Summary
Background
Early, sorting endosomes are a major crossroad of membrane traffic, at the intersection of the endocytic and exocytic pathways. The sorting of endosomal cargo for delivery to different subcellular destinations is mediated by a number of distinct coat protein complexes, including AP-1, AP-3 and GGAs. Ultrastructural studies suggest that these coats assemble onto tubular subdomains of the endosomal membrane, but the mechanisms of coat recruitment and assembly at this site remain poorly understood.
Results
Here we report that the endosomal Rab protein Rab4 orchestrates a GTPase cascade which results in the sequential recruitment of the Arf-like protein Arl1, the Arf-specific guanine nucleotide exchange factors BIG1 and BIG2, and the class I Arfs, Arf1 and Arf3. Knockdown of Arf1, or inhibition of BIG1/2 activity with Brefeldin A results in the loss of AP-1, AP-3 and GGA-3, but not Arl1, from endosomal membranes and the formation of elongated tubules. In contrast, depletion of Arl1 randomizes the distribution of Rab4 on endosomal membranes, inhibits the formation of tubular subdomains and blocks recruitment of BIG1/2, Arfs and adaptor complexes to the endosome.
Conclusion
Together these findings indicate that Arl1 links Rab4-dependent formation of endosomal sorting domains with downstream assembly of adaptor protein complexes that constitute the endosomal sorting machinery.
Introduction
Eukaryotic cells are divided into compartments by membranes with unique protein and lipid composition. Transport between these compartments is essential for the processing and delivery of newly synthesized proteins, and redistribution or degradation of proteins internalized from the cell surface. Integral membrane proteins are sorted and concentrated for transport by their interaction with coat protein complexes, which assemble onto the cytoplasmic surface of donor membranes [1]. These include COPI and COPII, which function primarily at the ER/Golgi, and clathrin, which functions at the Trans Golgi Network (TGN), at the plasma membrane and on endosomal compartments. In contrast to COPI and COPII, which bind cargo directly, clathrin requires the participation of adaptor proteins that link cargo to the clathrin coat.
In the endocytic pathway, early endosomes are a major sorting “hub” from which internalized cargo can be recycled back to the plasma membrane, to the TGN, or routed to lysosomes for degradation. Morphological studies have shown that cargos destined for recycling or retrograde transport to the TGN become concentrated in tubular extensions of the early endosomal membrane. Sorting of cargos within these tubules is mediated by the heterotetrameric adaptors AP-1 and AP-3 [2], the multimeric retromer complex [3], and monomeric adaptors of the GGA family [4].
It is well established that both the AP-1/AP-3 complexes and the GGAs are recruited to membranes through interactions with small G proteins of the ADP-ribosylation factor (Arf) family [5 and 6]. The six mammalian Arfs can be grouped into three classes based on sequence similarity, such that Arf1, Arf2, and Arf3 (the Arf2 gene is lost in humans) constitute class I, Arf4 and Arf5 constitute class II, and Arf6 represents class III [5]. Existing data suggest that class I Arfs are largely responsible for the recruitment of the AP complexes and the GGAs to the TGN [4 and 5], but whether the same Arfs act on endosomal compartments is not known.
Like other GTPases, the Arfs are activated by a family of guanine nucleotide exchange factors (GEFs) that catalyze their loading with GTP [6 and 7]. Two of these, BIG1 and BIG2, have been shown to localize to the TGN [8, 9 and 10] where they activate Arf1 and Arf3 [8 and 11]. The catalytic activity of BIG1 and BIG2 is inhibited by the fungal toxin Brefeldin A (BFA), which causes dissociation of AP-1 and GGAs from the TGN and extensive tubulation of TGN membranes. Although a third Arf GEF, GBF1, is also sensitive to BFA, simultaneous knockdown of both BIG1 and BIG2 phenocopies the effects of BFA on the TGN [12], suggesting that they act together to promote Arf activation at this site.
Adaptor protein assembly onto endosomal membranes is also sensitive to BFA, but the roles of BIG1 and BIG2 at the early endosome is less clear. Nakayama and colleagues reported that BIG2 is present at the TGN and on perinuclear Rab11-positive recycling endosomes, but not on early, peripheral, EEA1-positive endosomes [11]. Subsequent studies reported that knockdown of BIG1 had no effect on the Rab11-positive recycling endosomes compared to BIG2 knockdown which caused tubulation of this compartment [12]. However, whether the association of adaptor proteins with early endosomes requires BFA sensitive GEFs remains to be defined.
Recent studies indicate that GTPases often work together in signaling cascades, and crosstalk between Rabs and Arfs has also been reported. In the ER-Golgi intermediate compartment (ERGIC), Rab1 activity is required to promote recruitment of Arf GEF GBF1, which then activates Arf1 to drive recruitment of the COPI coat [13]. Of particular relevance to this study, recruitment of BIG1 and BIG2 to the TGN was recently reported to be mediated by the Arf-like protein Arl1 [14]. Whether Arl1 also recruits BIG1/2 to endosomes is not known.
Here we report that Arl1 is present on the Rab4 subdomain of early endosomes, where it is necessary for the recruitment of the BFA-sensitive Arf GEFs BIG1 and BIG2. The BIGs in turn activate Arf1 and Arf3 to promote assembly of the adaptor protein complexes AP-1 and AP-3 and the monomeric adaptor protein GGA-3 onto dynamic Rab4-enriched tubular endosomal domains. Depletion of Arl1 results in a complete loss of BIGs, Arfs and adaptor proteins, but not Rab4, while depletion of Rab4 prevents recruitment of Arl1 and all downstream events. Taken together, these data suggest the existence of a signaling cascade originating with Rab4 and mediated by Arl1 that drives the assembly of carrier vesicles emanating from the early endosomal compartment.
Results
Rab4 marks a tubular endosomal domain that contains multiple adaptor proteins
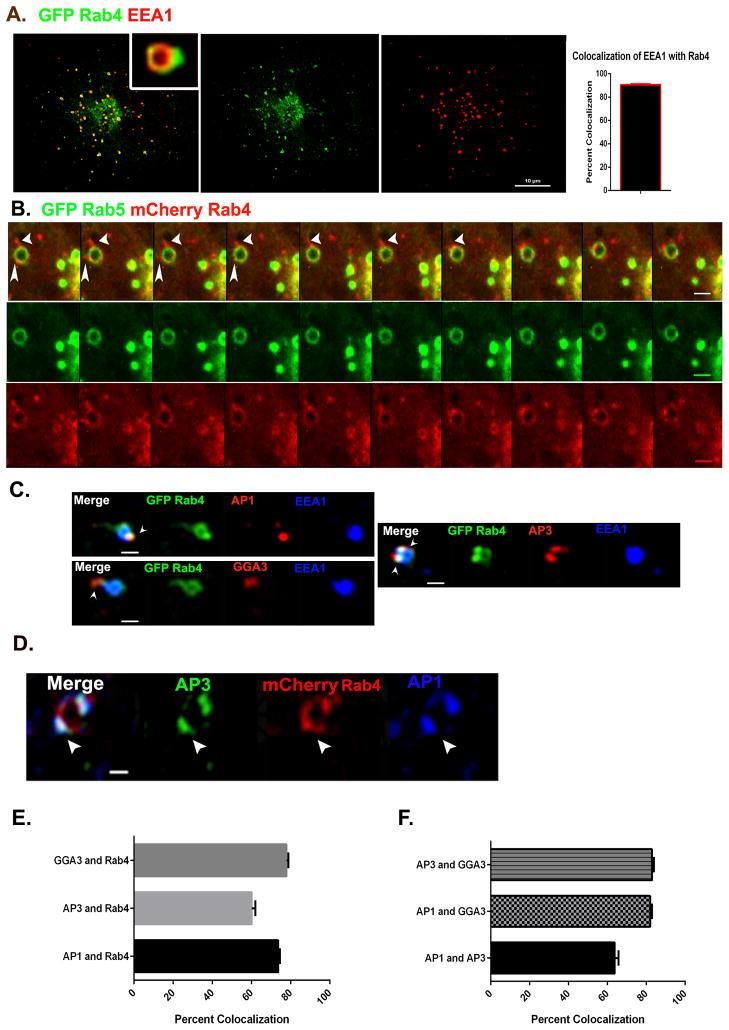
It has been known for many years that Rab4 and Rab5 occupy distinct domains on early endosomal membranes such that Rab5 marks the cisternal domain, while Rab4 concentrates in patches that undergo fission from the cisternae [15 and 16]. We confirmed that this is the case in Cos7 cells using the endogenous early endosomal marker EEA1 and GFP-Rab4a (Figure 1A). Colocalization analysis revealed that more than 90% of the endogenous EEA1-labeled endosomes colocalized with GFP-Rab4a (Figure 1A). Live cell imaging of Cos7 cells co- expressing GFP-Rab5 and mCherry-Rab4a demonstrated that the Rab4 domain is highly dynamic, giving rise to tubular extensions that often separate from the endosome before fragmenting into smaller structures (Figure 1B; Movie S1).
Figure 1. AP-1, AP-3 and GGA-3 adaptor proteins assemble on the Rab4 subdomain of early endosomes.
(A) Cos7 cells expressing GFP-Rab4a (green) were fixed and stained for endogenous EEA1. Note that Rab4 marks a distinct domain on the EEA1 positive endosomes. Scale bar = 10μm. Bar graph depicts the percentage overlap between EEA1 and Rab4. N=85 endosomes. (B) Time lapse series from Movie S1 showing emergence of Rab4 positive buds (red) from Rab5 positive (green) endosomes. Scale bar = 1μm. (C) Cos7 cells expressing GFP-Rab4a were fixed and stained for endogenous EEA1 (blue) and adaptor proteins (AP-1, AP-3 and GGA-3-red). Arrowheads indicate Rab4 positive buds lacking EEA1 but enriched in adaptor proteins. (D) mCherry-Rab4a and endogenous adaptor proteins AP-3 (green) and AP-1 (blue) colocalize on endosomal buds. Scale bar = 1μm. (E) Quantification of pairwise colocalization of GGA-3/Rab4, AP-3/Rab4 and AP-1/Rab4 based on Pearson’s coefficients. N=157 for GGA-3 and Rab4, 123 for AP-3 and Rab4 and 213 for AP-1 and Rab4 endosomes respectively. (F) Quantification of pairwise colocalization of AP-3/GGA-3, AP-1/GGA-3 and AP-1/AP-3 based on Pearson’s coefficients. N=73 for AP-1 v/s AP-3, 139 for GGA-3 v/s AP-3 and 93 for GGA-3 v/s AP-1 endosomes respectively. Error bars represent Mean±SEM. See also Movie S1.
Immunoelectron microscopy studies have reported the presence of clathrin, AP-1 and AP-3 adaptor protein complexes on tubular structures associated with early endosomes [17 and 18], although the endosomal domain where these adaptor proteins assemble has remained unclear. As shown in Figure 1C and E, we found that endogenous AP-1, AP-3 and GGA-3 all concentrated within the Rab4 subdomain of early endosomes and not on the cisternal subdomain (as identified by the presence of EEA1) and also observed a high degree of colocalization between adaptor proteins and Rab4-positive buds.
Because multiple Rab4 positive buds/tubules often emanated from a single endosome, we sought to resolve if different adaptor proteins localize to the same bud or on distinct buds on the same endosome. For this purpose cells expressing mCherry-Rab4a were fixed and immunostained for pairwise combinations of AP-1 and AP-3, AP-1 and GGA-3 or AP-3 and GGA-3. Quantitative analysis of confocal images revealed that approximately 80% of the buds that contained GGA-3 also contained AP-1 and/or AP-3, while AP-1 and AP-3 colocalized with each other on roughly 65% of the Rab4 positive buds (Figure 1D and F). These observations indicate that Rab4 marks a common site for the assembly of clathrin/adaptor protein complexes on early endosomal membranes.
Rab4 is required for adaptor protein assembly at early endosome exit sites
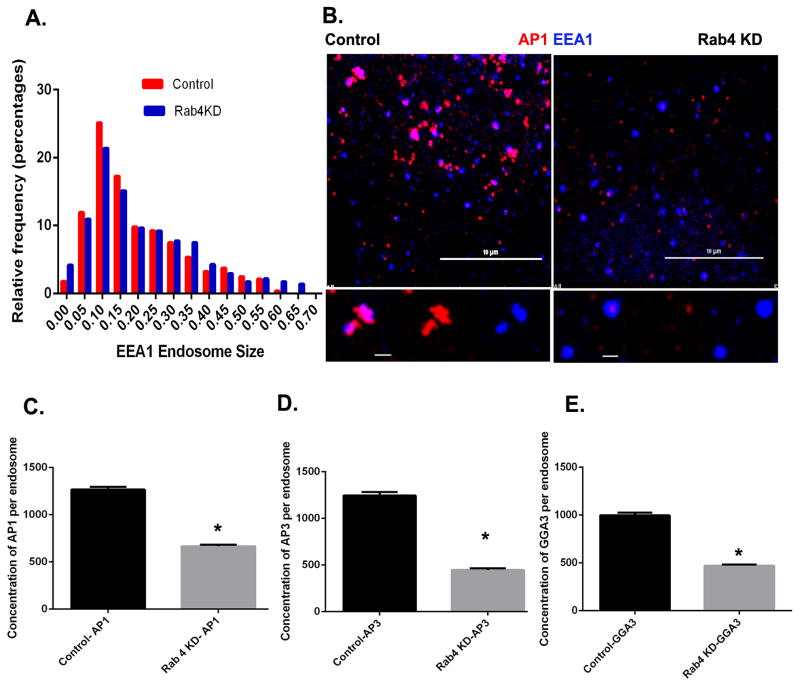
The selective assembly of three different adaptor protein complexes onto the Rab4 subdomain suggested that Rab4 nucleates formation of a sorting platform from which multiple classes of vesicular carriers emerge. To test this hypothesis we depleted cells of both Rab4a and Rab4b using two different pairs of shRNA-sequences (Figure S1A and B). Although we only show results from one pair of shRNAs, identical results were obtained with both sets of knockdowns. We then measured the association of AP-1, AP-3 and GGA-3 with the early endosomal compartment, as defined by the presence of endogenous EEA1. Knockdown of Rab4a/b did not result in a detectable change in size of early endosomes (Figure 2A), however association of all three adaptor proteins with EEA1-positive endosomes was siginificantly reduced; by 47% for AP-1, 64% for AP-3 and 52% for GGA-3 respectively (Figure 2B–E). We did observe residual adaptor protein puncta in Rab4a/b-depleted cells that were smaller in size, but were not associated with EEA1-labeled compartments. We interpret these to be recycling or late endosomes. Importantly, Rab4a/b knockdown did not affect adaptor protein assembly (AP-1, AP-3 and GGA-3) onto the TGN (Figure S1C). Together these data suggest that Rab4 is essential for the recruitment of adaptor proteins to early endosomal membranes.
Figure 2. Rab4a/b depletion affects adaptor protein localization to early endosomes.
(A) Endosome size was measured based on endogenous EEA1 staining in control and Rab4a/b depleted Cos7 cells. N for control = 564 and Rab4a/b KD = 1150 endosome respectively. There was no significant change in endosome size. (B) Control and Rab4a/b depleted Cos7 cells were fixed and stained for endogenous EEA1 (blue) and AP-1 (red). Scale bar on upper panel = 10μm and on lower panel = 1μm. (C–E) Quantification of endogenous AP-1 (C), AP-3 (D) and GGA-3 (E) fluorescence on early endosomes in control and Rab4a/b depleted cells. In (C), control, N=1510 and Rab4a/b KD, N=1438 endosomes respectively. In (D), control, N=805 and Rab4a/b KD, N=1068 endosomes respectively. In (E), control, N=861 and Rab4a/b KD, N=985 endosomes respectively. Error bars represent mean±SEM. ‘*’ indicates significantly different at p < 0.0001. Mann Whitney’s test was used to test for significance. See also Figure S1
Adaptor protein assembly on the Rab4 subdomain requires class I but not class II Arfs
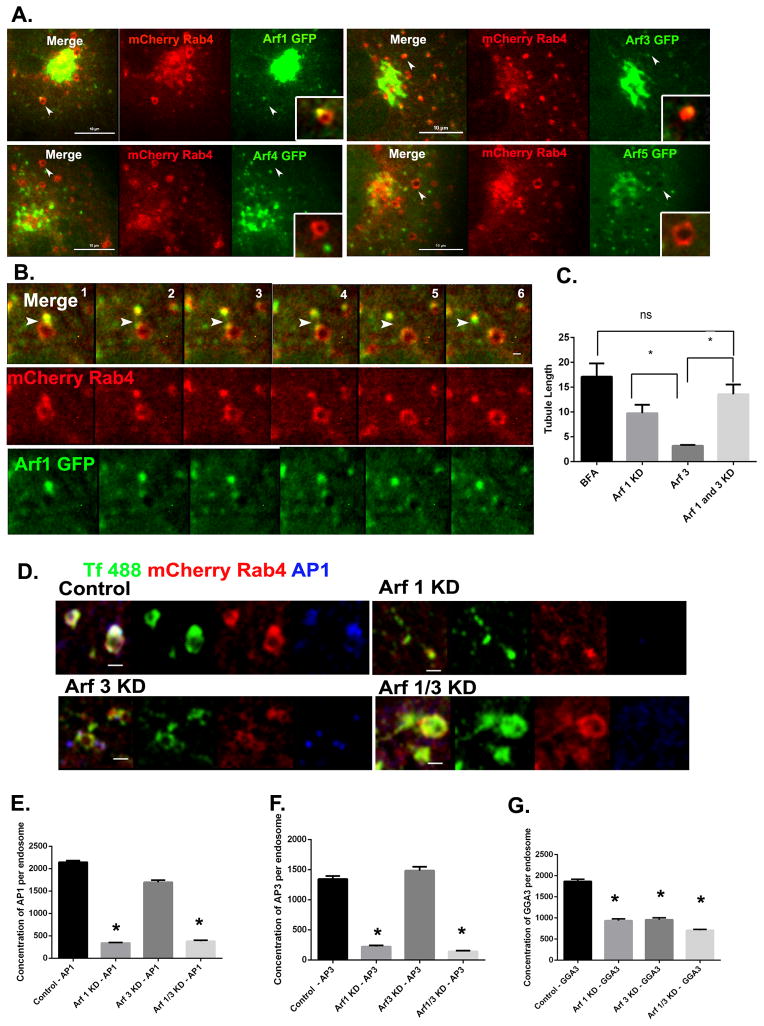
It is well established that recruitment of AP-1 and GGA adaptors to the TGN requires their interaction with one or more Arf family GTPases [5 and 6]. However, there are six mammalian Arfs (five in humans) and their respective roles in specific sorting events remains a topic of active investigation. To determine which Arfs are present on the endosomal Rab4 subdomain, we first imaged the recruitment of Arf1, Arf3, Arf4 and Arf5 to Rab4-enriched buds in live cells. As shown in Figure 3A, Arf1 was readily detectable on endosomal membranes where it colocalized strongly with Rab4-positive buds. Arf3 was also present in the Rab4 buds but at lower levels, despite the fact that it was abundant on larger perinuclear membranes (presumably the Golgi/TGN, Figure 3A). In contrast, neither Arf4 nor Arf5 was detectable on endosomal membranes (Figure 3A). These results are consistent with previous studies by Chun et al, who reported the presence of Arf4 and 5 at the ER-Golgi intermediate compartments [19]. Additionally, colocalization studies for Arf1, Rab4 and EEA1 revealed that Arf1 colocalized with the Rab4-positive buds and not with the Rab5/EEA1-positive subdomain. These Arf1/Rab4-positive buds were also positive for adaptor proteins (Figure S2D). Live cell imaging studies revealed that the intensity of Arf1 fluorescence on Rab4-positive buds appeared to increase as they matured over time (Figure 3B; Movie S2). Interestingly, both Arf1 and Rab4 remained associated with the severed buds that detached from endosomes.
Figure 3. Class I Arfs are required for adaptor protein recruitment to early endosomes.
(A) Still images from live cell videos of Cos7 cells expressing mCherry-Rab4a and Arf1/3/4/5-GFP respectively. Arrowheads indicate presence or absence of Arf (green) proteins on the Rab4a (red) bud. (B) Time lapse series from Movie S2 showing the emergence of a Rab4a-positive bud (red) containing Arf1 (green) as indicated by arrowheads. Scale bar = 1μm. (C) Quantification of endosomal tubule length in Cos7 cells depleted of Arf1/Arf3 singly or in pairs (see also Figure S2A–C) along with BFA treated cells as positive control. Student’s ‘t’ test was used to test for significance (D) Cos7 cells stably expressing mCherry-Rab4a were depleted of Arf1 or Arf3 singly or as a pair, loaded with transferrin (green), then fixed and stained for endogenous AP-1 (blue). Scale bar = 1μm. (E–G) Quantification of AP-1 (E) AP-3 (F) and GGA-3 (G) fluorescence on Rab4-positive buds. In (E), Control, N=649, Arf1 KD, N=982, Arf3 KD, N =518, Arf1/3 KD, N= 316 endosomes respectively. In (F), N for control = 326, Arf1 KD = 676 Arf3 KD = 330, Arf1/3 KD = 685 endosomes respectively. In (G), Control, N=356, Arf1 KD, N=198, Arf3 KD, N=311, Arf1/3KD, N=408 endosomes respectively. Unless specified, Mann Whitney’s test was used to test for significance. Error bars represent mean±SEM. ‘*’ indicates significantly different with respect to control at p < 0.0001. See also Figure S2A–D and Movie S2.
To determine if Arf1 and/or Arf3 are required for recruitment of adaptor proteins onto endosomal membranes, cells stably expressing mCherry-Rab4a were depleted of each Arf individually and together using siRNAs (Figure S2A–C). Cells were then loaded with Alexa488 labeled transferrin for 5 minutes to mark the early endosomal compartment, fixed and stained for AP-1, AP-3 and GGA-3, and imaged by confocal microscopy. Interestingly, knockdown of Arf1 resulted in formation of long tubules emanating from the Rab4 subdomain (Figure 3C). In contrast, knockdown of Arf3 did not induce endosomal tubulation, although shorter, less robust tubules were occasionally observed. We did observe a significant increase in tubule length when both Arf1 and 3 were knocked down, which was comparable to when cells were treated with BFA (Figure 3C). Arf1 knockdown resulted in an almost complete loss of both AP-1 and AP-3 from the endosomal membrane (Figure 3D, E and F). More importantly, Arf3 depletion did not significantly impair recruitment of AP-1 or AP-3 to endosomal membranes (Figure 3E and F). However, Arf3 depletion did reduce the level of GGA-3 on endosomal membranes by 50%, which was equivalent to the reduction observed upon knockdown of Arf1 (Figure 3G). Knockdown of both Arf1 and Arf3 had a small but statistically significant additive effect on GGA-3 recruitment (Figure 3G), suggesting that the two Arfs have overlapping but not completely redundant functions in this process.
Adaptor protein recruitment to the Rab4 subdomain is sensitive to BFA
Previous studies have revealed that the Rab4 domain of early endosomes is sensitive to BFA [15]. In this context, treatment with BFA results in tubulation of the Rab4 domain without affecting the cisternal Rab5 domain of the endosome [15]. However, recent studies have suggested that none of the BFA-sensitive Arf GEFs associate with early, EEA1-positive endosomes, although BIG2 was reported to associate with perinuclear recycling endosomes [11].
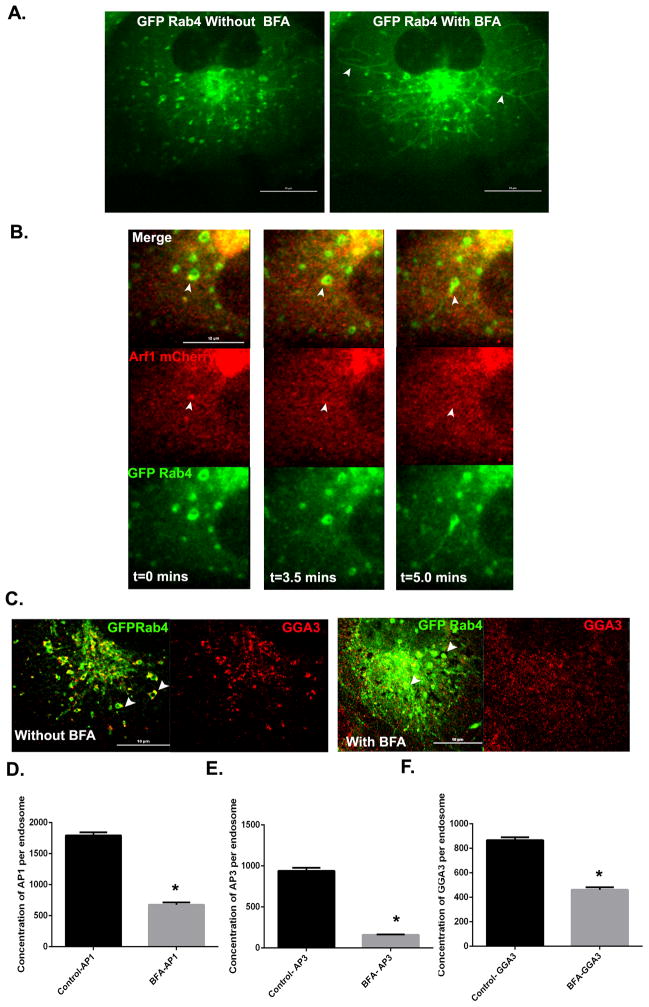
To confirm the BFA sensitivity of early endosomes, cells expressing GFP-Rab4a were incubated with 5μg/ml of BFA for 10 minutes and imaged live by confocal microscopy. Within minutes of BFA addition, tubules began to emerge from the Rab4 enriched endosomal buds, many of which extended for long distances across the cell (Figure 4A). These tubules remained Rab4 positive, indicating that association of Rab4 with tubule membranes is not intrinsically BFA sensitive. Extension of tubules was temporally preceded by dissociation of Arf1 (Figure 4B) from endosomal membranes (Movie S3 and S4). As predicted, BFA treatment also resulted in the loss of AP-1, AP-3 and GGA-3 from the endosomal membranes (Figure 4C–F). Taken together, these findings implicate a BFA sensitive Arf GEF in the recruitment of adaptor proteins to the early endosome.
Figure 4. Effect of BFA on the localization of class I Arfs and adaptor proteins at early endosomes.
(A) Still images from live cells expressing GFP-Rab4a before and after 10 minutes of BFA treatment. Arrowheads depict endosomal tubules formed after BFA treatment. Scale bar = 10μm. (B) Time lapse series from Movie S4 of GFP-Rab4a-positive endosomes (green) that lose Arf1-mCherry (red) followed by endosome tubulation (arrowheads) after BFA treatment. Scale bar = 10μm. (C) Cos7 cells expressing mCherry-Rab4a were treated with BFA or vehicle (DMSO) for 10 minutes, fixed and stained for endogenous GGA-3 (red). Arrowheads indicate presence or absence of GGA-3 in control or BFA treated cells respectively. Scale bar = 10μm. (D–F) Quantification of endogenous AP-1 (D), AP-3 (E) and GGA-3 (F) fluorescence on Rab4-positive buds in control and BFA treated cells. In (D), Control, N=252 and Treated, N=101 endosomes. In (E), Control, N=640 and Treated, N=277 endosomes. In (F), Control, N=476 and Treated, N=205 endosomes. Mann Whitney’s test was used to test for significance. Error bars represent mean±SEM. ‘*’ indicates significantly different at p < 0.0001. See also Movie S3 and S4.
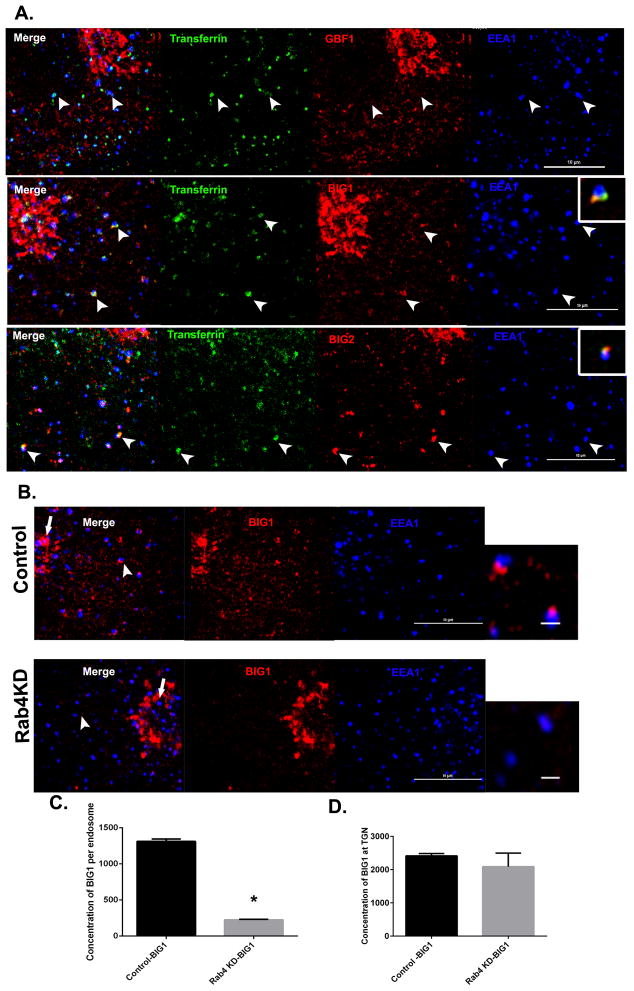
Rab4 is essential for recruitment of BIG1/2 to early endosomes
To identify the Arf GEFs associated with early endosomes, we loaded cells with transferrin for 5 minutes to mark the early endosomal compartment and stained for each of the three endogenous BFA-sensitive GEFs, GBF1, BIG1, BIG2 and for EEA1. As shown in Figure 5A, GBF1 localized to the Golgi and to dispersed puncta that were not associated with the transferrin containing compartment (presumably ERGIC). In contrast, both BIG1 and BIG2 were found to concentrate on the transferrin containing domain of the early endosome (Figure 5A, second and third panel). Exogenously expressed HA-tagged BIG1 and BIG2 also localized to the Rab4-positive buds (Figure S2E). Furthermore, knockdown of Rab4a/b resulted in 80% loss of endogenous BIG1 from endosomal membranes without affecting the TGN pool of BIG1 (Figure 5B–D). Thus we conclude that Rab4 is essential for the recruitment of BIG1 and BIG2 to the early endosome.
Figure 5. The Arf GEFs BIG1 and BIG2 are present at early endosomes.
(A) Cos7 cells were loaded with transferrin (green) for 5 minutes, fixed and stained for endogenous EEA1 (blue) and the endogenous Arf GEFs GBF1 (top panel, red), BIG1 (middle panel, red) or BIG2 (bottom panel, red). Arrowheads depict EEA1-positive endosomes containing transferrin, which are negative for GBF1 and positive for BIG1 and BIG2. Scale bar = 10 μm. (B) Cos7 cells were depleted of Rab4a/b, fixed and stained for BIG1 (red) and EEA1 (blue). Arrowhead depicts EEA1-positive endosomes enlarged at right. Note the absence of BIG1 in Rab4-depleted cells. Arrows represent the TGN pool of BIG1 that is unaffected by Rab4a/b depletion. Scale bar = 10μm on cells and 1μm on endosomes. (C) Quantification of BIG1 fluorescence at the EEA1-positive endosomes in control and Rab4a/b depleted cells. In Control, N=795, and Rab4a/b KD, N=691 endosomes respectively. Mann Whitney’s test was used to test for significance. (D) Quantification of BIG1 association with the TGN. N=4 cells in control and Rab4a/b KD cells respectively. Student’s ‘t’ test was used to test for significance. Error bars represent mean±SEM. ‘*’ indicates significantly different with respect to control at p < 0.0001. See also Figure S2E.
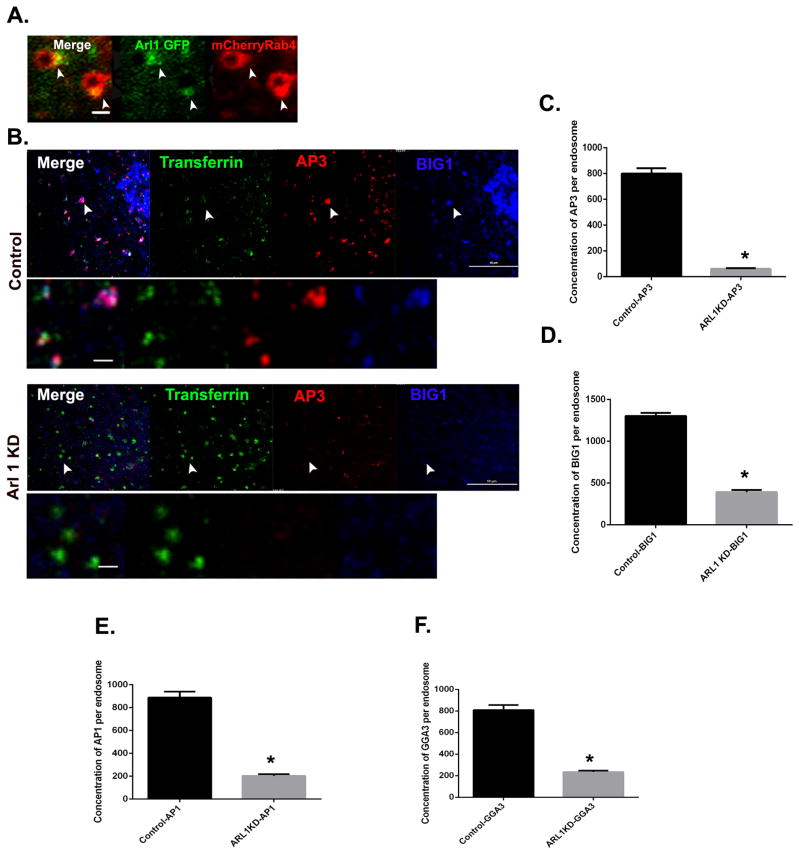
Arl1 mediates recruitment of BIG1 and BIG2 to early endosomes
Recent evidence has suggested that the Arf-like protein Arl1 facilitates recruitment of both BIG1 and BIG2 to the TGN through a direct interaction with the N-terminus of the BIGs [14]. As shown in Figure 6A, we found that Arl1 also associates with early endosomes, where it concentrates in the Rab4-positive buds. To determine if Arl1 is required for recruitment of BIGs to early endosomes, cells were depleted of Arl1 using two different shRNA sequences (Figure S3A and B). Control or Arl1-depleted cells were then loaded with Alexa488 transferrin for 5 minutes to mark early endosomes, fixed, and stained for endogenous BIG1 and AP-3. In control cells, BIG1 and AP-3 colocalized perfectly at the transferrin containing endosomes (Figure 6B, upper panel, arrowheads indicate endosomes). In contrast, recruitment of both BIG1 and AP-3 to the transferrin containing endosomes was greatly reduced in Arl1-depleted cells (Figure 6B, lower panel). We quantified the loss and found that there was a 70 % decrease of BIG1 (Figure 6D) and 92 % decrease of AP-3 (Figure 6C) from early endosomes after Arl1 knockdown. Parallel studies revealed that recruitment of AP-1 (73% decrease) and GGA-3 (71 % decrease) was similarly impaired by Arl1 knockdown (Figure 6E and F). We therefore conclude that Arl1 is required for the recruitment of BIG1 and adaptor proteins to the early endosomes.
Figure 6. Arl1 links Rab4 to the Arf GEFS BIG1/2 at early endosomes.
(A) Arl1 associates with the Rab4 subdomain. The image shown is from Cos7 cells expressing mCherry-Rab4a and Arl1-GFP. Arrowhead depicts Rab4-enriched buds (red) positive for Arl1 (green). Scale bar = 1μm. (B) Arl1 knockdown (see also Figure S3A and B) causes loss of both BIG1 and AP-3 from early endosomes. Control Cos7 cells or Arl1-depleted cells were loaded with transferrin (green), fixed and stained for endogenous BIG1 (blue) and AP-3 (red). Arrowheads depict transferrin containing endosomes that are positive for AP-3 and BIG1 in control but negative in Arl1-depleted cells respectively. Scale bar on cell = 10μm and 1μm on enlarged endosome images. (C– F) Quantification of endogenous AP-3 (C), BIG1 (D), AP-1 (E) and GGA-3 (F) fluorescence on EEA1/transferrin containing endosomes. In (C) Control, N = 252 and Arl1 KD, N=154 endosomes. In (D), Control, N=324 and Arl1 KD, N=116 endosomes. In (E), Control, N=207 and Arl1 KD, N=214 endosomes. In (F), Control, N=164 and Arl1 KD, N=195 endosomes. Mann Whitney’s test was used to test for significance. Error bars represent mean±SEM. ‘*’ indicates significantly different at p < 0.0001. See also Figure S3A–B.
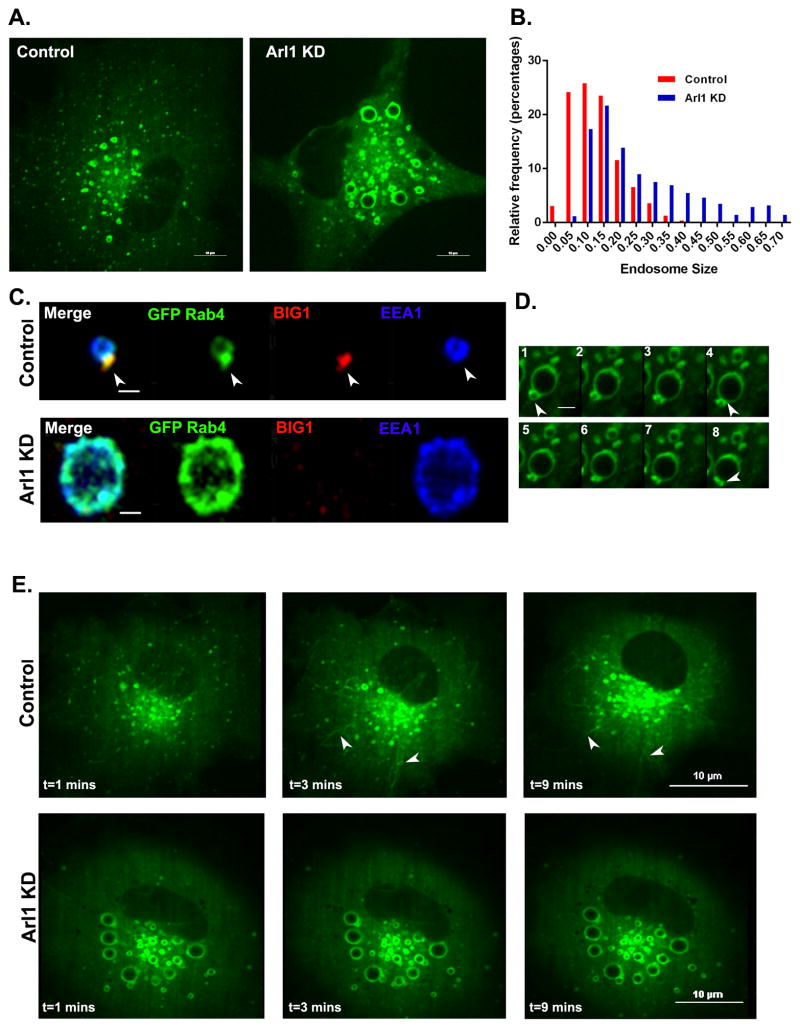
In contrast, Arl1 depletion did not impair recruitment of Rab4 to endosomal membranes (Figure 7A), suggesting that Arl1 acts downstream of Rab4. In the Arl1 depleted cells, the Rab4-positive endosomes remained positive for Rab5 (Figure S3C). We also observed a dramatic increase in endosome size, further suggesting that exit of cargo from sorting endosomes was significantly impaired. A similar increase in the size of EEA1 positive endosomes was also seen in the absence of exogenous GFP-Rab4a (Figure 7B), ruling out the possibility that this could be an artifact of Rab4 overexpression. However, Arl1 knockdown did randomize the distribution of Rab4 on the endosomal membrane, effectively preventing formation of the characteristic Rab4 subdomain containing BIG1 and adaptor proteins (Figure 7C). Live imaging of cells expressing GFP-Rab4a indicated that endosome-endosome fusion continued normally, but that Rab4 positive tubules did not extend from endosomal membranes after depletion of Arl1 (Figure 7D). Furthermore, treatment of Arl1-depleted cells with BFA did not result in endosomal tubulation, suggesting that one or more Arl1 effectors are necessary for the formation of BFA-induced tubules (Figure 7E; Movie S5 and S6). Live cell imaging also revealed the absence of Arf1 on Rab4-positive endosomes in Arl1 depleted cells (Figure S3D). Taken together, these observations indicate that Arl1 acts downstream of Rab4 to promote assembly of the early endosomal sorting machinery.
Figure 7. Arl1 depletion causes endosome enlargement and randomizes the distribution of Rab4 on the endosomal membrane.
(A) Cos7 cells stably expressing GFP-Rab4a were either mock-depleted or depleted of Arl1 (Arl1 KD). Scale bar =10μm. (B) Size distribution of EEA1-positive endosomes in control (red) and Arl1-depeleted cells (blue). Endosome size was measured based on endogenous EEA1 staining in control and Arl1-depleted cells. Control, N=778 and in Arl1 KD, N=364 endosomes respectively. There was a significant increase in endosome size at p < 0.0001. (C) Arl1 depletion randomizes the distribution of Rab4 on endosomal membranes. Cos7 cells stably expressing GFP-Rab4a and depleted of Arl1 (Arl1 KD) or not (control) were fixed and stained for endogenous BIG1 (red) and EEA1 (blue). Arrowheads indicate the presence of a Rab4-enriched bud containing BIG1 in control cells. Note the lack of Rab4 enriched foci in the Arl1 depleted cells. Scale bar =1μm. (D) Frames from a time lapse series of a GFP-Rab4a-positive endosome from Arl1 depleted cell showing that endosome fusion still occurs (indicated by arrowheads). (E) BFA treatment induces endosomal tubulation in control (Movie S5), but not Arl1-depleted cells (Movie S6). Cells stably expressing GFP-Rab4a were treated with BFA for 10 minutes. Scale Bar =10μm. See also Figure S3C–D, Movie S5 and MovieS6.
Discussion
It is well established that early endosomes are divided into vacuolar and tubular subdomains. Ultrastructural studies have demonstrated that the tubular subdomains are similar to the TGN in the sense that endosomal tubules give rise to multiple classes of carrier vesicles targeted to different destinations. For this reason these endosomal tubules are sometimes referred to as the Tubular Endosomal Network (TEN) [20].
Here we show that Rab4 marks one such tubular subdomain of the early endosome and orchestrates a GTPase cascade, in which Rab4 is essential for the recruitment of the Arf-like protein Arl1, which recruits the Brefeldin-sensitive Arf GEFs BIG1 and BIG2 to the Rab4-positive TEN. These GEFs in turn locally activate the class I Arfs, Arf1 and Arf3, which promote assembly of both tetrameric (AP-1 and AP-3) and monomeric (GGA-3) adaptors onto the tubular membranes, where they capture their respective cargos and initiate carrier vesicle formation. In the absence of Rab4, the TEN does not form, and neither Arl1 nor any of the downstream sorting machinery assembles onto the endosomal membrane. Thus Rab4 acts to nucleate assembly of multiple classes of carrier vesicles at the early endosome.
Non-redundancy of class I Arfs in adaptor protein recruitment
A particularly interesting result of this study is that Arf1 and Arf3, which are 97% identical at the amino acid level, appear to have overlapping but distinct functions in adaptor protein recruitment. Although both are present on the Rab4 subdomain, we found that knockdown of Arf3 has little effect on recruitment of either AP-1 or AP-3. This is in agreement with a recent report by Melancon and coworkers, who noted that Arf3 depletion did not displace AP-1 from the TGN [21]. Conversely, we show here that knockdown of Arf1 essentially abrogates recruitment of the tetrameric adaptors, resulting in the formation of long, Rab4-positive tubules similar to those induced by BFA. This finding in Cos7 cells contrasts with observations by Volpicelli-Daley et al, showing that endosomal tubulation required simultaneous knockdown of both Arf1 and Arf3 in Hela cells [22]. These differences may reflect the relative levels of endogenous Arf1 and Arf3 expression in different cell types. However, it should be noted that we did see an inhibitory effect of Arf3 knockdown on recruitment of GGA-3 to the TEN, suggesting that Arf3 functions selectively in GGA-dependent sorting events. Interestingly, knockdown of either Arf1 or Arf3 resulted in a 50% reduction in GGA-3 recruitment to early endosomes, and double knockdown did have a modestly additive effect. This suggests that the two Arfs may act in series, or that they act on distinct steps in the recruiting process. The remaining 50% of GGA-3 association may be mediated by Arl1, which has been shown to associate with the N-terminal VHS domain of GGAs, while Arfs associate with the more C-terminal GAT domain [23 and 24].
Arf activation at the TEN
The fungal toxin BFA inhibits the catalytic activity of a subset of Arf GEFs by stabilizing a conformational intermediate in the nucleotide exchange reaction [7 and 25]. Although it is well established that Rab4 subdomains of the early endosome are sensitive to BFA [15], the nature of the GEFs that activate Arfs at this site has remained elusive. Here we show that two of the three BFA-sensitive Arf GEFs, BIG1 and BIG2, are present on the Rab4 subdomain of Rab5/EEA1-positive sorting endosomes.
Previous studies have shown that BIG1 and BIG2 are abundant on the TGN [8, 9 and 10] whereas BIG2 is present on perinuclear recycling endosomes [11, 26 and 27]. More recent functional studies reported that knockdown of either BIG2 alone or both BIG1 and BIG2 resulted in tubulation of Rab11-positive recycling endosomes, but did not affect Rab4-positive sorting endosomes [12]. In contrast, our morphological evidence not only demonstrates colocalization of both BIGs with Rab4 on sorting endosomes, but also shows that Rab4 depletion results in a complete loss of BIGs from this compartment. One possible explanation for the discrepancy between our findings and those reported by Nakayama and colleagues could be the resolution used for imaging. The fluorescence intensity of early endosomal BIG staining is lower than it is on perinuclear compartments, and typically required longer exposure times to resolve. Importantly, Rab4 knockdown did not affect association of the BIGs with the TGN, suggesting the existence of an endosome specific mechanism of recruitment.
A role for Arl1 in Arf GEF recruitment to the TEN
How does Rab4 promote assembly of the BIGs onto endosomal membranes? Recent evidence indicates that recruitment of Sec71 (the single Drosophila ortholog of the mammalian BIGs) to the TGN requires a direct interaction with the Arf-like protein Arl1 [14]. Arl1 appears to interact with an N-terminal region of Sec71 containing the DCB (Dimerization and Cyclophilin Binding) domain and part of a second domain referred to as HUS (Homology Upstream of Sec7), both of which are conserved in mammalian BIG1 and BIG2. This region is both necessary and sufficient to target Sec71 to the TGN. Moreover, knockdown of Arl1 leads to displacement of both BIGs from the TGN in mammalian cells [14]. Here we found that Arl1 is also concentrated in the Rab4 subdomain of early endosomes, where it is similarly necessary for recruitment of BIG1 and BIG2.
Interestingly, we found that knockdown of Arl1 not only inhibited the recruitment of the BIGs, Arfs and adaptor proteins to the TEN, but also randomized the distribution of Rab4 on the surface of the endosome such that it was no longer concentrated in discrete foci. Arl1 is known to interact with several large Golgi-associated proteins (golgins), which are necessary for efficient export of specific cargos from the TGN in yeast [28] and mammalian cells [29]. A subset of golgins have been shown to bind Rab proteins directly [30], and it is plausible that golgin like proteins may help cluster Rab4 at endosome exit sites. Alternatively, yeast Arl1 has been reported to form a complex with the phospholipid flippase Drs2p and the yeast GBF1 ortholog Gea2p, in which the interaction with Gea2p is necessary for Drs2p activity [31]. If a similar complex exists in metazoans (containing Arl1, one of the several metazoan Drs1 orthologs and the BIGs), it could modulate phospholipid dynamics on endosomal membranes to promote Rab4 clustering.
Rab4 as part of a signaling cascade
It is increasingly apparent that GTPases often act in regulatory cascades [32]. For example, in the endocytic pathway, Rab5 recruits the Rab7 GEF sand1/mon1 to facilitate endosome maturation [33]. Among the Arfs, Arf6, Arf1 and Arl4 have all been shown to recruit the Arf GEF ARNO/cytohesin-2 to membranes [34 and 35], where it subsequently activates Arf1 [34]. In yeast, Arf1 was recently shown to participate in recruitment of the BIG ortholog Sec7p to the TGN, where it further stimulates Arf1 activation in a positive feedback loop [36].
Crosstalk between Rabs and Arfs has also been demonstrated. In the endocytic pathway, Rab35 and Arf6 reciprocally regulate each other; Rab35 binds the Arf6 GAP ACAP2 [37 and 38] while Arf6 recruits the Rab35 GAP EPI64B [39]. Here we show that Rab4 is necessary for recruitment of Arl1, which then promotes assembly of the Arf GEFs, Arfs and adaptor proteins that drive the sorting and packaging of endosomal cargo. However, it is not yet clear how Rab4 is coupled to Arl1. While it is plausible that Rab4 recruits an Arl1-GEF to the endosomal compartment, no such GEF has yet been identified in metazoans. Mon2/Ysl2 is a BIG-like protein that associates with Arl1 and contains all of the conserved domains present in BIG1/BIG2/GBF1, but lacks nucleotide exchange activity towards Arl1 [40]. Identification of a metazoan Arl1 GEF and determining how it is linked to upstream signaling events will be an important area of future research. Finally, it remains possible that the Arl1/BIG/Arf machinery requires Rab4 to initiate assembly of the TEN, but that its assembly onto the TEN occurs independently of Rab4. Presumably this would still require local activation of an Arl1 GEF, and defining how this is linked to TEN formation will require additional investigation.
Conclusions and Future Directions
It has been proposed that an ordered series of transition from one GTPase to another not only helps to activate/deactivate the subsequent GTPase, but also helps recruit other effector proteins [41]. We report that Rab4 initiates such a cascade that recruits Arl1 to the TEN where it recruits the Arf GEFs BIG1/2, which then activate class I Arfs for adaptor protein recruitment. We further hypothesize that this cascade may also help recruit other effectors contributing to vesicle formation and fission. Future experiments will be aimed at identifying the molecular link(s) between Rab4 and Arl1, how Arl1 contributes to Rab4 clustering on the endosomal membrane, and the identification of other effector proteins that contribute to vesicle formation at the TEN.
Experimental Procedures
For a list of reagents and details on methods, refer to supplemental experimental procedures.
Confocal Microscopy and Image Analysis
Images were captured after satisfying the Nyquist criteria for sampling using a 100X, 1.49 NA TIRF objective on a Nikon C1 Plus Confocal scanner. Z Sections of 0.25–0.5 microns were taken. For every experiment, a series of test images were taken to identify exposure gains that minimized oversaturation and this gain was subsequently used. When comparing two treatment groups, the same exposure gains were employed. 12 bit images were analyzed using NIKON Elements software. A single optical section was analyzed per cell. Three to ten cells from three independent experiments were analyzed for each experiment. The N in figure legends represents the total number of endosomes analyzed from these cells. For quantitation, an image segmentation tool was used to define endosomes based on signal intensity of either fluorescent transferrin, EEA1 or Rab4. The segmented endosomes were marked as regions of interest (ROI) and the fluorescent intensities for colocalizing proteins were measured in each ROI/endosome. Fluorescence intensities were calculated as mean fluorescence intensity or concentration (Integral Fluorescence intensity divided by the endosome area in pixels, which accounts for endosomal size) on a per endosome basis. Colocalization of adaptor proteins, EEA1 or GEFs on Rab4-positive endosomes were quantified as Pearson’s coefficient on a per endosome basis using the colocalization tools in Elements.
Statistical Analysis
Graphpad Prism6 software was used for all statistical analysis. In the course of analysis we found that the amount of endosome associated adaptor protein signal varied with endosome size, such that smaller endosomes were dimmer compared to larger ones. Since the endosomal adaptor protein fluorescence intensity follows a non-parametric distribution, Mann Whitney’s test was used to test for significance between two groups. In the case of multiple comparisons, Kruskal-Wallis one way analysis of variance was used along with Dunn’s multiple comparison tests. For all other statistical analysis, student’s ‘t’ test was used to test for significance.
Supplementary Material
Highlights.
Rab4 marks an early endosomal subdomain for localized adaptor protein recruitment.
Rab4 recruits Arl1, which recruits the Arf GEFs BIG1 and BIG2 to early endosomes.
BIG1/2 locally activate Class I Arfs, promoting adaptor recruitment to endosomes.
Arf1 and Arf3 have non-redundant functions in adaptor protein recruitment.
Acknowledgments
The authors thank David Castle and Bettina Winkler for critically reading the manuscript. The authors also wish to thank Ian Macara for providing access to his spinning disc confocal microscope. This work was supported by NIH grant GM078585 to JEC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:53–66. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 2.Robinson MS. Adaptable adaptors for coated vesicles. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Bonifacino JS, Hurley JH. Retromer. Curr Opin Cell Biol. 2008;20:427–436. doi: 10.1016/j.ceb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonifacino JS. The GGA proteins: adaptors on the move. Nat Rev Mol Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- 5.D’Souza -Schorey C, Chavrier P. Arf proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 6.Donaldson JG, Jackson CL. Arf family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casanova JE. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- 8.Mansour SJ, Skaug J, Zhao XH, Giordano J, Scherer SW, Melançon P. p200 Arf-GEP1: a Golgi-localized guanine nucleotide exchange protein whose Sec7 domain is targeted by the drug brefeldin A. Proc Natl Acad Sci U S A. 1999;96:7968–7973. doi: 10.1073/pnas.96.14.7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao X, Lasell TKR, Melancon P. Localization of large ADP-ribosylation factor- guanine nucleotide exchange factors to different golgi compartments: evidence for distinct functions in protein. Mol Cell Biol. 2002;13:119–133. doi: 10.1091/mbc.01-08-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinotsuka C, Waguri S, Wakasugi M, Uchiyama Y, Nakayama K. Dominant-negative mutant of BIG2, an Arf-guanine nucleotide exchange factor, specifically affects membrane trafficking from the trans-Golgi network through inhibiting membrane association of AP-1 and GGA coat proteins. Biochem Biophysc Res Commun. 2002;294:254–260. doi: 10.1016/S0006-291X(02)00456-4. [DOI] [PubMed] [Google Scholar]
- 11.Shin H, Morinaga N, Noda M, Nakayama K. BIG2, A guanine nucleotide exchange factor for ADP-ribosylation factors: Its localization to recycling endosomes and implication in the endosome Integrity. Mol Biol Cell. 2004;15:5283–5294. doi: 10.1091/mbc.E04-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishizaki R, Shin H, Mitsuhashi H, Nakayama K. Redundant roles of BIG2 and BIG1, guanine-nucleotide exchange factors for ADP-ribosylation factors in membrane traffic between the trans -Golgi network and endosomes. Mol Biol Cell. 2008;19:2650–2660. doi: 10.1091/mbc.E07-10-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monetta P, Slavin I, Romero N, Alvarez C. Rab1b Interacts with GBF1 and modulates both Arf1 dynamics and COPI association. Mol Biol Cell. 2007;18:2400–2410. doi: 10.1091/mbc.E06-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christis C, Munro S. The small G protein Arl1 directs the trans-Golgi-specific targeting of the Arf1 exchange factors BIG1 and BIG2. J Cell Biol. 2012;196:327–335. doi: 10.1083/jcb.201107115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sönnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149:901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 17.Stoorvogel W, Oorschot V, Geuze HJ. A novel class of clathrin-coated vesicles budding from endosomes. J Cell Biol. 1996;132:21–33. doi: 10.1083/jcb.132.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peden AA, Oorschot V, Hesser BA, Austin CD, Scheller RH, Klumperman J. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J Cell Biol. 2004;164:1065–1076. doi: 10.1083/jcb.200311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chun J, Shapovalova Z, Dejgaard SY, Presley JF, Melancon P. Characterization of class I and II ADP-ribosylation factors (Arfs) in live cells: GDP-bound class II Arfs associate with the ER-Golgi intermediate compartment independently of GBF1. Mol Cell Biol. 2008;19:3488–3500. doi: 10.1091/mbc.E08-04-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- 21.Manolea F, Chun J, Chen DW, Clarke I, Summerfeldt N, Dacks JB, Melancon P. Arf3 is activated uniquely at the trans-Golgi network by brefeldin A-inhibited guanine nucleotide exchange factors. Mol Biol Cell. 2010;21:1836–1849. doi: 10.1091/mbc.E10-01-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volpicelli-Daley LA, Li Y, Zhang C, Kahn RA. Isoform-selective effects of the depletion of ADP- ribosylation factors 1–5 on membrane traffic. Mol Biol Cell. 2005;16:4495–4508. doi: 10.1091/mbc.E04-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dell’Angelica EC, Puertollano R, Mullins C, Aguilar RC, Vargas JD, Hartnell LM, Bonifacino JS. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singer-Krüger B, Lasić M, Bürger AM, Hausser A, Pipkorn R, Wang Y. Yeast and human Ysl2p/hMon2 interact with Gga adaptors and mediate their subcellular distribution. EMBO J. 2008;27:1423–1435. doi: 10.1038/emboj.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson CL. Brefeldin A acts to stabilize an abortive Arf-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- 26.Shen X, Xu KF, Fan Q, Pacheco-Rodriguez G, Moss J, Vaughan M. Association of brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2) with recycling endosomes during transferrin uptake. Proc Natl Acad Sci U S A. 2006;103:2635–2640. doi: 10.1073/pnas.0510599103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boal F, Stephens DJ. Specific functions of BIG1 and BIG2 in endomembrane organization. PLoS One. 2010;5:e9898. doi: 10.1371/journal.pone.0009898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu YW, Lee SW, Lee FJS. Arl1p is involved in transport of the GPI-anchored protein Gas1p from the late Golgi to the plasma membrane. J Cell Sci. 2006;119:3845–3855. doi: 10.1242/jcs.03148. [DOI] [PubMed] [Google Scholar]
- 29.Lieu ZZ, Lock JG, Hammond LA, La Gruta NL, Stow JL, Gleeson PA. A trans-Golgi network golgin is required for the regulated secretion of TNF in activated macrophages in vivo. Proc Natl Acad Sci U S A. 2008;105:3351–3356. doi: 10.1073/pnas.0800137105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goud B, Gleeson PA. TGN golgins, Rabs and cytoskeleton: regulating the Golgi trafficking highways. Trends Cell Biol. 2010;20:329–336. doi: 10.1016/j.tcb.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Tsai PC, Hsu JW, Liu YW, Chen KY, Lee FJS. Arl1p regulates spatial membrane organization at the trans-Golgi network through interaction with Arf-GEF Gea2p and flippase Drs2p. Proc Natl Acad Sci U S A. 2013;110:e668–677. doi: 10.1073/pnas.1221484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stalder D, Antonny B. Arf GTPase regulation through cascade mechanisms and positive feedback loops. FEBS Lett. 2013;587:2028–2035. doi: 10.1016/j.febslet.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Kinchen JM, Doukoumetzidis K, Almendinger J, Stergiou L, Tosello-Trampont A, Sifri CD, Hengartner MO, Ravichandran KS. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol. 2008;10:556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen LA, Honda A, Varnai P, Brown FD, Balla T, Donaldson JG. Active Arf6 recruits ARNO/Cytohesin GEFs to the PM by binding their PH domains. Mol Cell Biol. 2007;18:2244–2253. doi: 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofmann I, Thompson A, Sanderson CM, Munro S. The Arl4 family of small G proteins can recruit the cytohesin Arf6 exchange factors to the plasma membrane. Curr Biol. 2007;17:711–716. doi: 10.1016/j.cub.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Richardson BC, McDonold CM, Fromme JC. The Sec7 Arf-GEF is recruited to the trans-Golgi network by positive feedback. Dev Cell. 2012;22:799–810. doi: 10.1016/j.devcel.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egami Y, Fukuda M, Araki N. Rab35 regulates phagosome formation through recruitment of ACAP2 in macrophages during FcγR-mediated phagocytosis. J Cell Sci. 2011;124:3557–3567. doi: 10.1242/jcs.083881. [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi H, Fukuda M. Rab35 regulates Arf6 activity through centaurin-β2 (ACAP2) during neurite outgrowth. J Cell Sci. 2012;125:2235–2243. doi: 10.1242/jcs.098657. [DOI] [PubMed] [Google Scholar]
- 39.Chesneau L, Dambournet D, Machicoane M, Kouranti I, Fukuda M, Goud B, Echard A. An Arf6/Rab35 GTPase cascade for endocytic recycling and successful cytokinesis. Curr Biol. 2012;22:147–153. doi: 10.1016/j.cub.2011.11.058. [DOI] [PubMed] [Google Scholar]
- 40.Mahajan D, Boh BK, Zhou Y, Chen L, Cornvik TC, Hong W, Lu L. Mammalian Mon2/Ysl2 regulates endosome-to-Golgi trafficking but possesses no guanine nucleotide exchange activity toward Arl1 GTPase. Sci Rep. 2013;3:3362. doi: 10.1038/srep03362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizuno-Yamasaki E, Rivera-Molina F, Novick P. GTPase networks in membrane traffic. Annu Rev Biochem. 2012;81:637–59. doi: 10.1146/annurev-biochem-052810-093700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.