Abstract
The pyruvate dehydrogenase complexes (PDCs) from all known living organisms comprise three principal catalytic components for their mission: E1 and E2 generate acetyl-coenzyme A, whereas the FAD/NAD+-dependent E3 performs redox recycling. Here we compare bacterial (Escherichia coli) and human PDCs, as they represent the two major classes of the superfamily of 2-oxo acid dehydrogenase complexes with different assembly of, and interactions among components. The human PDC is subject to inactivation at E1 by serine phosphorylation by four kinases, an inactivation reversed by the action of two phosphatases. Progress in our understanding of these complexes important in metabolism is reviewed.
Keywords: Covalent Regulation, Enzyme Catalysis, Protein-Protein Interaction, Pyruvate Dehydrogenase Complex (PDC), Pyruvate Dehydrogenase Kinase (PDC Kinase)
Introduction
The pyruvate dehydrogenase complex (PDC)3 catalyzes the oxidative decarboxylation of pyruvate with the formation of acetyl-CoA, CO2 and NADH (H+) (1–3). The PDC occupies a key position in the oxidation of glucose by linking the glycolytic pathway to the oxidative pathway of the tricarboxylic acid cycle. In mammals, PDC plays the role of a gatekeeper in the metabolism of pyruvate to maintain glucose homeostasis during the fed and fasting states. The flux through PDC is tightly regulated in tissues under different metabolic conditions. This is accomplished by covalent modification of the rate-limiting component of the PDC involving dedicated kinases and phosphatases (4–7). PDC is also implicated to play a role in degenerative neurological diseases, obesity, type 2 diabetes, and other diseases (8–11). More recently, PDC has emerged as an enzyme of interest in cancer biology because of a switch from oxidative metabolism to aerobic glycolysis in some cancers (12–14). The focus of this review is to present recent developments on structural aspects, as well as on structure-based catalytic mechanisms of the PDCs, illustrated with Escherichia coli PDC (PDCec) as the simplest form and human (mammalian) PDC (PDCh) as a highly evolved form of the complex for its regulation.
PDC Components and Organization of the Complexes
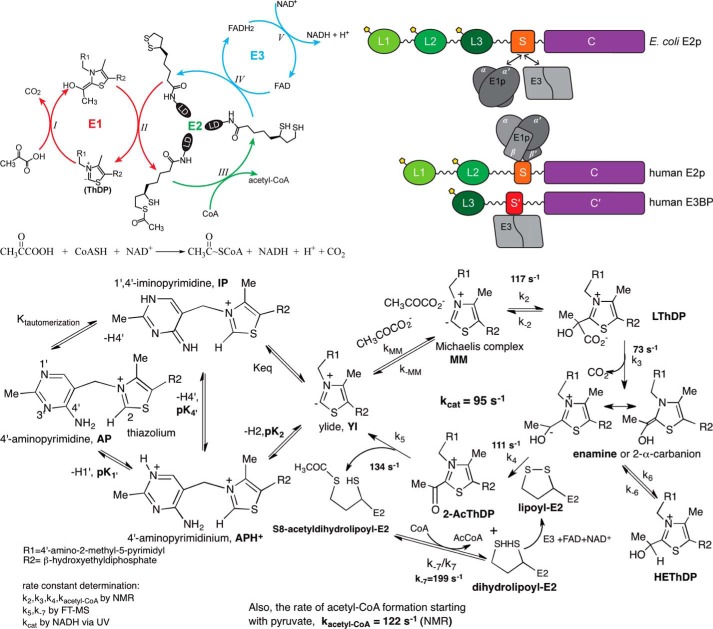
The PDCs in prokaryotes and eukaryotes are composed of multiple copies of three catalytic enzymes: pyruvate dehydrogenase (E1), dihydrolipoamide acetyltransferase (E2), and dihydrolipoamide dehydrogenase (E3) (Fig. 1, top left). These three catalytic components work sequentially, catalyzing the oxidative decarboxylation of pyruvate with the formation of acetyl-CoA, CO2 and NADH (H+). The E1 is a thiamin diphosphate (ThDP)-dependent enzyme and catalyzes two consecutive steps (refer to Fig. 1, bottom): (i) the decarboxylation of pyruvate to CO2 with the formation of C2α-hydroxyethylidene-ThDP (enamine) intermediate (k2, k3) and (ii) the reductive acetylation of the lipoyl groups covalently attached to the E2 (k4, k5). The E2 catalyzes the transfer of an acetyl moiety to CoA to form acetyl-CoA (k−7/k7). The transfer of electrons from the dihydrolipoyl moieties of E2 to FAD and then to NAD+ is carried out by E3. Higher eukaryotic PDCs have an additional structural component, dihydrolipoamide dehydrogenase-binding protein (E3BP), and two regulatory enzymes, pyruvate dehydrogenase kinase (PDK, four human isoforms) (15, 16) and pyruvate dehydrogenase phosphatase (PDP, two human isoforms) (6, 17, 18) totaling 11 proteins in PDCh with all isoforms included. Additionally, there are two isoforms of the α subunit of E1h that are encoded by separate genes in most mammals (19, 20). The X-linked gene (PDHA1 in human) encodes E1α subunit (PDHA1) present in all somatic tissues, whereas an autosomal, intronless gene (PDHA2 in human) is expressed only in the testis. E1h in this review refers to PDHA1 protein expressed in somatic cells.
FIGURE 1.
Overall PDC reactions, E2 and E3BP domain structures, and stepwise E1 reactions. Top left, reaction mechanism of the pyruvate dehydrogenase complex. Three catalytic components work sequentially, catalyzing the oxidative decarboxylation of pyruvate with the formation of acetyl-CoA, CO2, and NADH (H+). The reaction catalyzed by E1 is in red; the reaction catalyzed by E2 is in green; and that catalyzed by E3 is in blue. Top right, schematic representation of the domain structure of the E2ec, E2-h, and E3BP, comprising from the N-terminal end 1–3 LDs subunit-binding domain (PSBD or S) to which the E1 and E3 components are bound, and C-terminal catalytic domain (C or C′). Bottom, stepwise mechanism of PDHec reactions: right, tautomers and ionization states of ThDP; left, all kinetic constants were obtained for PDCec using methods listed in the lower left-hand side of the figure, and all rate constants are derived from pre-steady experiments with the exception of k7/k−7 (reproduced from Ref. 73).
The E2 in eukaryotes and prokaryotes has a multidomain structure, comprising from the N-terminal end 1–3 lipoyl domains (L1 and L2 in E2h; L1, L2, and L3 in E2ec; ∼80 amino acids each), the peripheral subunit-binding domain (PSBD, ∼45 amino acids) to which E1ec and E3ec bind in PDCec or the E1-binding domain to which E1h binds in PDCh, and a large C-terminal catalytic domain (∼250 amino acids) that forms the dodecahedral or cubic inner core and where acetyl-CoA is synthesized (Fig. 1, top right). Domains are connected by Ala- and Pro-rich hinge regions that are 20–30 residues in length. E3BP has a domain structure similar to that of E2h and is composed of one lipoyl domain, the E3-binding domain, and a catalytically incompetent C-terminal domain (Fig. 1, top right) (4, 21–23). PDKs are recruited to the E2h of PDCh by preferentially binding to the lipoyl domains (L1, L2) of the E2h and L3 of E3BP in the E2h/E3BP core (Fig. 1, top right) (17, 24).
PDC of most prokaryotes, including PDCec, is the simplest form of this complex. In PDCec, 24 copies of E2ec form a cubic core through the interaction of their catalytic domains. The multiple copies of E1ec (12 E1ec dimers) and E3ec (six E3ec dimers) are bound noncovalently to the PSBD of the E2ec core, and the entire complex with mass of 4.5 MDa exhibits octahedral symmetry (25, 26).
In mammalian PDCs, two models for E2-E3BP assembly have been proposed: the “addition” model, where 60 copies of E2h and 12 copies of E3BP form the E2h-E3BP core (27), and the “substitution” model, where 48 copies of E2h and 12 copies of E3BP (28) or 40 copies of E2h and 20 copies of E3BP form the E2h-E3BP core (29). Low-resolution structural studies of E2h-E3BP and cryo-electron microscopy reconstruction of E2h-E3BP and E2h-E3BP-E3 complex support the E2h-E3BP assembly via a substitution model (30). In mammalian PDC, 20–30 heterotetramers of E1 (α2β2) are bound to the PSBD of E2h; 6–12 homodimers of E3h are bound to the E3-binding domains (E3BD) of E3BP; and 1–3 homodimers of PDK and 2–3 heterodimers of PDP are bound to the lipoyl domains of E2h and/or E3BP. The entire complex (mass ∼9 MDa) exhibits icosahedral symmetry. Hence, PDCh represents a multienzyme assembly with greater complexity in function, structure, and regulation when compared with that of PDCec.
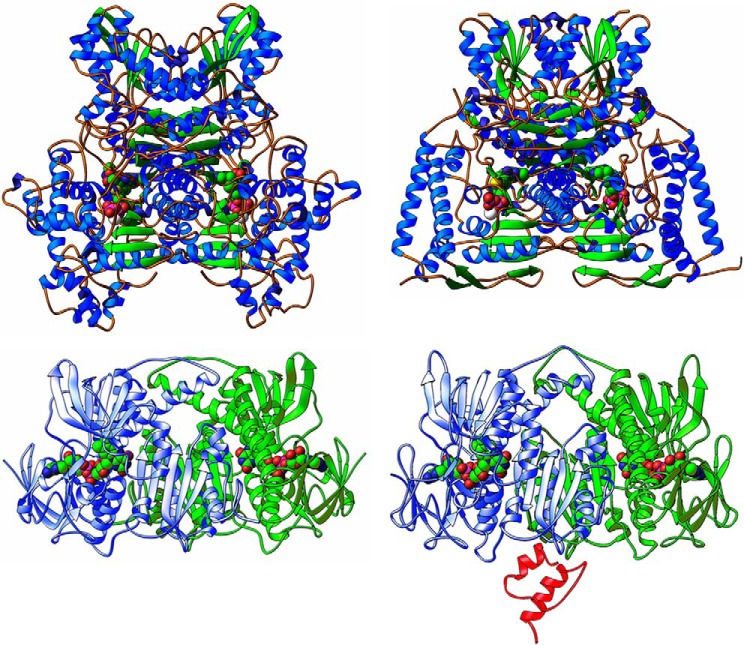
Detailed structures of individual components of PDCh (31–35) and PDCec (36–39) have been determined by x-ray crystallography (Fig. 2). The three-dimensional structure of the E2h catalytic domain (40), a full-length E2h-E3BP core, and full-length E2-E3BP-E3 complex have been reconstructed by cryo-electron microscopy (30).
FIGURE 2.
Crystal structures of E1 and E3 from E. coli and human. Top, ribbon diagrams illustrating structural differences between functional homodimeric and heterotetrameric E1 enzymatic assemblies from bacterial (E. coli), and mammalian (human) sources, respectively. Left, the homodimeric assembly from E. coli (reproduced from Ref. 34 with permission, Protein Data Bank (PDB) code 1L8A). Right, the heterotetrameric assembly from human (modified from Ciszak et al. (Ref. 31, PDB code 1NI4). The two figures are on the same scale and shown in the same orientation after least squares alignment based on the cofactors (ThDP) and structurally matching α carbons. The ThDP cofactors are shown in a space-filling representation. Bottom, ribbon diagrams illustrating structures of E3 functional dimers from bacterial E3ec (left) and human E3h (right) sources. In both the left and the right figures, one of the subunits is shown in blue and the other is shown in green, whereas the FAD cofactors are shown in a space-filling representation. The structure of the E3h dimer is shown in subcomplex with the E3-binding domain of E3 (in red). The bottom of this figure is reproduced from Ref. 39 for E3ec (PDB code: 4JDR) and from Ref. 33 for E3h (PDB code: 1ZY8).
E1 Component: Structure-based Studies of the Mechanism
Catalytic Intermediates
Using recently reported high-resolution structures of several components of the PDCs and novel methods developed for deciphering different steps in the five catalytic reactions carried out by three catalytic components of the PDCec, a deeper understanding of their mechanisms has emerged in recent years. These approaches and their outcomes are summarized in this section and refer to steps in Fig. 1, bottom. (a) The first kinetic step involves the rate of deprotonation of the weak acid C2H at the thiazolium ring of ThDP to generate the reactive ylide that is measured by monitoring the rate of solvent deuterium incorporation at C2 by 1H NMR (41). Both E1ec and E1h accelerate this rate substantially (42, 43). (b) There is evidence for the presence of three tautomeric and ionization states of enzyme-bound ThDP: the 4′-aminopyrimidine (AP form), the 1′,4′-iminopyrimidine (IP) tautomer (44–47) (Fig. 1, bottom left), and the protonated 4′-aminopyrimidinium (APH+) form, detected by CD and solid state NMR (48–50). (c) The presence of the pre-decarboxylation intermediate C2α-lactylThDP (LThDP) could be deduced by CD as an IP tautomer (49, 51–53). An NMR method could identify ThDP-bound covalent intermediates stable under acidic conditions (LThDP, HEThDP, 2-acetylThDP) from the chemical shifts of their C6′H proton resonances (54, 55). (d) Existence of the first post-decarboxylation intermediate, the enamine/C2α-carbanion, could be inferred from its acid quench conversion to HEThDP and its observation by NMR (54, 55). (e) The second post-decarboxylation intermediate, HEThDP on enzymes also exists as the IP tautomer, as do all ThDP-bound intermediates with tetrahedral substitution at C2α atom, and could be observed by CD (52). (f) 2-AcetylThDP, the oxidized form of the enamine, could be an intermediate on the PDC (56, 57) and was recently observed on E1ec (derived from fluoropyruvate) via a characteristic CD signal near 390 nm (58).(g) Reductive acetylation of E2 (information transfer from E1) and acetyl transfer between E2 domain are monitored by high-resolution MS methods (55), revealing that neither reductive acetylation nor interdomain acetyl transfer is rate-limiting through acetyl-CoA formation. (h) The formation of acetyl-CoA from [13C-C2]pyruvate could be monitored via NMR methods (55), also affirming the result in step g. As shown in Fig. 1, bottom, all rate constants could be assessed for PDCec with these methods.
Communication between Active Center ThDPs in the E1 Component
When aligned by least squares superposition and put into a common orientation, the overall structures appear very similar, although there are great differences between the structure of the enzymes from different species (Fig. 2, top). In both bacterial and human E1s, there are two distinct active centers capable of binding ThDP. In the structures of E1ec (α2,homodimer) in complex with ThDP (36) or its “transition-state analog” thiamin 2-thiothiazolone diphosphate (37), two ThDP molecules are bound at subunit-subunit interfaces forming two active centers. In the structure of E1h (α2β2), binding of two ThDP molecules involves the opposite pair of each heterodimer (such as the diphosphate-binding domain of one αβ heterodimer with aminopyrimidine-binding domain of the α′β′ heterodimer and vice versa) with flexibility required for movement (31). No structural evidence was observed of nonequivalence of two active centers.
Steady-state kinetic and spectroscopic observations of ThDP-dependent enzymes supported communication between active center ThDPs manifested by “half-of-the site” reactivity (or alternating active-site mechanism) (42, 43, 59, 60). Solution NMR evidence of nonequivalence of two active centers in E1h and E1ec was obtained from H/D exchange kinetics at C2-H of E1-bound ThDP (42, 43) and by analysis of covalent ThDP-bound intermediates in E1ec and E1h catalysis (54, 61). A “proton wire” mechanism (62, 63) with a hydrated “tunnel” of acidic residues and water molecules was suggested to enable direct communication for proton shuttling between two ThDP N1′ atoms. Experimental evidence of such a proton wire pathway was obtained for E1ec (43). No proton wire pathway was identified for E1h.
Structural Evidence for the Role of Phosphorylation Loops in E1h
In the E1h (α2β2), the α subunit is a target for phosphorylation/dephosphorylation by PDKs and PDPs (7, 16, 17, 64). There are three phosphorylation sites in E1h-α that could be phosphorylated independently, leading to PDC inactivation: site 1 at Ser-264-α, site 2 at Ser-271-α, and site 3 at Ser-203-α (65–68). In the E1h structure, site 1 and site 2 are located on a highly conserved phosphorylation loop A, whereas site 3 is on phosphorylation loop B (31, 33, 69). These loops are ordered on ThDP binding and disordered in its absence. On phosphorylation of site 1 (both Ser-264-α residues in the α2β2 heterotetramer were phosphorylated), the phosphorylation loops were disordered even in the presence of ThDP, leading to loss of PDC activity (67, 69). Apparently, upon Ser-264-α phosphorylation, the bulky phosphoryl group produces a steric clash that disrupts the H-bond network involving residues from phosphorylation loop A and Tyr-33-β′ from the E1p-β subunit, resulting in a disordered conformation of both phosphorylation loops (69). In E1h, the presence of both loops in the ordered conformation is a requirement for lipoyl domain recognition and substrate channeling to E2h (69). An alternative explanation based on the structure of E1h S264E (site 1 pseudo-phosphorylated) suggested that both steric and electrostatic factors affect substrate channeling between E1h and E2h (33).
The Active Center Loops in E1ec
In the structure of E1ec in complex with C2α-phosphonolactylthiamin diphosphate (PLThDP), a stable analog of the pre-decarboxylation intermediate LThDP, two disordered loops had become ordered and completed the active center: the inner (residues 401–413) and outer loop (residues 541–557) (38). Kinetic, spectroscopic, NMR, and structural studies demonstrated that His-407 from the inner loop and charged residues flanking His-407 have a role in stabilizing/ordering of the inner loop that is essential for substrate entry to the active site and for sequestering the active site from undesirable side reactions (38, 70–72). The E1ec variants with inner loop substitutions demonstrated greatly impaired rate of reductive acetylation of E2ec, suggesting that ordering of the inner loop plays a role in communication between the E1ec and E2ec (71, 72). For E1ec variants with E401K, H407A (inner loop), D549A (outer loop), and Y177A substitutions, the rate of LThDP formation was affected when the inner loop was disordered, also confirmed by x-ray structure of E401K and H407A E1ec variants. It was concluded that: (a) the rate of formation of the first C–C bond is affected when the inner loop is disordered and (b) loop dynamics controls covalent catalysis with ThDP (55).
Role of the E2 Component: Identification of the Interaction Loci between E1 and E2
Binding of E1h to the E1h-binding Domain of E2h
In icosahedral PDCh, the E2h-E3BP core provides the binding sites for E1h through the subunit-binding domain of the E2h. The structure of the E1 from Bacillus stearothermophilus (E1bs, α2β2) in complex with the E1/E3-binding domain of E2bs (E2bs forms dodecahedral core of PDCbs) was determined (74) and was employed as a model to identify residues important for interaction between E1-binding domain of the E2h and E1h (75). It was demonstrated that the E1bs (α2β2) binds to its cognate E2s through the C-terminal region of the β-subunits through electrostatic and hydrophobic interactions (74). Screening of the surface of the β subunit of E1h for electrostatic interactions revealed Asp-289 to be important in the formation of a salt bridge to Lys-276 on E2h (75).
Identification of the Interaction Loci of E2ec Core with E1ec and E3ec
In octahedral PDCs, the E1 binds to PSBD of E2 (PDCec) or to the E2 core domain (E. coli 2-ketoglutarate dehydrogenase) through their N-terminal regions. Studies of the E1ec variants with substitutions in the N-terminal region (residues 1–55) suggest that the entire N-terminal region of E1ec is responsible for binding to E2ec (76, 77). Studies of E2ec variants revealed that binding of E1ec and E3ec to E2ec was strongly affected by charge-reversed substitutions at Arg-129 and Arg-150 in PSBD (numbers correspond to 1-lip E2ec sequence), but not affected by substitutions at Lys-191 and Arg-202 in the N-terminal region of the E2ec core (76). With substitutions at Arg-129 more strongly affecting the binding of E3ec and substitutions at Arg-150 strongly affecting the E1ec binding to E2ec, the studies suggested that E1ec and E3ec bind to nonidentical but overlapping loci of PSBD (76). The R129E and R150E substitutions in E2ec also affected the rate of reductive acetylation of E2ec by E1ec and pyruvate, and suggest that functional communication between E1ec and E2ec was also affected. Evidence suggests that the lipoyl domain, in addition to PSBD, is also recruited into interaction with E1ec (76). No direct interaction of the N-terminal region of E1ec with the E2ec core domain was evident.
Role of the E3 Component: Interaction loci between the E3 and E2
The functional E3 from both sources is a homodimer with two identical active centers located at the interface between two subunits (32–34, 39). Each subunit is composed of four domains: FAD-binding domain, NAD+-binding domain, the central domain, and the interface domain. Two tightly bound FAD molecules in the E3 dimer are involved in electron transfer from dihydrolipoamide of E2 to NAD+ with involvement of an intramolecular disulfide bridge in each subunit (Fig. 2, bottom). Although the overall structures are similar for E3ec and E3h, their binding mode to the E2 core is different, and will be discussed below.
Interaction Loci of E3h-binding Protein (E3BP) with E3h
The E2h-E3BP core provides the binding sites for E3h through the E3BD of E3BP. Two x-ray structures of the human E3h-E3BD subcomplex were reported (Fig. 2, bottom) (33, 34), revealing that E3BD binds in the interface between two E3h subunits through a combination of hydrophobic and electrostatic interactions, and complexation with E3BD did not perturb the E3 structure (33, 34). The residues involved in the E3h-E3BD interaction were identified by site-directed substitutions using kinetic, spectroscopic, calorimetric, and surface plasmon resonance analysis of the binding constants (34, 75, 78). The Ile-157 in E3BD and Tyr-438 in E3h were found important for specificity of E3h recognition by E3BD (75).
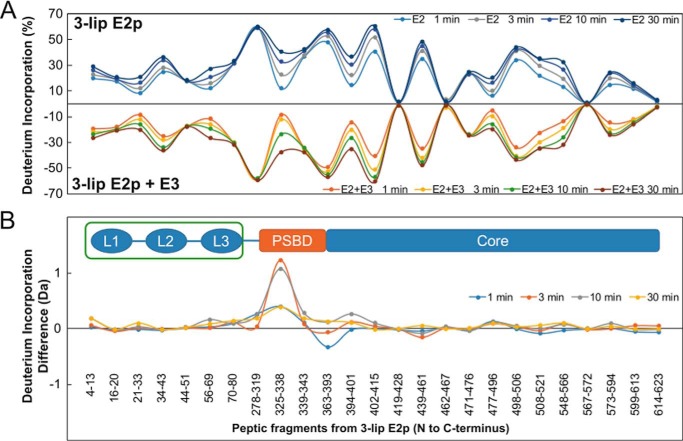
Interaction of the E2ec with E3ec Studied by Fourier Transform-Mass Spectroscopy
With the help of the known crystal structure of E3ec (39), the loci of interaction of E3ec with E2ec were identified by peptide-specific H/D exchange MS (Fig. 3). In the E2ec-E3ec subcomplex, two peptides from PSBD of 3-lip E2ec and three peptides from E3ec (two peptides from the FAD-binding domain and one from the interface domain) exhibited significant reduction in deuterium uptake on E3ec binding to 3-lip E2ec, when compared with deuterium uptake by the individual components (Fig. 3) (39). One of the two identified peptides in 3-lip E2ec (residues 325–338) is part of the α-helix (H1), which interacts with the E3ec dimer in all known structures with the exception of the 2-oxoglutarate dehydrogenase complex (79), and it provides electrostatic stabilization of the E2ec-E3ec complex. Residue Arg-333 in this peptide (corresponding to Arg-129 in 1-lip E2ec) was identified as a “hot spot” for interaction of E2ec with both E3ec and E1ec and is a highly conserved residue in all known PSBDs (80). In summary, in PDCec, the E1ec and E3ec components bind to nonidentical, but strongly overlapping epitopes of E2ec localized in the PSBD domain. Their binding does not require competition according to the model recently suggested for differential utilization of three chains of E2ec as a trimer unit on its assembly with E1ec and E3ec (81).
FIGURE 3.
Hydrogen deuterium exchange-MS analysis of the interaction loci of 3-lip E2ec and E3ec. Top figure, butterfly plot representing average relative deuterium incorporation percentage (y axis) (deuterons exchanged/maximum exchangeable amides × 100) of peptic fragments from 3-lip E2ec (x axis, listed from N to C terminus) in the absence of E3ec (top) and in the presence of E3ec (bottom). Bottom figure, difference plot showing deuterium incorporation changes in peptic fragments of 3-lip E2ec in the absence and presence of E3ec (reproduced from Ref. 39).
Regulation of PDCh by Phosphorylation
Regulation of mammalian PDC is important for the maintenance of glucose homeostasis during both the fed and the fasted states. This control is achieved primarily by the reversible phosphorylation of E1h, resulting in its inactivation and inhibition of PDCh activity. Phosphorylation status of E1h is highly regulated by activities of PDKs and PDPs at both the transcriptional and the post-translational levels (4, 6, 7). The three sites in E1h are phosphorylated in vivo at different rates and with different specificity by four PDKs (66, 67). In E1h (and also in mammalian PDCs), site 1 is preferentially phosphorylated and sites 2 and 3 are sequentially phosphorylated, leading to slower rates of the complex (64). Studies of E1h variants with only a single functional site for phosphorylation (with the other two phosphorylation sites converted to alanine) revealed that each of the three sites could be phosphorylated independently, resulting in E1h inactivation in each case (66, 67). Mammalian PDK1 is capable of phosphorylating all three sites in E1h, whereas PDKs 2–4 are able to phosphorylate only sites 1 and 2. The L1 and L2 of E2h and L3 of E3BP play an essential role in regulation of PDK activities (Table 1). Again, different binding preferences of four kinases toward the three lipoyl domains (L1, L2, and L3) provide flexibility for modulation of their activities. Binding of PDKs to the lipoyl domains of E2h and E3BP co-localizes to its substrate, E1h, bound to E2h, and also induces conformational changes in PDKs, resulting in variable degrees of activation, with PDK3 showing the highest degree of activation (Table 1). Interestingly, reduction by NADH and acetylation by acetyl-CoA of the lipoyl moiety result in variable degrees of stimulation of PDKs, with PDK2 being the most sensitive to this stimulation (Table 1). In contrast, high levels of pyruvate, ADP, NAD+, and CoA are inhibitory for PDK activities (16). The activity of PDC is tightly regulated by the cellular levels of PDKs and PDPs in tissues under different nutritional and disease states (7). Transcriptional modulation of these regulatory enzymes is under the influence of hormonal changes during starvation, diabetes, and hyperthyroidism (6, 7). PDK4 expression is regulated by fatty acids through peroxisome proliferator-activated receptor α (47) and also by glucocorticoids (48). Detailed accounts of transcriptional regulation of PDKs can be found in a recent comprehensive review (82, 83). Detailed aspects of transcriptional regulation of PDKs can be found in a recent, comprehensive review (11). Recent findings suggest a role of post-translational modifications in regulation of PDCh, including activation of PDK1 by tyrosine phosphorylation (85) and inhibition of PDP1 and E1h by lysine acetylation (86), that control PDC activity in cancer cells. These issues need more detailed elucidation.
TABLE 1.
Characteristics of mammalian PDK isoenzymes and PDP isoenzymes
The information presented in Table 1 is derived from Refs. 3, 6, 7, 11, 16, 24, 64, 65, 86, and 87.
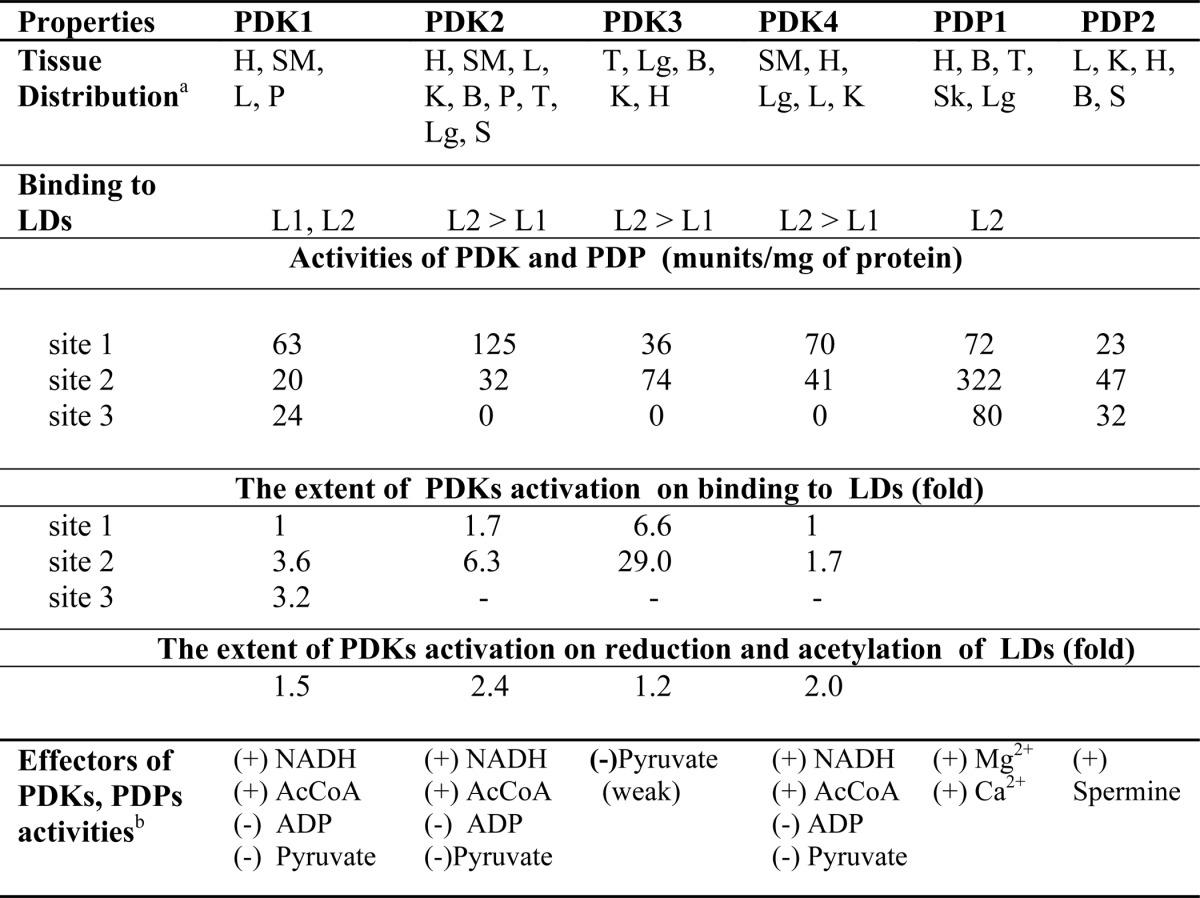
a H, heart; SM, skeletal muscles; L, liver; Lg, lungs; K, kidneys; B, brain; P, pancreas; T, testis; S, spleen.
b (+) activation; (−) inhibition.
The crystal structures of all four PDKs (either alone or in association with L2, ADP, or ATP) have been reported (87–93). Each PDK monomer has two domains of about equal size, the N-terminal domain and the C-terminal domain, and these two domains are connected by a flexible, poorly ordered loop. In dimeric PDK, the C-terminal tail of each monomer interacts with the lipoyl-binding pocket in the N-terminal domain of the other monomer. Monomers in the dimeric PDK are in a head-to-head orientation with the primary interaction between the C-terminal domains. In the absence of L2, PDK dimer forms a “closed” conformation, resulting in closing of the active-site cleft because of the disordered C-terminal tail of one monomer not interacting with the lipoyl-binding pocket of the opposite monomer (91–93). This conformation stabilizes the ATP lid and thus prevents dissociation of ADP, resulting in product inhibition. Upon binding of PDK3 to L2, the crossover configuration of the C-terminal tails results in an “open” conformation promoting the widening of the active-site cleft, causing disordering of the ATP lid and accelerating the release of trapped ADP (88, 89, 91). The two lipoyl-binding pockets are located on the outer surface of the PDK dimer in the opposite direction, suggesting that PDK may bind to two different lipoyl domains on PDC at a time. This arrangement is consistent with a “hand-on-hand” movement of PDK to serve many E1s in the complex (16).
PDP exists as two different forms, PDP1 (catalytic and regulatory subunits) and PDP2 (catalytic subunit only), with differing biochemical characteristics. PDP1c (catalytic) activity strongly depends on its binding to the lipoyl domain of E2h requiring Ca2+ (24, 94–96). In contrast, PDP2c requires neither for its activity (95). Both PDP1c and PDP2c are able to dephosphorylate all three phosphorylation sites on E1h. PDP1c exhibits a random mechanism of dephosphorylation (with relative rates of site 2 > site 3 > site 1), indicating a lack of site-site dependence for dephosphorylation. In contrast, PDP2c displays interdependence in dephosphorylation of site 1 with site 2 and site 3 separately, and no interaction between site 2 and site 3 (95). The crystal structure of PDP1c reveals a unique hydrophobic pocket on the surface to accommodate the lipoyl moiety of the lipoyl domain of E2 (97). It is proposed that the closure of the lipoyl moiety-binding site is achieved by the formation of the intermolecular (PDP1c/L2) Ca2+-binding site.
Concluding Remarks
More than 60 years have passed since lipoic acid was first isolated and characterized by Reed et al. (84), a seminal discovery in our understanding of the chemistry and core functions of these complexes. We now have x-ray structures of several PDC components. With advances in detecting ThDP-bound intermediates using spectroscopic methods, the rates of individual steps could be determined for the E. coli PDC, revealing that formation of the first covalent complex between pyruvate and ThDP is rate-limiting, and this step may be controlled by the mobility of active center loops. Remaining challenges include the need for a high-resolution structure of an intact E2 component, and of course of the entire complex. Renewed interest and enthusiasm for studying the human PDC are warranted by its recently revealed involvement in some forms of cancer, type 2 diabetes, and obesity, probably involving the interactions of the E2h-PDK subcomplex and providing new targets for rational drug design to regulate glucose metabolism in cancer, type 2 diabetes, obesity, and other diseases.
This work was supported, in whole or in part, by National Institutes of Health Grants DK 20478 (to M. S. P.), GM050380 (to F. J.), and GM061791 (to W. F.) and by the Veterans Affairs Merit Review (W. F.).
This article is dedicated to Dr. Richard W. Hanson of Case Western Reserve University School of Medicine for his devotion and enormous contributions in the area of regulation of phosphoenolpyruvate carboxykinase and gluconeogenesis, for passionate teaching of metabolism, and for mentoring the next generation of investigators in the area of metabolic regulation. One of us (M. S. P.) deeply acknowledges Dr. Hanson's generous mentorship, collegiality, and friendship over a period of 45 years.
- PDC
- pyruvate dehydrogenase complex
- E1
- pyruvate dehydrogenase
- E2
- dihydrolipoamide acetyltransferase
- E3
- dihydrolipoamide dehydrogenase
- E3BP
- E3-binding protein
- E3BD
- E3-binding domain
- ThDP
- thiamin diphosphate
- ec
- E. coli
- h
- human
- bs
- B. stearothermophilus
- LD
- lipoyl domain
- 3-lip E2ec
- wild type E2ec with three lipoyl domains
- 1-lip E2ec
- E2ec with one hybrid LD (LDh)
- PSBD
- the peripheral subunit-binding domain
- PDK
- pyruvate dehydrogenase kinase
- PDP
- pyruvate dehydrogenase phosphatase
- HEThDP
- C2α-hydroxyethylThDP
- LThDP
- C2α-lactylThDP
- IP
- iminopyrimidine
- c
- catalytic.
REFERENCES
- 1. Reed L. J. (2001) A trial of research from lipoic acid to α-keto acid dehydrogenase complexes. J. Biol. Chem. 276, 38329–38336 [DOI] [PubMed] [Google Scholar]
- 2. Perham R. N. (1991) Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry 30, 8501–8512 [DOI] [PubMed] [Google Scholar]
- 3. Patel M. S., Roche T. E. (1990) Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 4, 3224–3233 [DOI] [PubMed] [Google Scholar]
- 4. Patel M. S., Korotchkina L. G. (2003) The biochemistry of the pyruvate dehydrogenase complex. Biochem. Mol. Biol. Educ. 31, 5–15 [Google Scholar]
- 5. Korotchkina L. G., Sidhu S., Patel M. S. (2006) Characterization of testis-specific isoenzyme of human pyruvate dehydrogenase. J. Biol. Chem. 281, 9688–9696 [DOI] [PubMed] [Google Scholar]
- 6. Harris R. A., Bowker-Kinley M. M., Huang B., Wu P. (2002) Regulation of the activity of the pyruvate dehydrogenase complex. Adv. Enzyme Regul. 42, 249–259 [DOI] [PubMed] [Google Scholar]
- 7. Patel M. S., Korotchkina L. G. (2006) Regulation of pyruvate dehydrogenase complex. Biochem. Soc. Trans. 34, 217–222 [DOI] [PubMed] [Google Scholar]
- 8. Patel M. S., Harris R. A. (1995) Mammalian α-keto acid dehydrogenase complexes: gene regulation and genetic defects. FASEB J. 9, 1164–1172 [DOI] [PubMed] [Google Scholar]
- 9. Imbard A., Boutron A., Vequaud C., Zater M., de Lonlay P., Ogier de Baulny H., Barnerias C., Miné M., Marsac C., Saudubray J.-M., Brivet M. (2011) Molecular characterization of 82 patients with pyruvate dehydrogenase complex deficiency. Structural implications of novel amino acid substitutions in E1 protein. Mol. Gen. Metab. 104, 507–516 [DOI] [PubMed] [Google Scholar]
- 10. Patel K. P., O'Brien T. W., Subramony S. H., Shuster J., Stacpoole P. W. (2012) The spectrum of pyruvate dehydrogenase complex deficiency: clinical, biochemical and genetic features in 371 patients. Mol. Genet. Metab. 106, 385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jeoung N. H., Harris C. R., Harris R. A. (2014) Regulation of pyruvate metabolism in metabolic-related diseases. Rev. Endocr. Metab. Disord. 15, 99–110 [DOI] [PubMed] [Google Scholar]
- 12. Schulze A., Downward J. (2011) Flicking the Warburg switch-tyrosine phosphorylation of pyruvate dehydrogenase kinase regulates mitochondrial activity in cancer cells. Mol. Cell 44, 846–848 [DOI] [PubMed] [Google Scholar]
- 13. Kaplon J., Zheng L., Meissl K., Chaneton B., Selivanov V. A., Mackay G., van der Burg S. H., Verdegaal E. M. E., Cascante M., Shlomi T., Gottlieb E., Peeper D. S. (2013) A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature 498, 109–112 [DOI] [PubMed] [Google Scholar]
- 14. Sutendra G., Michelakis E. D. (2013) Pyruvate dehydrogenase kinase as a novel therapeutic target in oncology. Front. Oncol. 3, 38–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gudi R., Bowker-Kinley M. M., Kedishvili N. Y., Zhao Y., Popov K. M. (1995) Diversity of the pyruvate dehydrogenase kinase gene family in humans. J. Biol. Chem. 270, 28989–28994 [DOI] [PubMed] [Google Scholar]
- 16. Roche T. E., Baker J. C., Yan X., Hiromasa Y., Gong X., Peng T., Dong J., Turkan A., Kasten S. A. (2001) Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Prog. Nucleic Acid Res. Mol. Biol. 70, 33–75 [DOI] [PubMed] [Google Scholar]
- 17. Roche T. E., Hiromasa Y., Turkan A., Gong X., Peng T., Yan X., Kasten S. A., Bao H., Dong J. (2003) Essential roles of lipoyl domains in the activated function and control of pyruvate dehydrogenase kinases and phosphatase isoform 1. Eur. J. Biochem. 270, 1050–1056 [DOI] [PubMed] [Google Scholar]
- 18. Huang B., Gudi R., Wu P., Harris R. A., Hamilton J., Popov K. M. (1998) Isoenzymes of pyruvate dehydrogenase phosphatase: DNA-derived amino acid sequences, expression, and regulation. J. Biol. Chem. 273, 17680–17688 [DOI] [PubMed] [Google Scholar]
- 19. Brown R. M., Dahl H. H. M., Brown G. K. (1990) Pyruvate dehydrogenase E1α subunit genes in the mouse: mapping and comparison with human homologs. Somat. Cell. Mol. Genet. 16, 487–492 [DOI] [PubMed] [Google Scholar]
- 20. Dahl H.-H., Brown R. M., Hutchison W. M., Maragos C., Brown G. K. (1990) A testis-specific form of the human pyruvate dehydrogenase E1α subunit is coded by an intronless gene on chromosome 4. Genomics 8, 225–232 [DOI] [PubMed] [Google Scholar]
- 21. De Marcucci O., Lindsay J. G. (1985) Component X. An immunologically distinct polypeptide associated with mammalian pyruvate dehydrogenase multi-enzyme complex. Eur. J. Biochem. 149, 641–648 [DOI] [PubMed] [Google Scholar]
- 22. Jilka J. M., Rahmatullah M., Kazemi M., Roche T. E. (1986) Properties of a newly characterized protein of the bovine kidney pyruvate dehydrogenase complex. J. Biol. Chem. 261, 1858–1867 [PubMed] [Google Scholar]
- 23. Harris R. A., Bowker-Kinley M. M., Wu P., Jeng J., Popov K. M. (1997) Dihydrolipoamide-binding protein of the human pyruvate dehydrogenase complex: DNA-derived amino acid sequence, expression, and reconstitution of the pyruvate dehydrogenase complex. J. Biol. Chem. 272, 19746–19751 [DOI] [PubMed] [Google Scholar]
- 24. Chen G., Wang L., Liu S., Chuang C., Roche T. E. (1996) Activation function of the pyruvate dehydrogenase phosphatase through Ca2+-facilitated binding to the inner lipoyl domain of the dihydrolipoyl acetyltransferase. J. Biol. Chem. 271, 28064–28070 [DOI] [PubMed] [Google Scholar]
- 25. Reed L. J., Hackert M. L. (1990) Structure-function relationships in dihydrolipoamide acyltransferases. J. Biol. Chem. 265, 8971–8974 [PubMed] [Google Scholar]
- 26. Perham R. N. (2000) Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu. Rev. Biochem. 69, 961–1004 [DOI] [PubMed] [Google Scholar]
- 27. Sanderson S. J., Miller C., Lindsay J. G. (1996) Stoichiometry, organization and catalytic function of protein X of the pyruvate dehydrogenase complex from bovine heart. Eur. J. Biochem. 236, 68–77 [DOI] [PubMed] [Google Scholar]
- 28. Hiromasa Y., Fujisawa T., Aso Y., Roche T. E. (2004) Organization of the cores of the mammalian pyruvate dehydrogenase complex formed by E2 and E2 plus the E3-binding protein and their capacities to bind the E1 and E3 components. J. Biol. Chem. 279, 6921–6933 [DOI] [PubMed] [Google Scholar]
- 29. Brautigam C. A., Wynn R. M., Chuang J. L., Chuang D. T. (2009) Subunit and catalytic component stoichiometries of an in vitro reconstituted human pyruvate dehydrogenase complex. J. Biol. Chem. 284, 13086–13098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vijayakrishnan S., Kelly S. M., Gilbert R. J., Callow P., Bhella D., Forsyth T., Lindsay J. G., Byron O. (2010) Solution structure and characterization of the human pyruvate dehydrogenase complex core assembly. J. Mol. Biol. 399, 71–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ciszak E. M., Korotchkina L. G., Dominiak P. M., Sidhu S., Patel M. S. (2003) Structural basis for flip-flop action of thiamin pyrophosphate-dependent enzymes revealed by human pyruvate dehydrogenase. J. Biol. Chem. 278, 21240–21246 [DOI] [PubMed] [Google Scholar]
- 32. Brautigam C. A., Chuang J. L., Tomchick D. R., Machius M., Chuang D. T. (2005) Crystal structure of human dihydrolipoamide dehydrogenase: NAD+/NADH binding and the structural basis of disease-causing mutations. J. Mol. Biol. 350, 543–552 [DOI] [PubMed] [Google Scholar]
- 33. Ciszak E. M., Makal A., Hong Y. S., Vettaikkorumakankauv A. K., Korotchkina L. G., Patel M. S. (2006) How dihydrolipoamide dehydrogenase-binding protein binds dihydrolipoamide dehydrogenase in the human pyruvate dehydrogenase complex. J. Biol. Chem. 281, 648–655 [DOI] [PubMed] [Google Scholar]
- 34. Brautigam C. A., Wynn R. M., Chuang J. L., Machius M., Tomchick D. R., Chuang D. T. (2006) Structural insight into interactions between dihydrolipoamide dehydrogenase (E3) and E3 binding protein of human pyruvate dehydrogenase complex. Structure 14, 611–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seifert F., Ciszak E., Korotchkina L., Golbik R., Spinka M., Dominiak P., Sidhu S., Brauer J., Patel M. S., Tittmann K. (2007) Phosphorylation of serine 264 impedes active site accessibility in the E1 component of the human pyruvate dehydrogenase multienzyme complex. Biochemistry 46, 6277–6287 [DOI] [PubMed] [Google Scholar]
- 36. Arjunan P., Nemeria N., Brunskill A., Chandrasekhar K., Sax M., Yan Y., Jordan F., Guest J. R., Furey W. (2002) Structure of the pyruvate dehydrogenase multienzyme complex E1 component from Escherichia coli at 1.85 Å resolution. Biochemistry 41, 5213–5221 [DOI] [PubMed] [Google Scholar]
- 37. Arjunan P., Chandrasekhar K., Sax M., Brunskill A., Nemeria N., Jordan F., Furey W. (2004) Structural determinants of enzyme binding affinity: the E1 component of pyruvate dehydrogenase from Escherichia coli in complex with the inhibitor thiamin thiazolone diphosphate. Biochemistry 43, 2405–2411 [DOI] [PubMed] [Google Scholar]
- 38. Arjunan P., Sax M., Brunskill A., Chandrasekhar K., Nemeria N., Zhang S., Jordan F., Furey W. (2006) A thiamin-bound, pre-decarboxylation reaction intermediate analogue in the pyruvate dehydrogenase E1 subunit induces large scale disorder-to-order transformations in the enzyme and reveals novel structural features in the covalently bound adduct. J. Biol. Chem. 281, 15296–15303 [DOI] [PubMed] [Google Scholar]
- 39. Chandrasekhar K., Wang J., Arjunan P., Sax M., Park Y.-H., Nemeria N. S., Kumaran S., Song J., Jordan F., Furey W. (2013) Insight to the interaction of the dihydrolipoamide acetyltransferase (E2) core with the peripheral components in the Escherichia coli pyruvate dehydrogenase complex via multifaceted structural approaches. J. Biol. Chem. 288, 15402–15417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yu X., Hiromasa Y., Tsen H., Stoops J. K., Roche T. E., Zhou Z. H. (2008) Structures of the human pyruvate dehydrogenase complex cores: a highly conserved catalytic center with flexible N-terminal domains. Structure 16, 104–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kern D., Kern G., Neef H., Tittmann K., Killenberg-Jabs M., Wikner C., Schneider G., Hübner G. (1997) How thiamine diphosphate is activated in enzymes. Science 275, 67–70 [DOI] [PubMed] [Google Scholar]
- 42. Seifert F., Golbik R., Brauer J., Lilie H., Schröder-Tittmann K., Hinze E., Korotchkina L. G., Patel M. S., Tittmann K. (2006) Direct kinetic evidence for half-of-the-sites reactivity in the E1 component of the human pyruvate dehydrogenase multienzyme complex through alternating sites cofactor activation. Biochemistry 45, 12775–12785 [DOI] [PubMed] [Google Scholar]
- 43. Nemeria N. S., Arjunan P., Chandrasekhar K., Mossad M., Tittmann K., Furey W., Jordan F. (2010) Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for direct pathway between the 4′-aminopyrimidine N1′ atoms. J. Biol. Chem. 285, 11197–11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schellenberger A. (1998) Sixty years of thiamin diphosphate biochemistry. Biochim. Biophys. Acta 1385, 177–186 [DOI] [PubMed] [Google Scholar]
- 45. Jordan F., Mariam Y. H. (1978) N1′-Methylthiaminium diiodide: model study on the effect of a coenzyme bound positive charge on reaction mechanisms requiring thiamin pyrophosphate. J. Am. Chem. Soc. 100, 2534–2541 [Google Scholar]
- 46. Jordan F., Zhang Z., Sergienko E. (2002) Spectroscopic evidence for participation of the 1′,4′-imino tautomeric diphosphate in catalysis by yeast pyruvate decarboxylase. Bioorg. Chem. 30, 188–198 [DOI] [PubMed] [Google Scholar]
- 47. Jordan F. (1982) Role of the aminopyrimidine ring in thiamin-catalyzed reactions: II. Proton NMR evidence for high barriers to amino group rotation in 4-aminopyrimidines, including thiamin, at low pH in water. J. Org. Chem. 47, 2748–2753 [Google Scholar]
- 48. Nemeria N., Korotchkina L., McLeish M. J., Kenyon G. L., Patel M. S., Jordan F. (2007) Elucidation of the chemistry of enzyme-bound thiamin diphosphate prior to substrate binding: defining internal equilibria among tautomeric and ionization states. Biochemistry 46, 10739–10744 [DOI] [PubMed] [Google Scholar]
- 49. Nemeria N., Chakraborty S., Baykal A., Korotchkina L. G., Patel M. S., Jordan F. (2007) The 1′,4′-iminopyrimidine tautomer of thiamin diphosphate is poised for catalysis in asymmetric active centers on enzymes. Proc. Natl. Acad. Sci. U.S.A. 104, 78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nemeria N. S., Chakraborty S., Balakrishnan A., Jordan F. (2009) Reaction mechanisms of thiamin diphosphate enzymes: defining states of ionization and tautomerization of the cofactor at individual steps. FEBS J. 276, 2432–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jordan F., Nemeria N. S., Zhang S., Yan Y., Arjunan P., Furey W. (2003) Dual catalytic apparatus of the thiamin diphosphate coenzyme: acid-base via the 1′,4′-iminopyrimidine tautomer along with its electrophilic role. J. Am. Chem. Soc. 125, 12732–12738 [DOI] [PubMed] [Google Scholar]
- 52. Nemeria N., Baykal A., Joseph E., Zhang S., Yan Y., Furey W., Jordan F. (2004) Tetrahedral intermediates in thiamin diphosphate-dependent decarboxylations exist as a 1′,4′-imino tautomeric form of the coenzyme, unlike the Michaelis complex or the free coenzyme. Biochemistry 43, 6565–6575 [DOI] [PubMed] [Google Scholar]
- 53. Baykal A. T., Kakalis L., Jordan F. (2006) Electronic and nuclear magnetic resonance spectroscopic features of the 1′,4′-iminopyrimidine tautomeric form of thiamin diphosphate, a novel intermediate on enzymes requiring this coenzyme. Biochemistry 45, 7522–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tittmann K., Golbik R., Uhlemann K., Khailova L., Schneider G., Patel M. S., Jordan F., Chipman D. M., Duggleby R. G., Hübner G. (2003) NMR analysis of covalent intermediates in thiamin diphosphate enzymes. Biochemistry 42, 7885–7891 [DOI] [PubMed] [Google Scholar]
- 55. Balakrishnan A., Nemeria N. S., Chakraborty S., Kakalis L., Jordan F. (2012) Determination of pre-steady-state rate constants on the Escherichia coli pyruvate dehydrogenase complex reveals that loop movement controls the rate-limiting step. J. Am. Chem. Soc. 134, 18644–18655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Flournoy D. S., Frey P. A. (1986) Pyruvate dehydrogenase and 3-fluoropyruvate: chemical competence of 2-acetylthiamin pyrophosphate as an acetyl group donor to dihydrolipoamide. Biochemistry 25, 6036–6043 [DOI] [PubMed] [Google Scholar]
- 57. Gruys K. J., Datta A., Frey P. A. (1989) 2-Acetylthiamin pyrophosphate (acetyl-TPP) pH-rate for hydrolysis of acetyl-TPP and isolation of acetyl-TPP as a transient species in pyruvate dehydrogenase catalyzed reactions. Biochemistry 28, 9071–9080 [DOI] [PubMed] [Google Scholar]
- 58. Patel H., Nemeria N. S., Andrews F. H., McLeish M. J., Jordan F. (2014) Identification of charge transfer transitions related to thiamin-bound intermediates on enzymes provides a plethora of a signatures useful in mechanistic studies. Biochemistry 53, 2145–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jordan F., Nemeria N. S., Sergienko E. (2005) Multiple modes of active center communication in thiamin diphosphate-dependent enzymes. Acc. Chem. Res. 38, 755–763 [DOI] [PubMed] [Google Scholar]
- 60. Khailova L. S., Korochkina L. G. (1985) Half-of-the-site reactivity of the decarboxylating component of the pyruvate dehydrogenase complex from pigeon breast muscle with respect to 2-hydroxyethyl thiamin pyrophosphate. Biochem. Int. 11, 509–516 [PubMed] [Google Scholar]
- 61. Nemeria N., Tittmann K., Joseph E., Zhou L., Vazquez-Coll M. B., Arjunan P., Hübner G., Furey W., Jordan F. (2005) Glutamate 636 of the Escherichia coli pyruvate dehydrogenase-E1 participates in active center communication and behaves as an engineered acetolactate synthase with unusual stereoselectivity. J. Biol. Chem. 280, 21473–21482 [DOI] [PubMed] [Google Scholar]
- 62. Frank R. A., Titman C. M., Pratap J. V., Luisi B. F., Perham R. N. (2004) A molecular switch and proton wire synchronize the active sites in thiamin enzymes. Science 306, 872–876 [DOI] [PubMed] [Google Scholar]
- 63. Jordan F. (2004) How active sites communicate in thiamine enzymes. Science 306, 818–820 [DOI] [PubMed] [Google Scholar]
- 64. Reed L. J., Damuni Z., Merryfield M. L. (1985) Regulation of mammalian pyruvate α-keto acid dehydrogenase complexes by phosphorylation-dephosphorylation. Curr. Top. Cell Regul. 27, 41–49 [DOI] [PubMed] [Google Scholar]
- 65. Korotchkina L. G., Patel M. S. (1995) Mutagenesis studies of the phosphorylation sites of recombinant human pyruvate dehydrogenase: site-specific regulation. J. Biol. Chem. 270, 14297–14304 [DOI] [PubMed] [Google Scholar]
- 66. Kolobova E., Tuganova A., Boulatnikov I., Popov K. M. (2001) Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites. Biochem. J. 358, 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Korotchkina L. G., Patel M. S. (2001) Probing the mechanism of inactivation of human pyruvate dehydrogenase by phosphorylation of three sites. J. Biol. Chem. 276, 5731–5738 [DOI] [PubMed] [Google Scholar]
- 68. Korotchkina L. G., Patel M. S. (2001) Site specificity of four pyruvate dehydrogenase kinase isoenzymes toward the three phosphorylation sites of human pyruvate dehydrogenase. J. Biol. Chem. 276, 37223–37229 [DOI] [PubMed] [Google Scholar]
- 69. Kato M., Wynn R. M., Chuang J. L., Tso S.-C., Machius M., Li J., Chuang D. T. (2008) Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops. Structure 16, 1849–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kale S., Arjunan P., Furey W., Jordan F. (2007) A dynamic loop at the active center of the Escherichia coli pyruvate dehydrogenase complex E1 modulates substrate utilization and chemical communication with E2 component. J. Biol. Chem. 282, 28106–28116 [DOI] [PubMed] [Google Scholar]
- 71. Jordan F., Arjunan P., Kale S., Nemeria N. S., Furey W. (2009) Multiple roles of mobile active center loops in the E1 component of the Escherichia coli pyruvate dehydrogenase complex: linkage of protein dynamics to catalysis. J. Mol. Catal. B Enzym. 61, 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kale S., Jordan F. (2009) Conformational ensemble modulates cooperativity in the rate-determining catalytic step in the E1 component of the Escherichia coli pyruvate dehydrogenase multienzyme complex. J. Biol. Chem. 284, 33122–33129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang J., Nemeria N. S., Chandrasekhar K., Kumaran S., Arjunan P., Reynolds S., Calero G., Brukh R., Kakalis L., Furey W., Jordan F. (2014) Structure and function of the catalytic domain of the dihydrolipoyl acetyltransferase component in Escherichia coli pyruvate dehydrogenase complex. J. Biol. Chem. 289, 15215–15230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Frank R. A. W., Pratap J. V., Pei X. Y., Perham R. N., Luisi B. F. (2005) The molecular origins of specificity in the assembly of a multienzyme complex. Structure 13, 1119–1130 [DOI] [PubMed] [Google Scholar]
- 75. Patel M. S., Korotchkina L. G., Sidhu S. (2009) Interaction of E1 and E3 components with the core proteins of the human pyruvate dehydrogenase complex. J. Mol. Catal. B Enzym. 61, 2–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Park Y. H., Wei W., Zhou L., Nemeria N., Jordan F. (2004) Amino-terminal residues 1–45 of the Escherichia coli pyruvate dehydrogenase complex E1 subunit interact with the E2 subunit and are required for activity of the complex but not for reductive acetylation of the E2 subunit. Biochemistry 43, 14037–14046 [DOI] [PubMed] [Google Scholar]
- 77. Song J., Park Y.-H., Nemeria N. S., Kale S., Kakalis L., Jordan F. (2010) Nuclear magnetic resonance evidence for the role of the flexible regions of the E1 component of the pyruvate dehydrogenase complex from Gram-negative bacteria. J. Biol. Chem. 285, 4680–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Park Y. H., Patel M. S. (2010) Characterization of interactions of dihydrolipoamide dehydrogenase with its binding protein in the human pyruvate dehydrogenase complex. Biochem. Biophys. Res. Commun. 395, 416–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nakai T., Kuramitsu S., Kamiya N. (2008) Structural basis for the specific interactions between the E2 and E3 components of the Thermus thermophilus 2-oxo acid dehydrogenase complexes. J. Biochem. 143, 747–758 [DOI] [PubMed] [Google Scholar]
- 80. Mande S. S., Sarfaty S., Allen M. D., Perham R. N., Hol W. G. (1996) Protein-protein interactions in the pyruvate dehydrogenase multienzyme complex: dihydrolipoamide dehydrogenase complexed with the binding domain of dihydrolipoamide acetyltransferase. Structure 4, 277–286 [DOI] [PubMed] [Google Scholar]
- 81. Song J., Jordan F. (2012) Interchain acetyl transfer in the E2 component of bacterial pyruvate dehydrogenase suggests a model with different roles for each chain in a trimer of the homooligomeric component. Biochemistry 51, 2795–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Huang B., Wu P., Bowker-Kinley M. M., Harris R. A. (2002) Regulation of pyruvate dehydrogenase kinase expression by peroxisome proliferator-activated receptor-α ligands, glucocorticoids, and insulin. Diabetes 51, 276–283 [DOI] [PubMed] [Google Scholar]
- 83. Kwon H. S., Huang B., Unterman T. G., Harris R. A. (2004) Protein kinase B-α inhibits human pyruvate dehydrogenase kinase-4 gene induction by dexamethasone through inactivation of FOXO transcription factors. Diabetes 53, 899–910 [DOI] [PubMed] [Google Scholar]
- 84. Reed L. J., Debusk B. G., Gunsalus I. C., Hornberger C. S., Jr. (1951) Crystalline α-lipoic acid: a catalytic agent associated with pyruvate dehydrogenase. Science 114, 93–94 [DOI] [PubMed] [Google Scholar]
- 85. Hitosugi T., Fan J., Chuang T.-W., Lythgoe K., Wang X., Xie J., Ge Q., Gu T.-L., Polakiewicz R. D., Roesel J. L., Chen G. Z., Boggon T. J., Lonial S., Fu H., Khuri F. R., Kang S., Chen J. (2011) Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol. Cell 44, 864–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Fan J., Shan C., Kang H.-B., Elf S., Xie J., Tucker M., Gu T.-L., Aguiar M., Lonning S., Chen H., Mohammadi M., Britton L.-M., Garcia B. A., Alečković M., Kang Y., Kaluz S., Devi N., Van Meir E. G., Hitosugi T., Seo J. H., Lonial S., Gaddh M., Arellano M., Khoury H. J., Khuri F. R., Boggon T. J., Kang S., Chen J. (2014) Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol. Cell 53, 534–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Steussy C. N., Popov K. M., Bowker-Kinley M. M., Sloan R. B., Jr., Harris R. A., Hamilton J. A. (2001) Structure of pyruvate dehydrogenase kinase: novel folding pattern for a serine protein kinase. J. Biol. Chem. 276, 37443–37450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kato M., Chuang J. L., Tso S. C., Wynn R. M., Chuang D. T. (2005) Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex. EMBO J. 24, 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tso S. C., Kato M., Chuang J. L., Chuang D. T. (2006) Structural determinants for cross-talk between pyruvate dehydrogenase kinase 3 and lipoyl domain 2 of the human pyruvate dehydrogenase complex. J. Biol. Chem. 281, 27197–27204 [DOI] [PubMed] [Google Scholar]
- 90. Devedjiev Y., Steussy C. N., Vassylyev D. G. (2007) Crystal structure of an asymmetric complex of pyruvate dehydrogenase kinase 3 with lipoyl domain 2 and its biological implications. J. Mol. Biol. 370, 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kato M., Li J., Chuang J. L., Chuang D. T. (2007) Distinct structural mechanisms for inhibition of pyruvate dehydrogenase kinase isoforms by AZD7545, dichloroacetate, and radicicol. Structure 15, 992–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Green T., Grigorian A., Klyuyeva A., Tuganova A., Luo M., Popov K. M. (2008) Structural and functional insights into the molecular mechanisms responsible for the regulation of pyruvate dehydrogenase kinase 2. J. Biol. Chem. 283, 15789–15798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wynn R. M., Kato M., Chuang J. L., Tso S. C., Li J., Chuang D. T. (2008) Pyruvate dehydrogenase kinase-4 structures reveal a metastable open conformation fostering robust core-free basal activity. J. Biol. Chem. 283, 25305–25315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Turkan A., Gong X., Peng T., Roche T. E. (2002) Structural requirements within the lipoyl domain for the Ca2+-dependent binding and activation of pyruvate dehydrogenase phosphatase isoform 1 or its catalytic subunit. J. Biol. Chem. 277, 14976–14985 [DOI] [PubMed] [Google Scholar]
- 95. Karpova T., Danchuk S., Kolobova E., Popov K. M. (2003) Characterization of the isozymes of pyruvate dehydrogenase phosphatase: implications for the regulation of pyruvate dehydrogenase activity. Biochim. Biophys. Acta 1652, 126–135 [DOI] [PubMed] [Google Scholar]
- 96. Turkan A., Hiromasa Y., Roche T. E. (2004) Formation of a complex of the catalytic subunit of pyruvate dehydrogenase phosphatase isoform 1 (PDP1c) and the L2 domain forms a Ca2+ binding site and captures PDP1c as a monomer. Biochemistry 43, 15073–15085 [DOI] [PubMed] [Google Scholar]
- 97. Vassylyev D. G., Symersky J. (2007) Crystal structure of pyruvate dehydrogenase phosphatase 1 and its functional implications. J. Mol. Biol. 370, 417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]