Immunofluorescence
Neurosphere cells were collected by cytospin onto glass slides and fixed with 4% paraformaldehyde. Cells were permeabilized by Triton X-100 and immunostained with anti-HMMR antibody (Origene) and Alexa488-labelled secondary antibody. Images were taken and analyzed using ApoTome System (Zeiss).
Tumor xenografts
All animal protocols were approved by the Johns Hopkins School of Medicine Animal Care and Use Committee. SCID immunodeficient mice received 5,000 viable cells (determined by trypan blue staining) in 2 μl DMEM by stereotactic injection to the right caudate/putamen (AP=0mm, ML=-2.5mm, DV=-3.0mm). Mice were perfused with 4% paraformaldehyde. Maximum tumor volume on H&E-stained brain coronal sections was quantified using computer-assisted morphometry (MCID). Images of GFP- and RFP-labeled tumors were taken using FluoView confocal microscope (Olympus).
Quantitative real-time PCR (qRT-PCR)
RNA was extracted using RNeasy Mini kit (Qiagen). After reverse transcription using MuLV reverse transcriptase (Applied biosystems) and Oligo(dT) primer, qRT-PCR was performed using SYBR Green PCR Mix (Applied Biosystems) and IQ5 detection system (Bio-rad). Primer sequences are listed in Supplemental Table 1. Relative gene was normalized to 18S rRNA.
Statistical analysis
All results represent ≥3 independent replications. Statistical analysis was performed using Prizm software (GraphPad) and R/Bioconductor software package (39). Post-hoc tests included the Students t-test and Tukey multiple comparison tests as appropriate. All data are represented as mean value ± standard error of mean (SEM). Statistical significance in limiting dilution assay was determined by the extreme limiting dilution analysis (http://bioinf.wehi.edu.au/software/elda/) (40).
Gene expression data for human GBM samples (TCGA database) were normalized and summarized using RMA (41). STATA software (StataCorp) was used for generating HMMR decile. Following analyses (one-way ANOVA, linear trend and linear regression) were performed using Prizm software (GraphPad). The test for linear trend is a follow-up test after one-way ANOVA and asks whether the column means increase (or decrease) systematically as the columns go from left to right (42, 43).
Results
HMMR expression in GBM specimens and GSCs
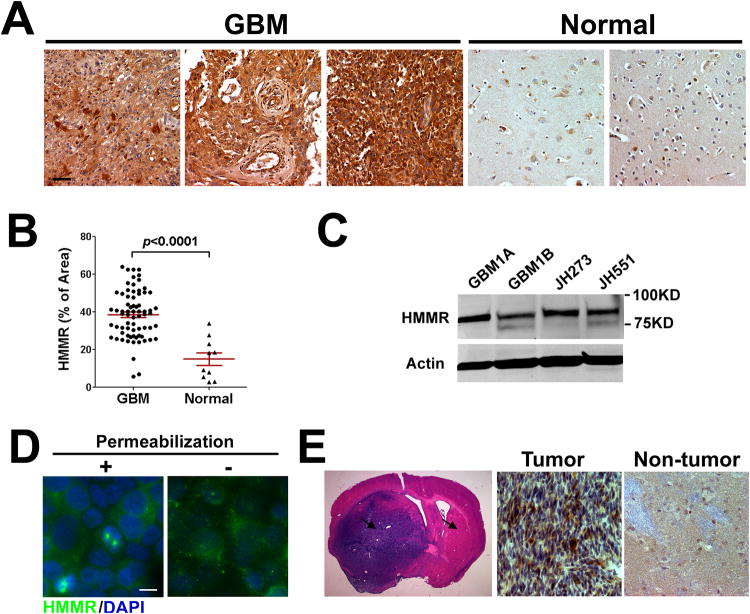
HMMR protein hyper-expression has been reported in a variety of human tumors including glioma (29). Here, we performed immunohistochemistry (IHC) on a paraffin-embedded human GBM tissue microarray. Representative results from 70 GBM and 10 normal brain specimens are shown in Fig. 1A, and negative control staining using rabbit IgG is shown in Supplemental Fig. 1. HMMR shows higher expression in GBM than normal brain specimens. HMMR expression was analyzed by semi-quantitative assessment of the HMMR IHC signal in GBM tissue array (see details in Materials and Methods). We found HMMR expression was 127% higher in GBM tumors than normal brain specimens (Figure 1B). We also analyzed HMMR mRNA expression in Repository for Molecular Brain Neoplasia Data (REMBRANDT) database (National Cancer Institute, https://caintegrator.nci.nih.gov/rembrandt/). HMMR expression is 4.9-fold, 2.8-fold and 2.1-fold higher in GBM, oligodendroglioma and astrocytoma, respectively, when compared to non-tumor samples (Supplemental Fig. 2). HMMR expression is also significantly up-regulated in GBM when compared with oligodendroglioma and astrocytoma (Supplemental Fig. 2).
Figure 1. HMMR expression in human GBM specimens, GSCs and GSC-derived xenografts.
(A) Representative photomicrographs from HMMR IHC staining in a GBM tissue array (70 GBM and 10 normal brain tissues) show HMMR+ cells in brown (bar = 20μm) with hematoxylin counterstaining.
(B) Semi-quantitative assessment of HMMR IHC signal in the GBM tissue array (red bar: Mean ± SEM, p<0.0001).
(C) Total cell lysates from GSC lines (GBM1A and GBM1B) and primary GSC cultures (JH273 and JH551) were blotted with HMMR antibody.
(D) GBM1A neurospheres with or without Triton X-100 permeabilization were immunostained against HMMR (Bar = 10 μm).
(E) H&E stained coronal brain sections (20 μm) show intracranial xenograft tumors from GBM1B cells (left panel). Arrows mark the regions for comparing HMMR expression by IHC staining. HMMR expression in tumors was higher than that in non-tumor tissues (middle and right panel).
Next, we examined HMMR expression in GBM-derived neurosphere lines (GBM1A and GBM1B) and low passage primary GBM-derived neurospheres (JH273 and JH551), both of which are enriched for GSCs. HMMR is ubiquitously expressed in various GSC cultures (Fig. 1C). We further examined HMMR expression using immunostaining in GSC cultures. Cytoplasmic and cell-surface HMMR expression was detected in permeabilized and nonpermeabilized cells, respectively (Fig. 1D). In addition, we performed HMMR IHC in intracranial tumor xenografts derived from human GBM-derived neurospheres. HMMR expression is higher in tumor xenografts when compared with non-tumor mouse brain tissues (Fig. 1E).
Targeting HMMR in GSCs impairs stem cell proliferation and self-renewal, and inhibits the expression of stemness markers and regulators
To study the requirement of HMMR in GSC maintenance, we transduced GBM-derived neurosphere cells with two distinct lentiviral shRNAs (HMMR shRNA1 and shRNA2) that can dramatically silence HMMR expression by >90% when compared to non-targeting (NT) shRNA control (Fig. 2A). We examined the effects of HMMR knockdown on the self-renewal and proliferation capacity of GSCs. Cell proliferation, as shown by growth curve, was inhibited by 55% after HMMR knockdown for 6 days (Fig. 2B). Cell proliferation, as determined by BrdU incorporation assay, was also decreased by 22% after HMMR knockdown (Supplemental Fig. 3). HMMR silencing did not significantly induce cell death (Fig. 2C). HMMR knockdown significantly reduced the efficiency of neurosphere formation, an in vitro marker of GSC self-renewal and proliferation capacity, by 69-88% in GBM neurosphere lines and primary GBM neurosphere culture (Figure 2D, 2E left panel and 2F). First passage GBM1A neurospheres with or without HMMR silencing were dissociated to form second passage neurospheres. Cells with HMMR knockdown failed to form neurospheres in the second passage (Figure 2E, right panel). These results support that HMMR expression is required for the self-renewal and proliferation of GSCs.
Figure 2. Silencing HMMR inhibits cell proliferation and sphere formation in GSCs.
(A, B and C) GBM1A cells were transduced with HMMR shRNAs or non-targeting (NT) shRNA. After 72h, total cellular proteins were blotted with HMMR antibody (A). HMMR fold expression normalized to β-actin is shown below each lane. All cells within cultures were stained with trypan blue and counted on the days shown to calculate cell number and viability (n=3 for each group). HMMR inhibition inhibited cell growth (B) but did not induce significant cell death (C).
(D, E and F) Equal numbers of viable cells after HMMR shRNA transduction were cultured for 6 days to form 1st passage neurospheres. Representative microscopic fields are shown to compare GBM1B neurospheres with NT or HMMR shRNA (D, Bar = 100μm). Neurospheres (>100μm diameter) were counted (E left panel and F). Equal numbers of viable cells from 1st passage GBM1A neurospheres were dissociated for 2nd passage neurosphere formation (E right panel).
Data represents Mean ± SEM; *: p<0.01 when compared with NT shRNA.
Next, we further examined the effects of HMMR silencing on the expression of stemness-associated markers in GSCs. CD133 expression has been shown to correlate with the tumor-initiating capacity of GSCs and is widely used as a GBM-stemness-associated marker (8, 44). The percentage of CD133+ cells in GBM1A and GBM1B cells was decreased by each of two HMMR shRNAs from 53%-63% to 27%-35% (Fig. 3A and 3B). It has been shown that GSCs are marked and regulated by various stem-cell-associated transcription factors, including SOX2 (45), BMI1 (46) and OLIG2 (47), both of which are essential for GSC maintenance. We found that HMMR silencing in GBM1A and GBM1B cells decreased the expression of these GSC markers/regulators (Fig. 3C). SOX4 is another stemness-regulating transcription factor that binds to the enhancer region of SOX2, promotes SOX2 expression and sustains the tumorgenicity of GSCs (48). HMMR silencing also suppressed the expression of SOX4 by ~60% (Fig. 3D).
Figure 3. HMMR knockdown inhibits the expression of stemness markers/regulators and the activation of ERK.

(A and B) GBM1A and GBM1B cells were transduced with HMMR shRNAs or NT shRNA. Cells were analyzed 6 days after transduction by flow cytometry using CD133 antibodies and isotype IgG control (n=3). Representative histogram and the percentage of CD133+ cells are shown.
(C and D) GBM1A and GBM1B cells were transduced with HMMR shRNAs or NT shRNA. After 6 days, total cell lysates were blotted against SOX2, OLIG2 and BMI1 (C). Relative expression of each protein (normalized to Actin) is shown below each band (expression of NT shRNA sample = 1). HMMR knockdown decreased SOX4 expression, as measured by qRT-PCR in GBM1B cells (D).
(E) GBM1B cells were transduced with HMMR shRNAs or NT shRNA. After 4 days, total cell lysates were blotted against phosphorylated ERK1/2 (p-ERK1/2), ERK1/2, phosphorylated MEK (p-MEK), MEK and β-actin. Relative expression of each protein (normalized to Actin) is shown below each band (expression of NT shRNA sample = 1).
Data represents Mean ± SEM; *: p<0.01 when compared with NT shRNA.
HMMR has been shown to interact with ERK and regulate ERK phosphorylation (49, 50). The ERK pathway is one of the pivotal transmitters of growth-factor signaling, which controls diverse cellular processes such as proliferation (51), differentiation (52) and motility (53), and also maintains stemness in human embryonic stem cells (54). Here, we found HMMR knockdown in GSCs attenuated ERK expression and phosphorylation but did not change MEK expression and phosphorylation (Fig. 3E).
Overall, these results support that HMMR silencing efficiently blocks GSC self-renewal and suggest that targeting HMMR may also inhibit GSC-initiated tumor growth in vivo.
Targeting HMMR suppresses GSC-derived tumor growth and extends the survival of mice bearing GSC xenografts
The most important property of GSCs is their ability to efficiently propagate tumors in vivo that recapitulate the parent GBM tumors (9). We examined the effects of HMMR silencing on the growth of intracranial (i.c.) xenografts established from GBM-derived neurospheres. GBM1A and GBM1B cells were infected with two HMMR shRNAs or non-targeting (NT) shRNA, and 24h after infection we transplanted 5,000 viable cells into the brains of immunocompromised mice to form xenograft tumors. Animals were sacrificed 60 days later, and mice implanted with HMMR-knockdown GBM1A cells showed significantly reduced tumor formation as compared with non-targeted controls (Fig. 4A and 4B, max tumor volume ± SEM (mm3): 19.1 ± 7.2 (HMMR shRNA1), 2.5 ± 1.1 (HMMR shRNA2), 149.2 ± 20.1 (Control shRNA)). Moreover, mice bearing intracranial xenografts derived from HMMR-shRNA1-infected GBM1A cells survived significantly longer than those with NT-shRNA-infected cells (median survival: 148 days vs 78 days, p=0.0018; Figure 4C). Although lentivirus used here for shRNA delivery consistently yields >90% transduction efficiency in GBM1A and GBM1B cells (data not shown), HMMR expression shows no obvious difference in tumors derived from either HMMR-knockdown or control GSCs (Supplemental Fig. 4), suggesting that HMMR-knockdown tumors with inhibited growth are derived from GSCs that escape from HMMR silencing.
Figure 4. Silencing HMMR suppresses the growth of GSC-derived tumor xenografts.
(A and B) GBM1A cells were infected with HMMR shRNAs or non-targeting (NT) control shRNA. 24h after infection, 5,000 viable cells were transplanted into the brains of NOD/SCID mice. H&E stained coronal brain sections (20 μm) obtained from post-implantation day 60 animals are shown (A, Bar = 1mm). Quantification of xenograft tumor volume shows that silencing HMMR suppressed xenograft growth (B, n=5, *: p<0.01 when compared to control shRNA).
(C) Kaplan-Meier survival curves of mice implanted with GBM1A cells expressing HMMR shRNA1 or NT shRNA (n=5, *: p=0.0018).
(D, E, F, G and H) Schematic of dual color competition assay in vivo (E). 5,000 cells from a 1:1 mixture of GFP-labeled HMMR knockdown cells (green) and RFP-labeled control cells (red) were implanted into mouse brains. Representative brain sections (20 μm) obtained from post-implantation day 60 animals (n=3 for each group) show tumor xenografts from GFP- and RFP-labeled GBM1B cells (E and G). Arrows indicate the regions shown in F and H (bar = 50 μm).
Next, we used the in vivo limiting dilution assay to further ask if targeting HMMR generates a significant disadvantage in tumor initiating potential. GBM1A cells were infected with HMMR shRNA2 or NT shRNA, and 24h after infection we transplanted 2,000 or 200 viable cells into mice. At 100 days post implantation, mice receiving control cells developed tumors (4/4 and 3/4 in mice receiving 2,000 and 200 cells, respectively). By contrast, none of the mice receiving HMMR-knockdown cells developed tumors (Supplemental Fig. 5A). Tumor initiation incidence from this experiment and Fig. 4A (summarized in Supplemental Fig. 5B) was subjected to the extreme limiting dilution analysis (40) to estimate the frequency of tumor-initiating cells. The frequency of tumor-initiating cells in HMMR-knockdown cells is significantly lower than that in control cells (estimate frequency: 1/5582 vs 1/144, χ2 = 19.7, df = 1, p<0.0001).
We also used dual-color competition assay to further confirm the effect of HMMR targeting on GSC-derived tumor formation. GBM1B cells were stably tranduced by lentivirus encoding green or red fluorescent protein (GFP or RFP) to establish green and red neurospheres (Supplemental Fig. 6). As shown in Fig. 4D, green and red cells were infected by HMMR shRNA or NT shRNA, respectively. 24h after lentivirus infection, 5,000 viable cells from a 1:1 mixture of green HMMR-knockdown cells and red control cells were implanted into mouse brains to perform dual-color competition assay in vivo (n=3 for each group, Figure 4E-4H shows representative results from one mouse). Green cells transduced with HMMR shRNA lost their tumor initiation capacity when compared to red cells transduced with control shRNA, as evidenced by the absence of green cells in tumor xenografts (Fig.4E and 4F). In a control experiment, green and red cells without shRNA transduction displayed no difference in tumor initiation capacity (Fig. 4G and 4H).
HMMR overexpression enhances GSC self-renewal and promotes GSC-derived tumor growth
HMMR overexpression has been reported to transform fibroblast cells and cause spontaneous metastases in the lung (30). Here, we address the effects of HMMR overexpression on stemness-associated phenotypes in GBM neurospheres. Two independent GBM neurosphere lines (GBM1A-HMMR and GBM1B-HMMR) were engineered to overexpress HMMR by transducing cells with a lentiviral vector encoding the human HMMR transgene. HMMR western blotting showed >4-fold overexpression of HMMR (Fig. 5A). HMMR overexpression significantly enhanced neurosphere formation in both GBM1A and GBM1B cells (Fig. 5B). The percentage of CD133+ cells in GBM1B cells was increased by HMMR overexpression from 50% to 63% (Fig. 5C). HMMR overexpression also elevated the expression of stemness-associated transcription factors, including SOX2, OLIG2, BMI1 and SOX4 (Fig. 5D and 5E).
Figure 5. HMMR overexpression promotes GSC neurosphere formation, marker expression and tumor formation.
(A) GBM1A and GBM1B cells were infected with lentivirus harboring cDNAs coding human HMMR or no cDNA insert as the control. Whole cell lysates were immunoblotted against HMMR. HMMR fold expression normalized to β-actin is shown below each lane.
(B) Neurospheres (>100μm diameter) from GBM1A-HMMR, GBM1B-HMMR and control cells were counted.
(C) GBM1B-HMMR and control cells were analyzed by flow cytometry using CD133 antibodies and isotype IgG control (n=3). Representative histogram and the percentage of CD133+ cells are shown.
(D and E) Total cell lysates from GBM1A-HMMR, GBM1B-HMMR and control cells were blotted against stemness markers/regulators (D). Relative expression of each protein (normalized to Actin) is shown below each band (expression of NT shRNA sample = 1). SOX4 expression was determined by qRT-PCR in GBM1B cells (E).
(F) 5,000 viable GBM1B-HMMR or control cells were transplanted into mice. H&E stained coronal brain sections (20 μm) from post-implantation day 40 animals are shown (Left panel, Bar = 1mm). Quantification of tumor volume shows that HMMR overexpression promoted tumor formation (right panel, n=5, *: p=0.0014 when compared with animals bearing control cells).
Data represents Mean ± SEM (n=3); *: p<0.01 when compared with control cells.
We further investigated the effects of HMMR overexpression on GSC-derived tumor propagation. GBM1B-HMMR cells or control cells with empty vector (5,000 viable cells per animal) were transplanted into mouse brains. Mice (n=5) were sacrificed 40 days post transplantation for tumor size measurement. Mice bearing GSCs with HMMR overexpression showed significantly enhanced tumor growth when compared with control mice (Fig. 5F, max tumor volume ± SEM (mm3): 66.0 ± 7.4 vs 27.4 ± 3.3).
These results support that HMMR overexpression promotes GSC self-renewal and tumor propagation.
HMMR expression correlates with the expression of stemness markers and regulators in human GBM specimens
Our HMMR knockdown and overexpression experiments in GSC models demonstrate that HMMR expression in GSCs drives stem cell self-renewal and the expression of stemness markers and regulators. Next, we asked whether HMMR level correlates with the expression of GBM stemness markers and regulators in clinical GBM specimens. Total RNA were isolated from 20 human GBM tumors. HMMR expression and the expression of a panel of markers and regulators were analyzed by qRT-PCR. Linear regression analyses at 95% confidence interval showed that HMMR positively correlated with the expression of PROM1 (CD133), SOX2, SOX4 and BMI1, respectively, in human GBM specimens (Fig. 6A-6D). HMMR expression did not correlate with OLIG2 expression (Supplemental Fig. 7). These results are consistent with a role of HMMR in regulating GBM cell stemness not only in GSC culture models but also in human GBM tumors.
Figure 6. HMMR expression correlates with the expression of stemness markers and regulators in human GBM tumors.
Total RNA from 20 human GBM tumors were examined for the expression of HMMR, PROM1 (CD133), BMI1, SOX2 and SOX4. (A-D) The scatter plots represent the levels of HMMR and individual stemness markers and regulators as determined by qRT-PCR and normalized to 18S rRNA. The correlation coefficient (R2) and p values, as calculated by linear regression (95% confidence interval) are shown inside each panel.
To further validate these results, we analyzed the expression of HMMR and GBM stemness markers/regulators in 414 GBM samples using The Cancer Genome Atlas (TCGA) database. We ranked GBM samples by HMMR expression and grouped samples to establish HMMR deciles according to HMMR level (Fig. 7A, left panel, mean expression values of each decile normalized to Decile 1 are shown in the table). HMMR expression in Decile 10 (HMMR High) and Decile 5 (HMMR Medium) were 62-fold and 5.1-fold, respectively, higher than that in Decile 1 (HMMR Low). The expression of PROM1 (CD133), SOX4, BMI1 and OLIG2 in Decile 10 were 3.8-fold, 2.5-fold, 1.6-fold and 1.5-fold, respectively, of those in Decile 1; and in Decile 5, they were 4.3-fold, 2.3-fold, 1.1-fold and 0.68-fold of those in Decile 1 (Fig. 7B-7E, left panel). One-way ANOVA with the post test for linear trend (see details in Materials and Methods) revealed that the expression of HMMR, PROM1 (CD133), SOX4, BMI1 and OLIG2 expression increase with a significant linear trend from HMMR Low decile to HMMR High decile (p<0.01, individual p value marked inside the left panels of Fig. 7B-7E). There is no significant linear trend between SOX2 expression and HMMR deciles (p=0.31, Fig. 7F left panel). Linear regression analyses with a 95% confidence interval revealed significant correlations between HMMR expression and the expression of PROM1 (CD133), SOX4 and BMI1 (Fig. 7B-7D, right panel with r2 and p value marked inside), consistent with the results from linear trend analyses. OLIG2 and SOX2 show no significant correlation with HMMR in this analysis (Fig. 7E and 7F).
Figure 7. The correlation between HMMR and stemness markers/regulators in TCGA GBM dataset.
(A-F, left panel) A decile analysis was performed by grouping 414 GBM patient samples into 10% categories based on HMMR level to establish HMMR decile. The bar graphs represent the Mean ± SEM expression of individual genes in each HMMR decile. Decile 1 to 10 corresponds to HMMR low to high, respectively. Tables below the graphs show the mean gene expression values normalized to the value of Decile 1. The p values marked inside each panel are calculated by one-way ANOVA with the post test for linear trend.
(A-F, right panel) The medium expression of HMMR in each HMMR decile was plotted with the medium expression of each stemness gene. The r2 and p values marked inside each panel were calculated using test of linear regression with a 95% confidence interval.
These results using two independent sources of human GBM specimens show that HMMR levels correlate with some stemness markers and regulators in GBM tumors.
Discussion
The advances in GBM therapies over the past decade provide only modest survival improvements in GBM patients. One of the explanations for the failure of GBM therapies is the incomplete elimination of tumor-initiating GSCs, a cell population that harbors stem-cell-like characteristics (stemness) and promotes therapeutic resistance and tumor recurrence. Identifying therapeutic targets in GSCs should lead to effective therapies against GSCs and improve GBM prognosis particularly when combining with current cytotoxic therapies. In this study, we identified HMMR as a promising target for the development of anti-GSC therapeutic agents.
By analyzing GBM tissue array and mining glioma gene expression database, we show HMMR hyper-expression in human GBM tumors when compared with low-grade brain tumors and non-malignant brain tissues. These results support HMMR as a GBM-tumor-associated protein, whose functions in regulating GBM tumor stemness and propagation are largely unknown. We present the novel finding that HMMR regulates GBM tumor stemness. Silencing HMMR in GSCs impairs stem cell proliferation and self-renewal, and inhibits the expression of stemness markers and regulators, including CD133, SOX2, SOX4, BMI1 and OLIG2. Additionally, HMMR overexpression in GSCs enhances self-renewal and increases the expression of stemness markers and regulators. These results from a combination of gain- or loss-of-function assays support a positive correlation between HMMR and GSC stemness. Stemness-regulating transcription factors, such as SOX2, SOX4, BMI1 and OLIG2, function as integral components of the core regulatory circuitry in both cancer stem cells and normal pluripotent/multipotent cells (45-48). The regulatory role of HMMR on stemness markers and regulators suggests the feasibility to blocking GSC stemness via HMMR-targeted strategies. Although the exact signaling pathway linking HMMR and stemness-regulating network is yet to be determined, it is likely to involve the HMMR-ERK signaling pathway that can be activated by extracellular stimuli such as hyaluronan and platelet-derived growth factor (PDGF) (49, 50). Hyaluronan has been identified as a potential niche matrix for supporting the long-term self-renewal of embryonic stem cells (ESCs) and neural stem cells (55, 56). Silencing hyaluronan receptor HMMR in human ESCs results in loss of cell pluripotency and viability (57), which is consistent with our finding that GSCs with HMMR silencing lost their self-renewal capability. Meanwhile, we found that HMMR silencing in GSCs attenuates ERK expression and phosphorylation, which is consistent with the evidence that targeted inactivation of ERK using pharmacological inhibitors or siRNAs inhibited GSC self-renewal and reduced the expression of stemness regulators including BMI1 and SOX2 (58). Recent studies focusing on the genome-wide ERK-chromatin interaction in human embryonic stem cells revealed that ERK binds to the promoters of stemness-regulating genes, such as SOX2 (54), which further supports ERK as a potential mediator linking HMMR and stemness regulators in GSCs. Because HMMR is a multifunctional oncogenic protein that also associates with centrosome and regulates mitotic spindle formation during cell division (34), we cannot rule out the possibility that HMMR might modulate GSC stemness by affecting centrosome structure and cell division, both of which are involved in stem cell maintenance (59, 60).
The translational significance of our findings stems from the potential for targeting HMMR to inhibit GSC-initiated tumor propagation. In our intracranial xenograft models, HMMR knockdown significantly inhibited GSC-derived tumor growth and extended the survival of mice bearing GSC xenografts. Although being effectively inhibited, tumors derived from HMMR-silenced GSCs still expressed HMMR, suggesting that incomplete virus infection or silencing of lentiviral transgenes, which has also been reported in similar gene knockdown experiments using lentiviral shRNAs (61, 62), contributed at least in part to tumor formation in the HMMR-silenced xenografts. In comparison, we found that HMMR-silenced GSCs dramatically lost their tumor-initiating capacity when they were co-implanted with non-silenced control cells. It is possible that competition from control GSCs inhibits the tumor-initiation capability of HMMR-silenced cells. In addition, this result also suggests that, although HMMR can be exported and thereby modify signaling properties of cell surface receptors (e.g. CD44) (33, 63), HMMR expressing GSCs do not efficiently rescue the tumor-initiating defect in HMMR-silenced cells.
A clinically relevant finding in this paper is the association between HMMR expression and stemness markers and regulators (hereby referred to as “HMMR-stemness association”) in GSC models and human GBM specimens. In GSC models, HMMR-stemness association is supported by the positive correlation between HMMR and various stemness markers and regulators including CD133, SOX2, SOX4, OLIG2 and BMI1. This HMMR-stemness association was further studied in human GBM clinical specimens. We validated the correlation between HMMR and a subset of stemness markers and regulators (CD133, SOX4 and BMI1) in two independent sources of human GBM specimens. SOX2 expression was found to correlate with HMMR in one group of GBM samples but not in the TCGA dataset. Recent findings from multiple laboratories have shown that the tumor-initiating stemness phenotype can be induced in non-stem cancer cells in response to either genetic mutations or environmental cues, and this induction of stemness further leads to therapeutic resistance and tumor recurrence (64-68). The HMMR-stemness association we identified here suggests that HMMR-targeted strategies may offer effective therapies against GBM stemness, which is a dynamically regulated phenotype in tumor hierarchy (64).
In summary, we found that HMMR maintains GSC stemness. Targeting HMMR efficiently inhibits GSC stemness and tumorigenicity, suggesting HMMR as a new therapeutic target in GBM.
Supplementary Material
Acknowledgments
Financial support: This work was supported by American Brain Tumor Association Discovery Grant (M.Y.), Maryland Stem Cell Research Fund (M.Y. and J.L.), NIH R01NS076759 R01NS073611 (J.L.), NIH 5R01NS070024 (A.Q.H.) and the James S. McDonnell Foundation (J.L.).
We thank Dr. Angelo Vescovi for providing human GBM-derived stem cell lines.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors indicate no potential conflicts of interest.
References
- 1.McGirt MJ, Than KD, Weingart JD, Chaichana KL, Attenello FJ, Olivi A, et al. Gliadel (BCNU) wafer plus concomitant temozolomide therapy after primary resection of glioblastoma multiforme. J Neurosurg. 2009;110:583–8. doi: 10.3171/2008.5.17557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Park CH, Bergsagel DE, McCulloch EA. Mouse myeloma tumor stem cells: a primary cell culture assay. J Natl Cancer Inst. 1971;46:411–22. [PubMed] [Google Scholar]
- 5.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 6.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–99. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 7.Ward RJ, Dirks PB. Cancer stem cells: at the headwaters of tumor development. Annu Rev Pathol. 2007;2:175–89. doi: 10.1146/annurev.pathol.2.010506.091847. [DOI] [PubMed] [Google Scholar]
- 8.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 9.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 10.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 11.Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6:425–36. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- 12.Stiles CD, Rowitch DH. Glioma stem cells: a midterm exam. Neuron. 2008;58:832–46. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 13.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 14.Stockhausen MT, Kristoffersen K, Poulsen HS. The functional role of Notch signaling in human gliomas. Neuro Oncol. 2010;12:199–211. doi: 10.1093/neuonc/nop022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaur B, Khwaja FW, Severson EA, Matheny SL, Brat DJ, Van Meir EG. Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. Neuro Oncol. 2005;7:134–53. doi: 10.1215/S1152851704001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–72. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Sullenger BA, Rich JN. Notch signaling in cancer stem cells. Adv Exp Med Biol. 2012;727:174–85. doi: 10.1007/978-1-4614-0899-4_13. [DOI] [PubMed] [Google Scholar]
- 20.Heddleston JM, Hitomi M, Venere M, Flavahan WA, Yang K, Kim Y, et al. Glioma stem cell maintenance: the role of the microenvironment. Curr Pharm Des. 2011;17:2386–401. doi: 10.2174/138161211797249260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lathia JD, Heddleston JM, Venere M, Rich JN. Deadly teamwork: neural cancer stem cells and the tumor microenvironment. Cell Stem Cell. 2011;8:482–5. doi: 10.1016/j.stem.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maxwell CA, McCarthy J, Turley E. Cell-surface and mitotic-spindle RHAMM: moonlighting or dual oncogenic functions? J Cell Sci. 2008;121:925–32. doi: 10.1242/jcs.022038. [DOI] [PubMed] [Google Scholar]
- 23.Telmer PG, Tolg C, McCarthy JB, Turley EA. How does a protein with dual mitotic spindle and extracellular matrix receptor functions affect tumor susceptibility and progression? Commun Integr Biol. 2011;4:182–5. doi: 10.4161/cib.4.2.14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giannopoulos K, Li L, Bojarska-Junak A, Rolinski J, Dmoszynska A, Hus I, et al. Expression of RHAMM/CD168 and other tumor-associated antigens in patients with B-cell chronic lymphocytic leukemia. Int J Oncol. 2006;29:95–103. [PubMed] [Google Scholar]
- 25.Toole BP. Hyaluronan: from extracellular glue to pericellular cue. Nat Rev Cancer. 2004;4:528–39. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 26.Turley EA, Noble PW, Bourguignon LY. Signaling properties of hyaluronan receptors. J Biol Chem. 2002;277:4589–92. doi: 10.1074/jbc.R100038200. [DOI] [PubMed] [Google Scholar]
- 27.Wang C, Thor AD, Moore DH, 2nd, Zhao Y, Kerschmann R, Stern R, et al. The overexpression of RHAMM, a hyaluronan-binding protein that regulates ras signaling, correlates with overexpression of mitogen-activated protein kinase and is a significant parameter in breast cancer progression. Clin Cancer Res. 1998;4:567–76. [PubMed] [Google Scholar]
- 28.Pujana MA, Han JD, Starita LM, Stevens KN, Tewari M, Ahn JS, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 2007;39:1338–49. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 29.Akiyama Y, Jung S, Salhia B, Lee S, Hubbard S, Taylor M, et al. Hyaluronate receptors mediating glioma cell migration and proliferation. J Neurooncol. 2001;53:115–27. doi: 10.1023/a:1012297132047. [DOI] [PubMed] [Google Scholar]
- 30.Hall CL, Yang B, Yang X, Zhang S, Turley M, Samuel S, et al. Overexpression of the hyaluronan receptor RHAMM is transforming and is also required for H-ras transformation. Cell. 1995;82:19–26. doi: 10.1016/0092-8674(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 31.Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254–67. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 32.Anido J, Saez-Borderias A, Gonzalez-Junca A, Rodon L, Folch G, Carmona MA, et al. TGF-beta Receptor Inhibitors Target the CD44(high)/Id1(high) Glioma-Initiating Cell Population in Human Glioblastoma. Cancer Cell. 2010;18:655–68. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 33.Hamilton SR, Fard SF, Paiwand FF, Tolg C, Veiseh M, Wang C, et al. The hyaluronan receptors CD44 and Rhamm (CD168) form complexes with ERK1,2 that sustain high basal motility in breast cancer cells. J Biol Chem. 2007;282:16667–80. doi: 10.1074/jbc.M702078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maxwell CA, Keats JJ, Crainie M, Sun X, Yen T, Shibuya E, et al. RHAMM is a centrosomal protein that interacts with dynein and maintains spindle pole stability. Mol Biol Cell. 2003;14:2262–76. doi: 10.1091/mbc.E02-07-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodwin CR, Lal B, Zhou X, Ho S, Xia S, Taeger A, et al. Cyr61 mediates hepatocyte growth factor-dependent tumor cell growth, migration, and Akt activation. Cancer Res. 2010;70:2932–41. doi: 10.1158/0008-5472.CAN-09-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–21. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 37.Ying M, Wang S, Sang Y, Sun P, Lal B, Goodwin CR, et al. Regulation of glioblastoma stem cells by retinoic acid: role for Notch pathway inhibition. Oncogene. 2011;30:3454–67. doi: 10.1038/onc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ying M, Sang Y, Li Y, Guerrero-Cazares H, Quinones-Hinojosa A, Vescovi AL, et al. Kruppel-like family of transcription factor 9, a differentiation-associated transcription factor, suppresses Notch1 signaling and inhibits glioblastoma-initiating stem cells. Stem Cells. 2011;29:20–31. doi: 10.1002/stem.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 41.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 42.Sheskin DJ. Handbook of Parametric and Nonparametric Statistical Procedures. (Fourth) 2007 [Google Scholar]
- 43.Altman DG. Practical Statistics for Medical Research. 1990 [Google Scholar]
- 44.Bidlingmaier S, Zhu X, Liu B. The utility and limitations of glycosylated human CD133 epitopes in defining cancer stem cells. J Mol Med (Berl) 2008;86:1025–32. doi: 10.1007/s00109-008-0357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, et al. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27:40–8. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- 46.Abdouh M, Facchino S, Chatoo W, Balasingam V, Ferreira J, Bernier G. BMI1 sustains human glioblastoma multiforme stem cell renewal. J Neurosci. 2009;29:8884–96. doi: 10.1523/JNEUROSCI.0968-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–17. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–14. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 49.Hatano H, Shigeishi H, Kudo Y, Higashikawa K, Tobiume K, Takata T, et al. RHAMM/ERK interaction induces proliferative activities of cementifying fibroma cells through a mechanism based on the CD44-EGFR. Lab Invest. 2011;91:379–91. doi: 10.1038/labinvest.2010.176. [DOI] [PubMed] [Google Scholar]
- 50.Zhang S, Chang MC, Zylka D, Turley S, Harrison R, Turley EA. The hyaluronan receptor RHAMM regulates extracellular-regulated kinase. J Biol Chem. 1998;273:11342–8. doi: 10.1074/jbc.273.18.11342. [DOI] [PubMed] [Google Scholar]
- 51.Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 52.Lai CF, Chaudhary L, Fausto A, Halstead LR, Ory DS, Avioli LV, et al. Erk is essential for growth, differentiation, integrin expression, and cell function in human osteoblastic cells. J Biol Chem. 2001;276:14443–50. doi: 10.1074/jbc.M010021200. [DOI] [PubMed] [Google Scholar]
- 53.Viala E, Pouyssegur J. Regulation of tumor cell motility by ERK mitogen-activated protein kinases. Ann N Y Acad Sci. 2004;1030:208–18. doi: 10.1196/annals.1329.027. [DOI] [PubMed] [Google Scholar]
- 54.Goke J, Chan YS, Yan J, Vingron M, Ng HH. Genome-wide Kinase-Chromatin Interactions Reveal the Regulatory Network of ERK Signaling in Human Embryonic Stem Cells. Mol Cell. 2013;50:844–55. doi: 10.1016/j.molcel.2013.04.030. [DOI] [PubMed] [Google Scholar]
- 55.Gerecht S, Burdick JA, Ferreira LS, Townsend SA, Langer R, Vunjak-Novakovic G. Hyaluronic acid hydrogel for controlled self-renewal and differentiation of human embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:11298–303. doi: 10.1073/pnas.0703723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Preston M, Sherman LS. Neural stem cell niches: roles for the hyaluronan-based extracellular matrix. Front Biosci (Schol Ed) 2011;3:1165–79. doi: 10.2741/218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choudhary M, Zhang X, Stojkovic P, Hyslop L, Anyfantis G, Herbert M, et al. Putative role of hyaluronan and its related genes, HAS2 and RHAMM, in human early preimplantation embryogenesis and embryonic stem cell characterization. Stem Cells. 2007;25:3045–57. doi: 10.1634/stemcells.2007-0296. [DOI] [PubMed] [Google Scholar]
- 58.Sunayama J, Matsuda K, Sato A, Tachibana K, Suzuki K, Narita Y, et al. Crosstalk between the PI3K/mTOR and MEK/ERK pathways involved in the maintenance of self-renewal and tumorigenicity of glioblastoma stem-like cells. Stem Cells. 2010;28:1930–9. doi: 10.1002/stem.521. [DOI] [PubMed] [Google Scholar]
- 59.Cheng J, Turkel N, Hemati N, Fuller MT, Hunt AJ, Yamashita YM. Centrosome misorientation reduces stem cell division during ageing. Nature. 2008;456:599–604. doi: 10.1038/nature07386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–74. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 61.Guryanova OA, Wu Q, Cheng L, Lathia JD, Huang Z, Yang J, et al. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer Cell. 2011;19:498–511. doi: 10.1016/j.ccr.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lathia JD, Gallagher J, Heddleston JM, Wang J, Eyler CE, Macswords J, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–32. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tolg C, Hamilton SR, Nakrieko KA, Kooshesh F, Walton P, McCarthy JB, et al. Rhamm-/- fibroblasts are defective in CD44-mediated ERK1,2 motogenic signaling, leading to defective skin wound repair. J Cell Biol. 2006;175:1017–28. doi: 10.1083/jcb.200511027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y, Laterra J. Cancer stem cells: distinct entities or dynamically regulated phenotypes? Cancer Res. 2012;72:576–80. doi: 10.1158/0008-5472.CAN-11-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghisolfi L, Keates AC, Hu X, Lee DK, Li CJ. Ionizing radiation induces stemness in cancer cells. PLoS One. 2012;7:e43628. doi: 10.1371/journal.pone.0043628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y, Li A, Glas M, Lal B, Ying M, Sang Y, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci U S A. 2011;108:9951–6. doi: 10.1073/pnas.1016912108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CM, et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71:4640–52. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Po A, Ferretti E, Miele E, De Smaele E, Paganelli A, Canettieri G, et al. Hedgehog controls neural stem cells through p53-independent regulation of Nanog. EMBO J. 2010;29:2646–58. doi: 10.1038/emboj.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.