To target a specific region of a protein, monoclonal antibodies (mAbs) are often developed against peptide fragments representative of the target epitope on the intact protein. We report on a chemical strategy for targeting epitopes near phosphorylation sites; we developed a synthetic capture agent against the C-terminal epitope of Akt2 (or protein kinase B (PKB)), which contains the Ser474 residue. Akt, which collectively refers to three isoforms (Akt1, Akt2, and Akt3), is a serine/threonine protein kinase.[1, 2] Akt plays a central regulatory role in growth factor signaling and serves as a key node in phosphatidylinositol 3-kinase (PI3K) signaling. Over-expression and/or hyperactivation of Akt is associated with many cancers,[3–6] thus making Akt a drug and diagnostic target.[7, 8] Ser474 in the Akt2 protein (Ser473 in Akt1) is located in the hydrophobic motif (HM) of the C-terminal tail, and is phosphorylated by the Rictor–mTOR complex.[9] The phosphorylated HM allosterically activates Akt2 by binding to a hydrophobic groove in the N-lobe of Akt2 and enhancing the kinase activity 10-fold.[10, 11] In the PKB crystal structure, the electron density for residues 442–481 is not resolved, suggesting a disordered C terminus.[11] This epitope thus provides a challenging target, but it is also a potentially attractive drug target site. Adenosine triphosphate (ATP)-competitive inhibitors of Akt2 can cause hyperphosphorylation of the protein.[12, 13]
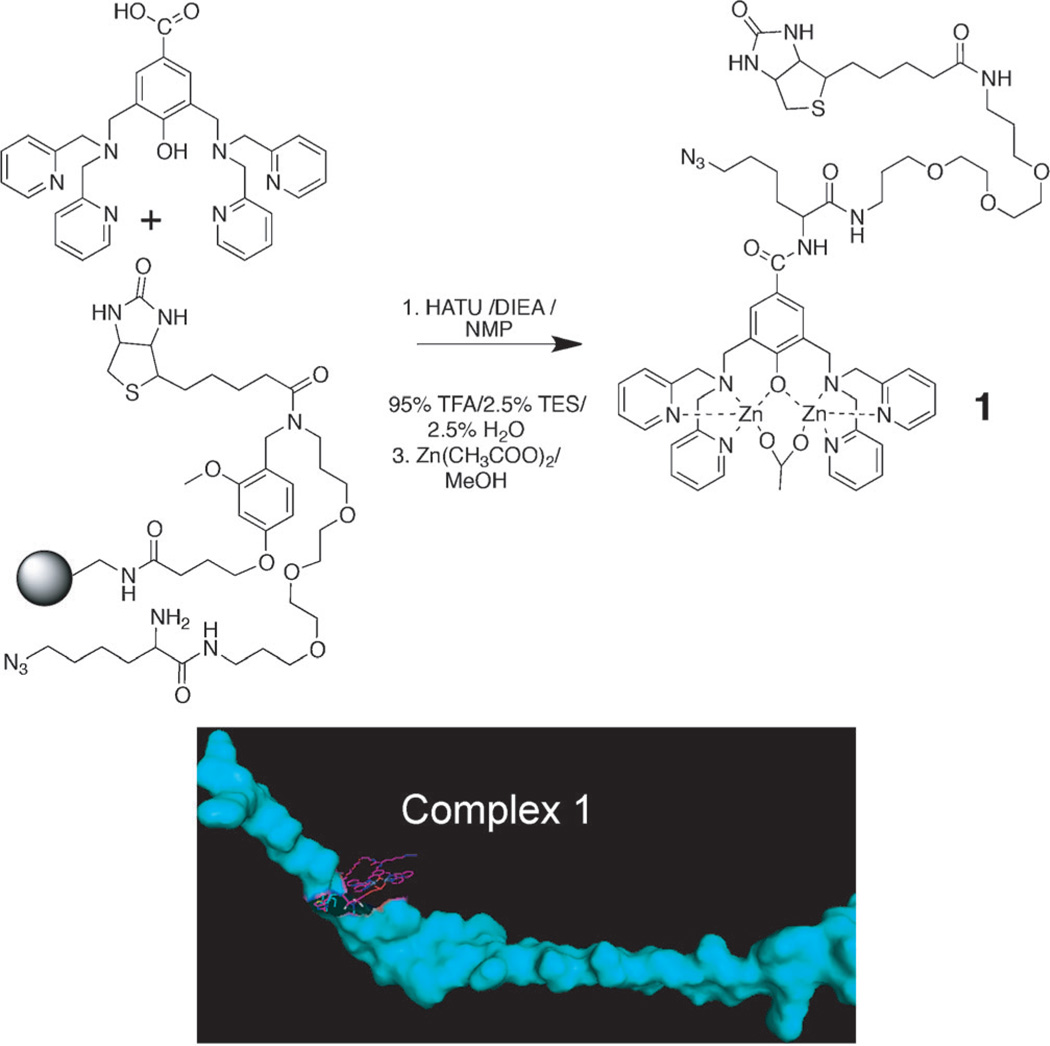
Our approach, which produces what we call protein catalyzed capture (PCC) agents,[14] starts with the synthesis of the epitope. We prepared the 32 amino acid long C-terminal polypeptide fragment of Akt2 (amino acids 450–481) that contains the phosphorylated serine 474 residue (pS474). The fragment length was chosen to represent the C terminus of Akt2 relative to the Akt1 and Akt3 isoforms, while being readily prepared by using standard peptide synthesis chemistry. To make this peptide into an anchor for screening based on in situ click chemistry, we exploited the selective binding of dinuclear zinc(II) complexes containing a dipicolylamine (DPA)-type ligand to phosphate groups[15] (Figure S1 in the Supporting Information) to chemically modify the pS474 residue so that a biotin label and an azide group were presented near (but not on) the pS474 site. This leads to the formation of complex 1 (Scheme 1). The azide group is an anchor site for an in situ click reaction,[16,17] the biotin provides an assay handle, and the 32-mer polypeptide serves as the catalyst.
Scheme 1.
The preparation of 1; a divalent zinc chelator presenting both biotin and azide moieties. Complex 1 is comprised of 1 bonded to the pSer474 of the 32 amino acid peptide fragment of Akt2. The graphic of complex 1 (bottom right; constructed using PYMOL[18] and ChemBio3D Ultra) illustrates the scale of 1 (purple) bonded to pS474 on the polypeptide (blue).
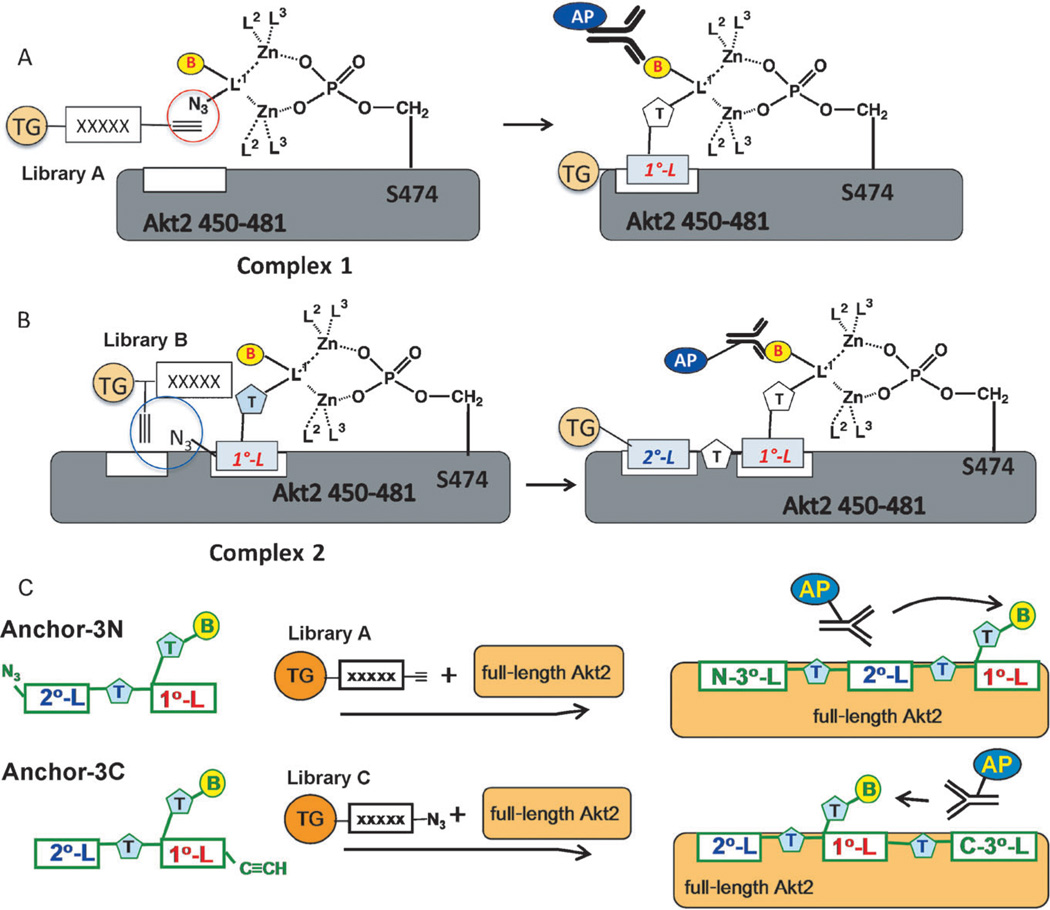
Complex 1 was subjected to an in situ click screen (Scheme 2A) against a large one-bead-one-compound (OBOC) library (library A) of acetylene-presenting hexameric peptides to identify an initial (1°) peptide ligand. See Table S1 in the Supporting Information for all OBOC libraries used. The peptides were built from d-stereoisomers of the natural amino acids to ensure protease stability in the final capture agent. The selectivity of this multistep screen was such that it produced two hit beads that sequenced to produce the same peptide (d-Pra)-wkvkl (d-Pra=d-propargylglycine). This 1° ligand, mono-L (Figure S2A in the Supporting Information), was found to have an approximately 3 µm affinity for Akt2 by single-component immunoassay and to have sufficient selectivity to immunoprecipitate Akt from OVCAR3 cancer cell lysate when immobilized on streptavidin–agarose resin.
Scheme 2.
Screening strategy for developing a PCC agent targeted near Ser474 of Akt2. A) Identification of a 1° ligand. Zinc chelator 1 couples to the Akt2 C-terminal fragment through the pS474 phosphate group to create complex 1. Complex 1 is screened against library A to yield 1° ligand candidates, from which a consensus 1° ligand (mono-L) is identified. B) Complex 2 is prepared from 1, mono-L, and the 32-mer pS474 containing polypeptide. Complex 2 is screened against library B to identify candidate 2° ligands, from which two variations of the consensus biligand (bi-L) were prepared. C) The first variation, anchor-3N, was screened against library A in the presence of full-length Akt2 to identify candidate 3° ligands, from which a consensus N-terminal triligand, N-tL, was identified. Anchor-3C was similarly utilized to identify a consensus C-terminal triligand, C-tL. (TG=tentagel; B=biotin; AP=alkaline phosphatase; T=triazole linker; no-L=no ligand; L1, L2=parts of the DPA-type ligand).
We iterated this strategy[14, 19] to identify candidate 2° ligands. Complex 2 was prepared from the zinc chelator 1, the 1° ligand sequence, and the 32-mer polypeptide (Scheme 2B and Figure S2B). Multistep screens against library B were carried out to eliminate background binders and to identify 2° ligand candidates. The motif hnGxx-d-Pra was observed across several candidate 2° ligands (See Tables S2 and S3 and Figure S3 in the Supporting Information for a listing and statistical analysis of these hit sequences). Biligand candidates were synthesized by linking the 1° and 2° peptides using copper(I)-catalyzed click chemistry, and were then tested for selectivity (Figure S4 in the Supporting Information). The best performing biligand, bi-L (Figure S2C), exhibited an approximately 1 µm affinity for Akt2, and higher selectivity than mono-L for immunopreciptating Akt from OVCAR3 cell lysate.
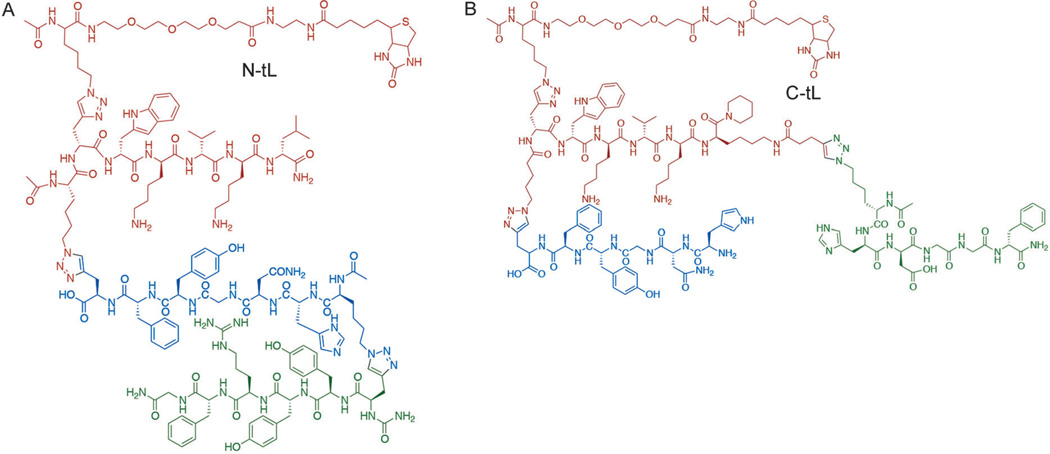
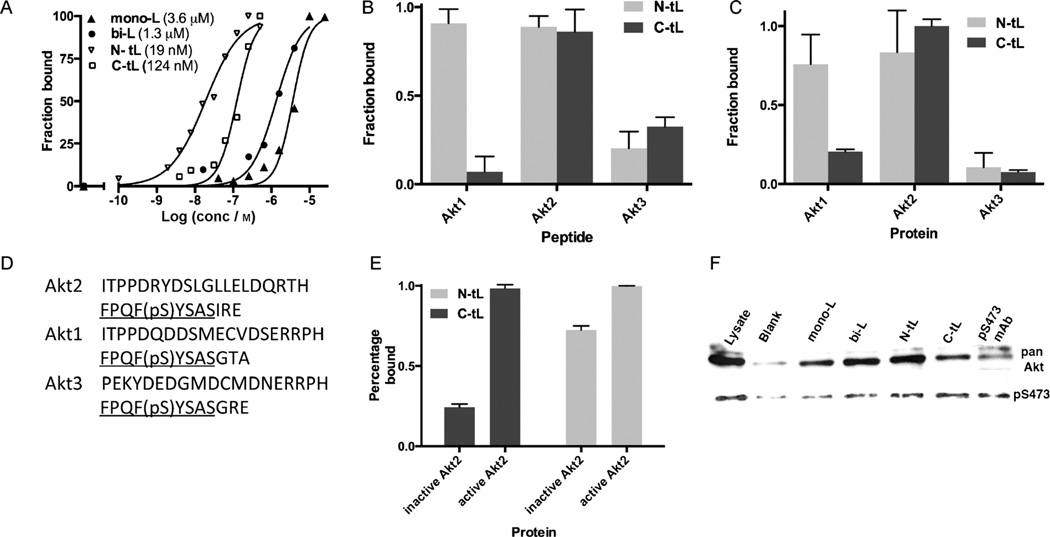
Starting from the biligand, we obtained two triligands by using in situ click screens that utilized full-length active Akt2 (pAkt2) and two biligand-derived anchors, anchor-3N (Figure S2D) and anchor-3C (Figure S2E), but no Zn chelator (Scheme 2C). An N-terminal triligand (N-tL; Figure 1A) was developed by screening for 3° ligands that click to the N terminus of anchor-3N, while a C-terminal triligand (C-tL; Figure 1B) was similarly developed by identifying 3° ligands that click to the C terminus of anchor-3C. All hit sequences and assays used to guide the selection of the two best triligands are provided in Tables S4–S6 and Figures S5 and S6 in the Supporting Information. The best 3° ligand for the C-tL (hdGGf) is somewhat similar to the 2° ligand sequence, perhaps indicating competition for the same site on the protein. The N-tL and C-tL yielded EC50 values from single-component ELISA assays of 19 and 124 nm, respectively (Figure 2A). The EC50 value for the N-tL (19 nm) compares well with the dissociation constant (KD) value (25 nm) obtained using surface plasmon resonance (Figure S7 in the Supporting Information).
Figure 1.
Molecular structures of the N-tL (A) and C-tL (B) developed against the C-terminal epitope of Akt2 near the pS474 residue. The 1° and 2° ligand branches are common to both PCC agents, and are drawn in red and blue, respectively. The poly (ethylene glycol)-linked biotin groups were included in the development process and thus do not represent interfering perturbations.
Figure 2.
A) ELISA assays in which the PCC agents are utilized either as the surface-localized capture agent (N-tL and C-tL) or as the detection agent (mono-L and bi-L). Relative affinity measurements are given as EC50 values in the key. B) The C-tL has significant selectivity for the Akt2 epitope (450–481) compared to the corresponding regions in Akt1 (449–480) and Akt3 (448–479). C) The C-tL distinguishes the full-length Akt2 from the Akt1 and Akt3 isoforms. D) The 32 amino acid long C-terminal fragment from Akt2, Akt1, and Akt3. The hydrophobic motif is underlined. pS=phosphoserine. E) The C-tL is selective for pAkt2 over inactive Akt2. F) Selectivity assays: Biotinylated ligands or biotinylated pS473 antibody are immobilized on streptavidin–agarose and treated with OVCAR3 cell lysate. The eluents are stained with antibodies against pan-Akt or pS473 Akt. The N-tL immunoprecipitates more Akt overall but the C-tL and N-tL immunoprecipitate comparable amounts of active pS473 Akt.
We tested both triligands for binding selectivity by comparing them against the C-terminal 32-mer fragments of Akt1, Akt2, and Akt3, as well as the full-length proteins (Figure 2B,C). ELISA-type assays were done in triplicate against a single concentration of the His6-tagged peptide or the His6-tagged full protein. Akt1, Akt2 and Akt3 are more than 85% identical in the kinase domain[20] and are highly homologous near residue S474 (Figure 2D). Both triligands exhibit selectivity at both the peptide and full protein level, with the C-tL exhibiting 5:1 and 10:1 selectivity for full-length Akt2 over Akt1 and Akt3, respectively. This result confirms the selectivity of the epitope targeting approach. We also tested the selectivity of the triligands for pAkt2 relative to inactive Akt2, and found a high (5:1) selectivity only for the C-tL (Figure 2E). A similar selectivity was also observed in immunoprecipitation assays. For these assays, either the biotinylated ligands or the biotinylated pS473 mAb were immobilized on streptavidin–agarose resin and treated with OVCAR3 cell lysate overnight. After stringent washing, the samples were eluted from the beads and run on a gel. When the eluted proteins (Coomassie stain in Figure S8 in the Supporting Information) were treated with a pan-Akt antibody that detects all Akt isoforms, an increased capture efficiency from mono-L to N-tL is evident.[14] The C-tL gives lower signal than the N-tL. However, when the eluent is treated with an antibody specific for phosphorylated S473 (S474 in Akt2), the C-tL shows relatively increased pAkt immunoprecipitation and hence an increased selectivity for the phosphorylated protein.
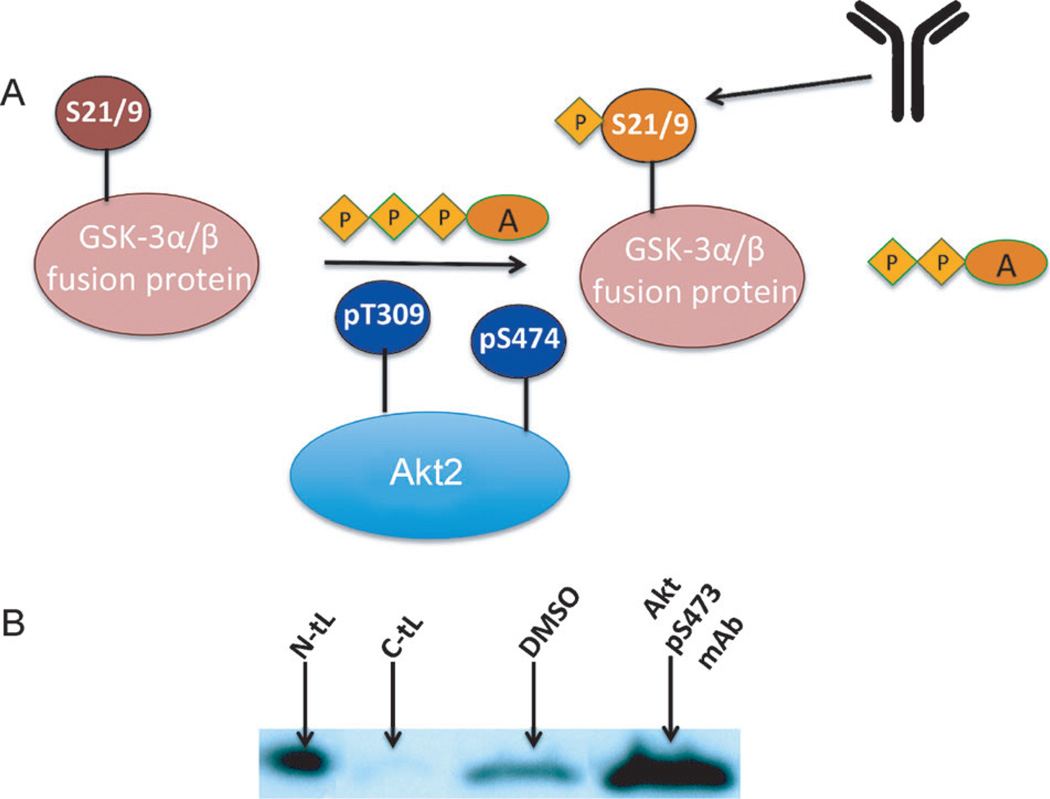
In cancer cell lines[21, 22] and tissue samples[23], the constitutively active Akt2 is phosphorylated at Ser474. Over-expression and/or activation of Akt can increase the resistance of tumors to chemo-[24] or radio-[21] therapies. The importance of Akt2 as a therapeutic target has prompted development of both ATP-competitive and allosteric inhibitors.[25] Most allosteric inhibitors bind to the interface between the membrane docking pleckstrin homology domain (PHD) and the kinase domain.[26] Given that the phosphorylated hydrophobic motif around S474 at the C terminus of Akt2 has been shown to act as an allosteric activator, we hypothesized that peptides that bind to the S474 epitope might influence the Akt2 kinase activity. We utilized a nonradioactive Akt2 kinase assay kit to estimate the influence of the ligands on the kinase activity of Akt2. For this assay, active Akt2 is combined with its substrate peptide glycogen synthetase kinase (GSK)-3α/β and ATP. To this solution was also added one of the peptide ligands or DMSO as a control. The level of phosphorylation of GSK-3α/β provides a readout for Akt2 kinase activity (Figure 3A). The N-tL was found to activate Akt2, as did the commercial pS473 antibody (Figure 3B). By contrast, the C-tL inhibits Akt2, with an EC50 of around 4 µm, as estimated from the Western blotting assays of the Akt2-induced phosphorylation of the GSK-3α/β substrate at varying C-tL concentrations (Figure S9 in the Supporting Information). Thus, the enzymatic activity of Akt2 is hypersensitive to perturbations near the C terminus. This delicately balanced structure–function relationship between S474 and Akt enzymatic activity apparently works in both directions: ATP-competitive inhibitors like A-443654 cause paradoxical hyperphosphorylation of Akt at both Thr308 and Ser473, thus leading to protein reactivation after inhibitor dissociation.[13]
Figure 3.
The influence of the PCC agents on the enzymatic activity of active Akt2. A) The activity of Akt2 directly influences the phosphorylation of the GSK-3α/β peptide (fused with glutathione S-transferase) at Ser9/Ser21. The level of phosphorylated GSK-3α/β thus provides a readout for the activation or inhibition of Akt2 by a molecular probe. B) Western blot analysis of p-GSK-3α/β levels shows that, relative to the DMSO control, both the N-tL and the commercial anti-pS473 antibody activate Akt above baseline. The C-tL inhibits Akt2 kinase activity. (A=adenosine; P=phosphate).
We report on an epitope targeting strategy that, when combined with sequential in situ click chemistry, permitted the development of a series of peptide multiligands (PCC agents) that are targeted near a key phosphorylation site of Akt2. This strategy relies on the use of a dinuclear zinc(II) complex that binds to the phosphorylated residue of interest, and presents a biotin label and an azide functionality near that site. The strategy is initiated using only a fragment representing the phosphorylated epitope of the target. This synthetic approach permits stringent chemical characterization of the chemical species employed. The strategy was used to develop an initial PCC agent monoligand, a PCC agent biligand, and two PCC agent triligands. The strongest binding triligand (the N-tL) exhibits an approximately 20 nm affinity for Akt2. Epitope and protein isoform specificity is shown for both triligands.
The C terminus of Akt2 was explored because phosphorylation of S474 leads to allosteric activation of Akt2, thus making this site a potential drug target. However, this unstructured epitope has no binding pocket, and so it is not obviously targetable by traditional small-molecule inhibitors. Indeed, the PCC agents developed against this epitope exhibit both inhibitory and activating characteristics depending upon the PCC agent structure. Additional advantages relative to the mAb include the well-defined chemical structure of the PCC agents developed, as well as their stability against proteases and physical stresses.[27] Delivery of these or similarly targeted ligands into live cells to explore their potential as therapeutic inhibitors of Akt2 is under investigation.
Supplementary Material
Footnotes
This work was supported by the Institute for Collaborative Biotechnologies through grant W911NF-09-0001 from the U.S. Army Research Office. Additional personnel or facilities support from the National Cancer Institute (5U54 CA119347: J.R.H. PI), the Grand Duchy of Luxembourg, and the Bill and Melinda Gates Foundation is acknowledged.
Supporting information (including characterization data for all the molecules used) for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201305882.
References
- 1.Vivanco I, Sawyers CL. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 2.Manning BD, Cantley LC. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellacosa A, Defeo D, Godwin AK, Bell DW, Cheng JQ, Altomare DA, Wan MH, Dubeau L, Scambia G, Masciullo V, Ferrandina G, Panici PB, Mancuso S, Neri G, Testa JR. Int. J. Cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 4.Cheng JQ, Godwin AK, Bellacosa A, Taguchi T, Franke TF, Hamilton TC, Tsichlis PN, Testa JR. Proc. Natl. Acad. Sci. USA. 1992;89:9267–9271. doi: 10.1073/pnas.89.19.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altomare DA, Tanno S, De Rienzo A, Klein-Szanto A, Skele KL, Hoffman JP, Testa JR. J. Cell. Biochem. 2003;88:470–476. doi: 10.1002/jcb.10287. [DOI] [PubMed] [Google Scholar]
- 6.Tanno S, Yanagawa N, Habiro A, Koizumi K, Nakano Y, Osanai M, Mizukami Y, Okumura T, Testa JR, Kohgo Y. Cancer Res. 2004;64:3486–3490. doi: 10.1158/0008-5472.CAN-03-1788. [DOI] [PubMed] [Google Scholar]
- 7.Lindsley CW, Barnett SF, Layton ME, Bilodeau MT. Curr. Cancer Drug Targets. 2008;8:7–18. doi: 10.2174/156800908783497096. [DOI] [PubMed] [Google Scholar]
- 8.Thomas GV, Horvath S, Smith BL, Crosby K, Lebel LA, Schrage M, Said J, De Kernion J, Reiter RE, Sawyers CL. Clin. Cancer Res. 2004;10:8351–8356. doi: 10.1158/1078-0432.CCR-04-0130. [DOI] [PubMed] [Google Scholar]
- 9.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 10.Pearl LH, Barford D. Curr. Opin. Struct. Biol. 2002;12:761–767. doi: 10.1016/s0959-440x(02)00386-x. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Cron P, Good VM, Thompson V, Hemmings BA, Barford D. Nat. Struct. Biol. 2002;9:940–944. doi: 10.1038/nsb870. [DOI] [PubMed] [Google Scholar]
- 12.Luo Y, Shoemaker AR, Liu XS, Woods KW, Thomas SA, de Jong R, Han EK, Li TM, Stoll VS, Powlas JA, Oleksijew A, Mitten MJ, Shi Y, Guan R, McGonigal TP, Klinghofer V, Johnson EF, Leverson JD, Bouska JJ, Mamo M, Smith RA, Gramling-Evans EE, Zinker BA, Mika AK, Nguyen PT, Oltersdorf T, Rosenberg SH, Li Q, Giranda VL. Mol. Cancer Ther. 2005;4:977–986. doi: 10.1158/1535-7163.MCT-05-0005. [DOI] [PubMed] [Google Scholar]
- 13.Okuzumi T, Fiedler D, Zhang C, Gray DC, Aizenstein B, Hoffman R, Shokat KM. Nat. Chem. Biol. 2009;5:484–493. doi: 10.1038/nchembio.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millward SW, Henning RK, Kwong GA, Pitram S, Agnew HD, Deyle KM, Nag A, Hein J, Lee SS, Lim J, Pfeilsticker JA, Sharpless KB, Heath JR. J. Am. Chem. Soc. 2011;133:18280–18288. doi: 10.1021/ja2064389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakamoto T, Ojida A, Hamachi I. Chem. Commun. 2009:141–152. doi: 10.1039/b812374h. [DOI] [PubMed] [Google Scholar]
- 16.Lewis WG, Green LG, Grynszpan F, Radic Z, Carlier PR, Taylor P, Finn MG, Sharpless KB. Angew. Chem. 2002;114:1095–1099. doi: 10.1002/1521-3773(20020315)41:6<1053::aid-anie1053>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2002;41:1053–1057. doi: 10.1002/1521-3773(20020315)41:6<1053::aid-anie1053>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 17.Grimster NP, Stump B, Fotsing JR, Weide T, Talley TT, Yamauchi JG, Nemecz A, Kim C, Ho K-Y, Sharpless KB, Taylor P, Fokin VV. J. Am. Chem. Soc. 2012;134:6732–6740. doi: 10.1021/ja3001858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The PyMOL Molecular Graphics System. Version 1.5.0.4. Schrödinger, LLC: [Google Scholar]
- 19.Agnew HD, Rohde RD, Millward SW, Nag A, Yeo W-S, Hein JE, Pitram SM, Tariq AA, Burns VM, Krom RJ, Fokin VV, Sharpless KB, Heath JR. Angew. Chem. 2009;121:5044–5048. doi: 10.1002/anie.200900488. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2009;48:4944–4948. doi: 10.1002/anie.200900488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindsley CW, Zhao Z, Leister WH, Robinson RG, Barnett SF, Defeo-Jones D, Jones RE, Hartman GD, Huff JR, Huber HE, Duggan ME. Bioorg. Med. Chem. Lett. 2005;15:761–764. doi: 10.1016/j.bmcl.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Brognard J, Clark AS, Ni YC, Dennis PA. Cancer Res. 2001;61:3986–3997. [PubMed] [Google Scholar]
- 22.Sun M, Wang G, Paciga JE, Feldman RI, Yuan ZQ, Ma XL, Shelley SA, Jove R, Tsichlis PN, Nicosia SV, Cheng JQ. Am. J. Pathol. 2001;159:431–437. doi: 10.1016/s0002-9440(10)61714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan ZQ, Sun M, Feldman RI, Wang G, Ma XL, Jiang C, Coppola D, Nicosia SV, Cheng JQ. Oncogene. 2000;19:2324–2330. doi: 10.1038/sj.onc.1203598. [DOI] [PubMed] [Google Scholar]
- 24.Clark AS, West K, Streicher S, Dennis PA. Mol. Cancer Ther. 2002;1:707–717. [PubMed] [Google Scholar]
- 25.Cheng JQ, Lindsley CW, Cheng GZ, Yang H, Nicosia SV. Oncogene. 2005;24:7482–7492. doi: 10.1038/sj.onc.1209088. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Gray NS. Nat. Chem. Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 27.Pfeilsticker JA, Umeda A, Farrow B, Hsuch CL, Deyle KM, Kim JT, Lai BT, Heath JR. PLoS One. 2013;8:e76224. doi: 10.1371/journal.pone.0076224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.