Background: β-Primeverosidase hydrolyzes disaccharide conjugates in tea leaves and releases aromatic volatiles.
Results: Crystal structures of β-primeverosidase in complex with disaccharide analogues were determined.
Conclusion: β-Primeverosidase has a disaccharide-binding site, and the molecular architecture for disaccharide recognition was elucidated.
Significance: β-Primeverosidase adapts the active site to the 6-O-β-d-xylopyranosyl modification of β-glucoside and contributes multiple volatile emissions in tea leaves.
Keywords: Enzyme Structure, Glycoconjugate, Glycoside Hydrolase, Oligosaccharide, Plant Defense, β-Primeverosidase, Disaccharide, Glycosylamidine, Tea, Volatile Emission
Abstract
β-Primeverosidase (PD) is a disaccharide-specific β-glycosidase in tea leaves. This enzyme is involved in aroma formation during the manufacturing process of oolong tea and black tea. PD hydrolyzes β-primeveroside (6-O-β-d-xylopyranosyl-β-d-glucopyranoside) at the β-glycosidic bond of primeverose to aglycone, and releases aromatic alcoholic volatiles of aglycones. PD only accepts primeverose as the glycone substrate, but broadly accepts various aglycones, including 2-phenylethanol, benzyl alcohol, linalool, and geraniol. We determined the crystal structure of PD complexes using highly specific disaccharide amidine inhibitors, N-β-primeverosylamidines, and revealed the architecture of the active site responsible for substrate specificity. We identified three subsites in the active site: subsite −2 specific for 6-O-β-d-xylopyranosyl, subsite −1 well conserved among β-glucosidases and specific for β-d-glucopyranosyl, and wide subsite +1 for hydrophobic aglycone. Glu-470, Ser-473, and Gln-477 act as the specific hydrogen bond donors for 6-O-β-d-xylopyranosyl in subsite −2. On the other hand, subsite +1 was a large hydrophobic cavity that accommodates various aromatic aglycones. Compared with aglycone-specific β-glucosidases of the glycoside hydrolase family 1, PD lacks the Trp crucial for aglycone recognition, and the resultant large cavity accepts aglycone and 6-O-β-d-xylopyranosyl together. PD recognizes the β-primeverosides in subsites −1 and −2 by hydrogen bonds, whereas the large subsite +1 loosely accommodates various aglycones. The glycone-specific activity of PD for broad aglycone substrates results in selective and multiple release of temporally stored alcoholic volatile aglycones of β-primeveroside.
Introduction
β-Primeverosidase (PD)2 (EC 3.2.1.149) is a diglycosidase (disaccharide-specific β-glycosidase) isolated from an important cash crop for tea production, Camellia sinensis (1, 2). PD hydrolyzes aroma precursors of β-primeverosides (6-O-β-d-xylopyranosyl-β-d-glucopyranosides, Fig. 1A, 1) and liberates alcoholic aroma compounds during the manufacturing process of oolong tea and black tea. Aromatic alcohols (Fig. 1A, A1, 2-phenylethanol; and A2, benzyl alcohol) and monoterpene alcohols (A3, linalool; and A4, geraniol) are naturally occurring aglycones of β-primeverosides in C. sinensis, and are the major aroma constituents of oolong tea and black tea (3, 4). PD was identified in C. sinensis as the specific enzyme to hydrolyze β-primeverosides (5).
FIGURE 1.
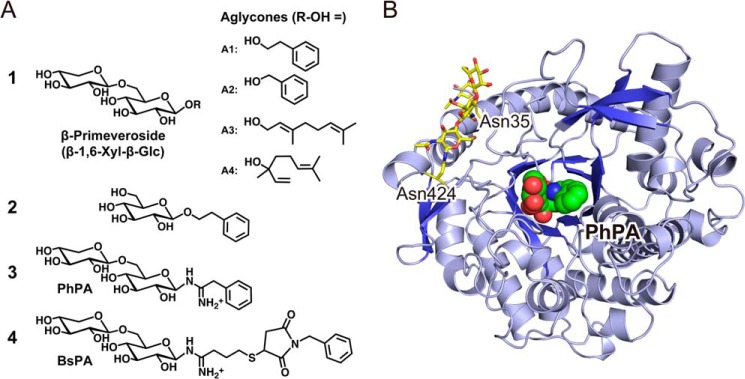
Overview of chemicals and the protein structure. A, chemical structures of substrates and substrate analogue inhibitors for β-primeverosidase (PD). 1, β-primeveroside and four aglycones (R-OH: A1, 2-phenylethanol; A2, benzyl alcohol; A3, geraniol; A4, linalool) are natural substrates for PD. 2, 2-phenylethyl β-d-glucopyranoside is a very poor substrate for PD. 3, PhPA, a N-β-primeverosylamidine derivative with a small aromatic aglycone. 4, BsPA, a derivative with a large aglycone. B, crystal structure of the PD substrate analogue complex. A ribbon representation of PD in complex with PhPA. The green-carbon sphere represents a PhPA molecule in the substrate-binding site at the center of the (β/α)8 barrel. The N-glycosylation at Asn-35 and Asn-424 are shown by the yellow sticks of the carbohydrate chains.
Aglycones of β-primeverosides are benzenoids and monoterpenoids, which are volatile organic compounds with a well known role in the defense of plants against stress and herbivore attacks (6–8). Benzyl alcohol, 2-phenylethanol, linalool, and geraniol have antimicrobial activities against Gram-positive and Gram-negative bacteria and fungi (9–14). Wounding and drought stress of tea leaves raise the levels of these compounds to ∼10- to 40-fold (15). Plucking, withering, and shaking procedures during the manufacture of oolong tea wounds the fresh leaves to secrete volatile compounds as sweet, flower, and honey aromatic flavors. During this process PD hydrolyzes β-primeverosides and releases aromatic aglycones as volatile defensive compounds against stress.
The hydrolytic activity of PD is specific to β-primeverosides, i.e. highly glycone-specific for 6-O-β-d-xylopyranosyl-β-d-glucopyranosyl moiety (β-1,6-Xyl-β-Glc, Fig. 1A, 1). PD has a strict requirement for the β-1,6-Xyl moiety as the β-1,6-linked sugar of the disaccharide glycoside (16). PD barely hydrolyzes other β-glycosides, in which the β-1,6-Xyl group is absent or substituted by other sugars, such as monosaccharide glycosides (β-d-glucopyranoside and β-d-xylopyranoside, etc.) or other disaccharide glycosides, β-vicianosides (6-O-α-l-arabinopyranosyl-β-d-glucopyranoside), β-gentiobiosides (6-O-β-d-glucopyranosyl-β-d-glucopyranoside), and β-acuminosides (6-O-β-d-apiofuranosyl-β-d-glucopyranoside). PD exhibits only one five-hundredth of activity to 2-phenylethyl β-d-glucopyranoside (Fig. 1A, 2) compared with activity to 2-phenylethyl β-primeveroside.
In contrast to strict glycone selectivity, PD broadly accepts various aglycones, 2-phenylethanol, benzyl alcohol, linalool, and geraniol. This loose aglycone specificity is a trait of PD, in contrast to aglycone-specific β-glucosidases in the glycoside hydrolase family 1 (GH1) such as strictosidine β-glucosidase (17, 18) and dhurrin β-glucosidase (19). Molecular cloning and amino acid sequence alignment demonstrated that PD conserves two catalytic glutamates of GH1 β-glucosidases: Glu-203 (acid/base) and Glu-416 (nucleophile), and also conserves 9 residues responsible for β-d-glucopyranoside recognition (2). However, the amino acids crucial for disaccharide specificity and broad aglycone specificity of PD have not been identified from the primary structure.
The substrate specificity of PD raises two basic questions: how does PD recognize the β-1,6-Xyl-β-Glc glycone for specific hydrolysis of β-primeverosides, and how does PD accept four structurally varied aglycones? X-ray crystallography of PD should provide crucial information to elucidate the molecular mechanism of the substrate specificity of PD. We have established large scale expression using a baculovirus-insect cell system to obtain a sufficient amount of PD for crystallization (20). PD is a 61-kDa glycoprotein and its cDNA encodes secretion of a signal peptide of 28 residues at the N-terminal. Recombinant PD has been shown to be active and stable with post-translational modification that involves glycosylation and secretion with cleavage of the signal peptide. A synthetic affinity adsorbent was prepared for purification of PD using N-β-primeverosylamidines as the core structure of the ligand. The affinity chromatography yielded large amounts of PD with a purity sufficiently high for crystallographic study.
N-β-Primeverosylamidine is a derivative of β-glycosylamidine (21–23), which has the same sugar glycone as the natural substrate. N-β-Primeverosylamidines are suitable substrate-analogue inhibitors for probing the substrate recognition of PD because the inhibitory activity of N-β-primeverosylamidines exhibits glycone selectivity and broad aglycone specificity consistent with the substrate specificity of PD. N-β-Primeverosylamidines with two aglycones, 2-phenyl-N-(6-O-β-d-xylopyranosyl-β-d-glucopyranosyl)ethylamidine (Fig. 1A, 3: PhPA) and 4-(N-benzylsuccinimide-3-sulfanyl)-N-(6-O-β-d-xylopyranosyl-β-d-glucopyranosyl)butylamidine (Fig. 1A, 4: BsPA) inhibit PD at Ki = 140 and 26 μm, respectively (20). The monosaccharide glycoside analogue, β-glucosylamidine, has no inhibitory activity at 500 μm, consistent with the fact that PD barely hydrolyzes β-d-glucopyranosides. Therefore, x-ray crystal analysis of PD in complex with N-β-primeverosylamidine should clarify the mechanisms of the unique disaccharide glycone-aglycone recognition of PD.
This article describes studies on three-dimensional structures of PD in apo and complex forms with N-β-primeverosylamidines. We crystallized PD after deglycosidated treatment to avoid heterogeneity, and obtained high-quality crystals. We then elucidated the structural basis of the disaccharide specific and aglycone nonspecific recognition mechanisms of PD in comparison with monosaccharide-specific β-glucosidase structures.
EXPERIMENTAL PROCEDURES
Chemicals
N-β-Primeverosylamidines and affinity adsorbent were prepared by the method as described previously (20, 21).
Production and Purification of PD
The full-length cDNA of PD (GenBankTM, AB088027) including the N-terminal signal sequence was cloned and expressed in a baculovirus-insect cell system as described previously (20). PD was purified by SP-Sepharose cation exchange chromatography and ConA-Sepharose. Affinity chromatography with N-β-primeverosylamidine adsorbent was performed according to the procedure described previously (20). In brief, this included adsorption with 10 mm Tris-HCl (pH 7.0), washing with the same buffer, and elution with 10 mm acetate buffer (pH 4.0). The purified PD (10 mg/ml) was prepared as a stock solution in 10 mm sodium acetate (pH 4.0) at 4 °C for use. The purified PD was treated with α-mannosidase before crystallization.
Crystallization
In initial screening, PD yielded only low quality needle crystals due to structural heterogeneities of post-translational glycosylation. We performed partial digestion of glycans with α-mannosidase for further crystallization trials (20). Deglycosylated PD apo-crystals were grown by the sitting-drop vapor diffusion method at 20 °C, using a mixture of 1.5 μl of protein (10 mg/ml in 10 mm sodium acetate buffer) and 1.5 μl of the reservoir solution containing 0.01% Triton X-100, 21% PEG 4000, 0.3 m ammonium acetate, and 0.1 m sodium citrate (pH 5.5). Cryoprotection was achieved by soaking in 27.5% (w/v) PEG 4000, 0.3 m ammonium acetate, 0.125 m sodium citrate (pH 6.5) for 1 h, followed by freezing in liquid nitrogen. To prepare the PD complexes, the grown crystals were soaked in 30 mm PhPA or 3 mm BsPA of the cryoprotectant solution as described above.
Data Collection and Refinement
Data were collected using an in-house x-ray generator, RIGAKU FR-E equipped with a RAXIS IV detector. Data collection was performed at the wavelength 1.5418 Å, and 0.5° oscillation images of 300-s exposure were collected for 90° (180 images) under 100 K cryo-cooling. Image processing and integrations were performed with MOSFLM (24) and scaled with SCALA, part of the CCP4 package (25). Molecular replacement was performed by MOLREP (26) using the crystal structure of the β-glucosidase of white clover (PDB code 1CBG) as the search model, and two monomers were observed in the asymmetric unit. ARP/wARP (27) was used for automatic model building, with subsequent refinement using the maximum likelihood method in REFMAC5 (28) and PHENIX (29), using hydrogen riding model and isotropic B-factor refinement. Visual inspection of the models was performed using COOT (30). The data collection and refinement statistics are summarized in Table 1. In the refinement process, N- or C-terminal residues and side chains with missing electron density were deleted. One inhibitor was found in each active site, and one PhPA molecule was incidentally found in the solvent region of the PhPA complex crystal. The Ramachandran plot was assessed using RAMPAGE (31). Superposed structures were calculated by LSQKAB of CCP4 (25). The figures were prepared using PyMOL (Schrödinger, LLC) (32).
TABLE 1.
Data quality and refinement statistics for the crystal structure of apo-PD, complex with PhPA, and complex with BsPA
| Apo form | Complex with PhPA | Complex with BsPA | |
|---|---|---|---|
| Date collection | |||
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 59.6, 88.8, 195.2 | 60.0, 88.2, 195.6 | 59.1, 89.9, 195.2 |
| α = β = γ (°) | 90 | 90 | 90 |
| Resolution range (Å) | 40.4–1.9 (2.0–1.9) | 50.0–1.8 (1.9–1.8) | 50–1.8 (1.9–1.8) |
| Rmerge (%) | 6.6 (27.1) | 5.4 (28.7) | 6.8 (25.7) |
| I/σI | 16.4 (4.3) | 18.9 (4.7) | 15.7 (4.6) |
| Completeness (%) | 99.6 (100) | 96.8 (94.3) | 100 (100) |
| Multiplicity | 3.5 (3.5) | 3.7 (3.7) | 3.6 (3.5) |
| Refinement | |||
| Resolution (Å) | 1.9 | 1.8 | 1.8 |
| R/Rfree | 0.159/0.195 | 0. 170/0.192 | 0.155/0.183 |
| No. of atoms | |||
| Protein | 7563 | 7589 | 7612 |
| Inhibitor | 90 | 82 | |
| N-Glycan | 70 | 81 | 105 |
| Water | 1295 | 1390 | 1781 |
| Average B-factors | |||
| Overall | 21.7 | 20.2 | 15.6 |
| Protein | 19.1 | 18.2 | 12.7 |
| Inhibitor | 18.6, 27.0a | 14.5 | |
| N-Glycan | 43.2 | 41.7 | 35.5 |
| Water | 31.0 | 30.1 | 26.8 |
| Ramachandran plot analysis | |||
| Favored | 922 | 923 | 924 |
| Allowed | 23 | 22 | 23 |
| Outliers | 0 | 0 | 0 |
| Root mean square deviation | |||
| Bond length (Å) | 0.007 | 0.006 | 0.010 |
| Bond angles (°) | 1.05 | 1.02 | 1.12 |
| PDB code | 3WQ4 | 3WQ5 | 3WQ6 |
a The B-factor of the PhPA molecule in the solvent region.
RESULTS
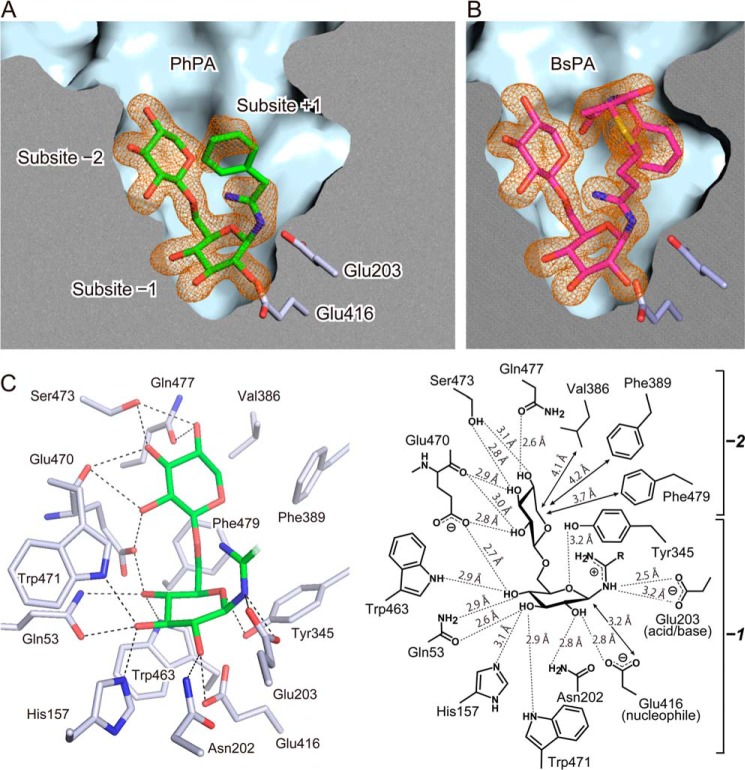
The crystal structures of PD were determined in apo and in complexes with N-β-primeverosylamidine PhPA and BsPA at resolutions of 1.9, 1.8, and 1.8 Å, respectively. The protein-fold of PD is a classical (β/α)8 barrel, and PhPA/BsPA were located in the substrate-binding site at the center of the barrel (Fig. 1B). The electron densities of PhPA and BsPA were observed in both complex crystals (Fig. 2, A and B), including the β-1,6-Xyl-β-Glc moiety as well as a large aglycone moiety of BsPA. The phenyl group (aglycone) of PhPA showed ambiguous Fo − Fc electron densities, and therefore the phenyl group was put in a position that partially fitted to the electron densities. The 2Fo − Fc map of the phenyl ring was ambiguous even after refinement, and the ambiguity was the same in each monomer in the asymmetric unit. There was no significant change in the (β/α)8-fold and loops among the apo and two complex structures.
FIGURE 2.
Tight binding of the disaccharide in the deep active site. A and B, the substrate-binding site composed of three subsites (−2, −1, and +1) in complex with the substrate analogue inhibitors of PhPA and BsPA with electron density. Cross-section of the substrate-binding site with the protein surface. The green and magenta sticks indicate the bound PhPA (panel A) and BsPA (panel B), respectively. The catalytic acid/base (Glu-203) and nucleophile (Glu-416) residues are shown as white sticks. The Fo − Fc omit map electron densities of PhPA and BsPA are in orange mesh contoured at 3.0 σ. C, disaccharide glycone recognition architecture in subsites −2 and −1. The model represents β-1,6-Xyl-β-Glc and amidine moiety (green stick) of PhPA. Side chains of amino acids responsible for disaccharide recognition are shown by the white stick model, whereas the dashed line represents hydrogen bonds between amino acids and PhPA. Schematic diagram represents contacts and distance between BsPA and amino acids.
Electron densities of post-translationally modified glycans were found at two NX(S/T) sites, Asn-35 and Asn-424. One GlcNAc was attached to Asn-424 by N-β-glycosidic bonds, whereas dimeric GlcNAc with β-1,4-link were attached to Asn-35. The core GlcNAc on Asn-35 was further modified with a α-1,3-linked fucose by the N-glycosylation system of secretion proteins in insect cells (33).
Architectures of the Substrate-binding Site
The substrate-binding site can be partitioned into three subsites (Fig. 2A); binding sites for the β-1,6-Xyl moiety (subsite −2), β-Glc moiety (subsite −1), and aglycone (subsite +1). The substrate-binding site was shaped like a funnel and was ∼18 Å deep, with the entrance of the pocket being 14 Å. The pocket had a large entrance at the surface of PD, and its inner space gradually narrowed toward the bottom. PhPA and BsPA were bound in the pocket by a sharp bend of the β-Glc moiety at the bottom of the pocket, with the β-1,6-Xyl moiety extending toward the entry of the pocket. The catalytic resides Glu-203 (acid/base) and Glu-416 (nucleophile) were located at the bottom of the pocket.
The structure of PhPA and BsPA showed well defined hydrogen bonding networks in subsites −2 and −1 (Fig. 2C). The heteroatoms of N-β-primeverosylamidines, oxygen of the β-1,6-Xyl-β-Glc moiety, and nitrogen of the amidino group had well defined electron densities. All the hydroxy groups of the β-1,6-Xyl-β-Glc moiety formed hydrogen bonds with amino acids in subsites −2 and −1. All contacts and their distance are shown in a schematic diagram (Fig. 2C). The conformation of β-1,6-Xyl was in a relaxed 4C1 chair in subsite −2, whereas that of β-Glc in subsite −1 was distorted. The β-d-glucopyranose ring was in an 4E-like conformation between 4C1 and 4E, in which the anomeric carbon C1 was close to the plane of the glucopyranose ring. This distortion was probably induced by the 3.2-Å contact of the nucleophilic oxygen of Glu-416. Thus, this conformation is likely to reflect an early state structure of the ES complex formation, preceding the nucleophilic attack occurring in the 4E transition state.
Disaccharide Glycone Recognition in Subsite −2 and Subsite −1
Subsite −2 held the β-1,6-Xyl moiety by six amino acids, Glu-470, Ser-473, and Gln-477 with hydrogen bonds and Val-386, Phe-389, and Phe-479 with hydrophobic contacts (Fig. 2C). These six residues were almost certainly responsible for the strict recognition of the β-1,6-Xyl moiety in the disaccharide specificity of PD. Glu-470 made hydrogen bonds with the carboxylate oxygen of the side chain and the amide carbonyl oxygen of the main chain to 2-OH and 3-OH of the β-1,6-Xyl moiety. Ser-473 was hydrogen bonded to 3-OH and 4-OH by a side chain hydroxy at 3.1 and 2.8 Å distance, respectively, whereas Gln-477 was hydrogen bonded with 4-OH by the side chain oxygen. Val-386, Phe-389, and Phe-479 covered the hydrophobic surface (C5) of the β-1,6-Xyl moiety with van der Waals interactions at 4.1, 4.2, and 3.7 Å, respectively. Other interactions, such as stacking with aromatic amino acid residues, was not observed in subsite −2.
Subsite −1, the binding site for the β-Glc moiety, consisted of 8 amino acid residues including the catalytic glutamates, Glu-203 and Glu-416 as the acid/base and nucleophile, respectively (Fig. 2C). The side chains of Gln-53, His-157, Asn-202, Tyr-345, Glu-416, Trp-463, Glu-470, and Trp-471 were hydrogen bonded with the hydroxy groups of the β-Glc moiety. Asn-202 and Glu-416 were bonded with 2-OH at a distance of 2.7 and 3.2 Å, and Gln-53 and His-157 with 3-OH at a distance of 2.6 and 3.0 Å, respectively. Gln-53 also formed a hydrogen bond between its NH and 4-OH at 2.9 Å. Trp-463, Glu-470, and Trp-471 were bonded with 4-OH at hydrogen bonding distances. The aromatic face of Trp-463 was adjacent to the C3 and C5 of β-Glc. These interactions were almost conserved to that reported in GH1 β-glucosidases, except for Glu-470. Glu-470 was located between subsites −2 and −1, and its oxygen atom was hydrogen bonded, not only with 4-OH of β-Glc but also with 2-OH of β-1,6-Xyl in subsite −2.
Interactions between disaccharide and amino acids in subsites −2 and −1 were identical in the complex structures of PhPA and BsPA. Subsites −2 and −1 recognize the β-1,6-Xyl-β-Glc moiety in the same manner, independent of the aglycone structures. The amidino group of N-β-primeverosylamidines and Glu-203 formed electrostatic interactions (Fig. 2C), which probably enhanced the affinities of N-β-primeverosylamidines in addition to the hydrogen bonds of the β-1,6-Xyl-β-Glc moiety.
The Binding of Aglycone in Subsite +1
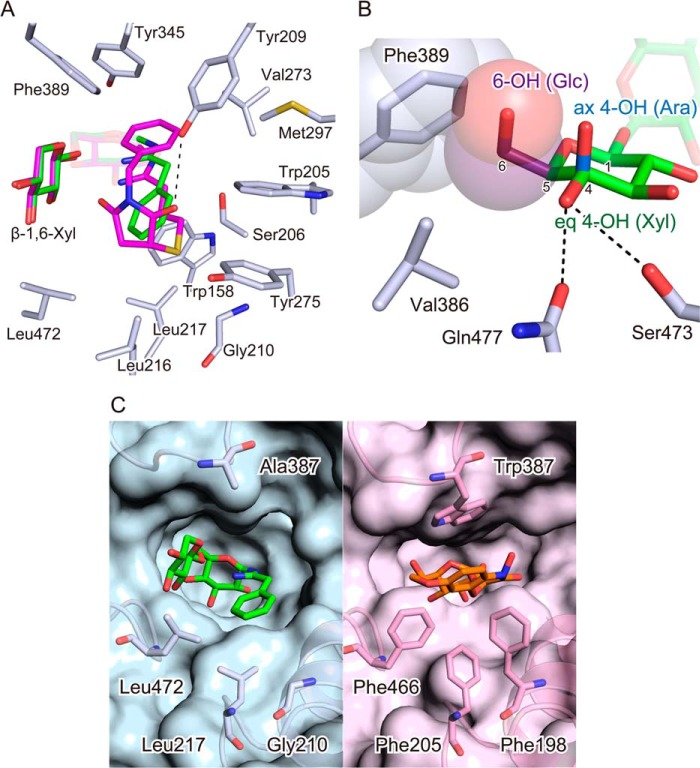
The aglycone moiety of BsPA showed clear electron density in subsite +1 (Fig. 2B). Subsite +1 formed a sufficiently large space to accept the bulky, artificial aglycone of BsPA. The aglycone includes spacer (CH2)3S and linker (N-benzyl succinimide) structures as synthetic connections used in the preparation on the affinity absorbent (21) and is therefore larger than the natural aglycones of β-primeverosides. In subsite +1, the spacer (CH2)3S led to the entry of the substrate binding pocket, with the N-benzyl turned into the pocket by bending at the succinimide. The cavity of subsite +1 was filled with the large aglycone of BsPA.
The aglycone of BsPA was surrounded by 13 amino acid residues of subsite +1 at distances within 4.5 Å; Trp-158, Trp-205, Ser-206, Tyr-209, Gly-210, Leu-216, Leu-217, Val-273, Tyr-275, Met-297, Tyr-345, Phe-389, and Leu-472 (Fig. 3A). The hydrophobic side of the pyranose ring of β-1,6-Xyl in subsite −2 faces subsite +1. The mostly hydrophobic amino acids in subsite +1 and the hydrophobic side of β-1,6-Xyl ring form a highly hydrophobic environment in subsite +1 to accept hydrophobic natural monoterpene and aromatic aglycones of β-primeverosides. In contrast to the aglycone of BsPA that fills subsite +1, there was a large gap around the phenyl aglycone of PhPA, suggesting that the binding of the small aglycone is loose. The carbonyl oxygen of the succinimide moiety of BsPA formed a hydrogen bond with Tyr-209, consistent with the higher inhibitory activity of BsPA compared with PhPA (20).
FIGURE 3.
Aglycones and β-1,6-Xyl at the entry of the active site. A, broad aglycone recognition residues of subsite +1 with superimposed PhPA and BsPA. The amino acids of subsite +1, which are close to the aglycone moiety of BsPA within 4.5 Å, are represented by the white stick model. The green and magenta sticks represent PhPA and BsPA, respectively. The dashed line represents a hydrogen bond between Tyr-209 and the succinimide moiety of BsPA. B, disaccharide specificity determinant residues in subsite −2 with bound β-1,6-Xyl moiety of PD complex structure accompanied with modeled 6-O-α-l-arabinopyranosyl (Ara) and 6-O-β-d-glucopyranosyl (Glc). The green stick represents the β-1,6-Xyl of the bound BsPA in the complex structure. The hydrogen bonds of equatorial 4-hydroxy of β-1,6-Xyl moiety (eq 4-OH) with Ser-423 and Gln-427 are represented by the dashed line. The blue stick is the axial 4-hydroxy of α-1,6-l-Ara (ax 4-OH) in β-vicianoside. The purple stick is the 5-hydroxymethyl of β-1,6-Glc (5-CH2OH) in β-gentiobioside. The sphere models indicate steric hindrance of 5-CH2OH by less distance than van der Waals radii to Phe-389. C, wide and narrow subsite +1 in aglycone-nonspecific PD and aglycone-specific DIMBOA β-glucosidase, respectively. The accessible surface area of substrate-binding sites in crystal structures of PD (cyan, left panel) and DIMBOA (2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one) β-glucosidase (pink, right panel) are shown, with important amino acid resides in subsite +1 shown by the stick model. The bound PhPA is represented by the green stick model. The DIMBOA β-glucoside is shown by the orange stick.
DISCUSSION
The substrate specificity of PD has three properties (16): (i) strict selectivity for β-1,6-Xyl as a β-1,6-linked sugar moiety of β-primeveroside, (ii) broad aglycone specificity, and (iii) rather low activities for β-glucosides. In this study, we determined the crystal structure of PD in complexes with substrate analogues, and investigated the substrate-binding site to determine the amino acid residues responsible for substrate recognition. Based on the crystal structures, we formulated a structural basis for the substrate specificities of PD.
The Selective Binding of β-1,6-Linked Sugar Moiety of Disaccharide Glycosides in Subsite −2
The β-1,6-Xyl of N-β-primeverosylamidine was bound to subsite −2 and its three hydroxy groups formed hydrogen bonds with Glu-470, Ser-473, and Gln-477. In addition, Val-386, Phe-389, and Phe-479 formed hydrophobic interactions with C5 of the xylopyranose ring. These interactions were consistent with the high activity of PD for β-primeveroside and consistent with the lower or absence of activity for other disaccharide glycosides with different β-1,6-linked sugars. PD exhibits one-fifth of the activity to β-vicianoside compared with β-primeveroside, with the Km value for β-vicianoside being seven times greater than that for β-primeveroside (16). The structural difference between β-primeveroside and β-vicianoside is the stereochemistry of the β-1,6-linked sugars, 6-O-β-d-xylopyranosyl and 6-O-α-l-arabinopyranosyl (α-1,6-l-Ara). That is, these two disaccharide-glycosides are R/S-isomers on the C4 carbon of the pyranose ring, and the isomeric C4 results in different orientations of 4-OH in a 4C1 chair conformation; equatorial in β-1,6-Xyl and axial in α-1,6-l-Ara. The crystal structure in the complex with N-β-primeverosylamidine showed that the equatorial 4-OH of the β-1,6-Xyl moiety formed hydrogen bonds with Ser-473 and Gln-477 in the α-face of the pyranose ring (Fig. 3B). In contrast, the axial 4-OH of α-1,6-l-Ara would locate away from the α-face of the 4C1 pyranose ring, suggesting that Ser-473 and Gln-477 do not form hydrogen bonds with 4-OH of α-1,6-l-Ara. The lack of interactions between Ser-473, Gln-477, and the 4-OH of α-1,6-l-Ara reduce the affinity for β-vicianosides.
PD exhibits 1/20th of the activity to β-gentiobioside compared with β-primeveroside (16). β-Gentiobioside has 6-O-β-d-glucopyranosyl (β-1,6-Glc) as its β-1,6-linked sugar and is therefore the 5-hydroxymethyl (5-CH2OH) derivative of β-primeveroside. In the crystal structure, the C5 of the β-1,6-Xyl moiety was covered by Val-386, Phe-389, and Phe-479 in subsite −2. These hydrophobic residues would not be suitable for accepting the hydrophilic 5-CH2OH of β-1,6-Glc, and simultaneously, the carbon atom of the 5-CH2OH crashes with Phe-389 because it overlaps with Phe-389 at 3.1 Å in our model (3.4 Å is the limit of the van der Waals radius) (Fig. 3B). This steric hindrance probably causes the low activity of PD for β-gentiobioside.
Loose Aglycone Recognition in Subsite +1
The crystal structure in the complex with BsPA showed that subsite +1 forms a large hydrophobic cavity to accept naturally occurring aglycones. To investigate the structural basis and differences between loose aglycone recognition of PD and specific aglycone recognition of GH1 β-glucosidases, we compared our crystal structure with β-glucosidase from Zea mays (DG) because aglycone binding of this enzyme is well studied in complexes with DIMBOA (2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one) β-d-glucopyranoside (PDB code 1E56) (34). The barrel-fold of PD was very similar to that of DG with an overall root mean square deviation of 1.06 Å in the superimposed structure. The subsite +1 of DG has Phe-198, Phe-205, Trp-378, and Phe-466 as crucial residues for aglycone binding. These sites are also involved in aglycone binding of dhurrinase reported with high resolution structures (19). The corresponding amino acid residues of PD were investigated in the crystal structure of subsite +1.
Gly-210, Leu-217, Ala-387, and Leu-472 were identified in PD, and corresponded to the aglycone-recognizing residues of DG, Phe-198, Phe-205, Trp-378, and Phe-466 (34), respectively (Fig. 3C). The residues in PD (Gly, Leu, and Ala) were sterically smaller than the corresponding residues of DG (Phe, Trp), and these smaller residues formed a large cavity in subsite +1 of PD. In particular, Ala-387 is remarkable because the corresponding amino acid in DG is Trp, a residue that is conserved in almost all GH1 β-glucosidases (35) and directly interacts with aglycones in crystal structures of plant GH1 β-glucosidases (17–19, 34, 36). These results suggest that the large cavity with the smaller residue is the basis for the broad aglycone specificity of PD, and simultaneously suggest that the large cavity allows acceptance of aglycones with bulky disaccharide-glycone, which could not be accommodated in the narrow entry cleft of DG.
Substrate Recognition of PD
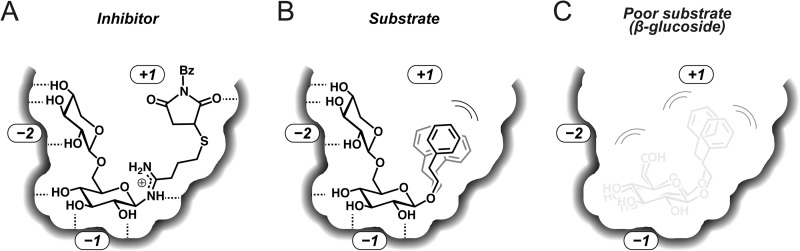
A scheme for the substrate recognition mechanism indicated by the crystal structure is summarized in Fig. 4. PD recognizes the β-1,6-Xyl-β-Glc (β-primeverosyl) moiety of subsites −2 and −1 by hydrogen bonds with polar residues (Fig. 4A). Subsite −2 is attached tightly to the β-1,6-Xyl moiety and disfavors other β-1,6-linked sugars. Subsite +1 is sufficiently large to accept various naturally occurring aglycones of β-primeverosides, and also accommodates the extremely bulky artificial aglycone of BsPA in the crystal structure.
FIGURE 4.
Schematic representations of substrate recognition with BsPA as a bulky substrate analogue inhibitor, 2-phenylethyl β-primeveroside as a small aglycone substrate, and 2-phenylethyl β-d-glucopyranoside as a monosaccharide poor substrate. A, subsite −2 and −1 hold the disaccharide glycone of N-β-primeverosylamidine tightly with many hydrogen bonds (dashed line), and subsite +1 being sufficiently large to accommodate the bulky aglycone of BsPA. B, subsite −2 and −1 hold the glycone of β-primeveroside, and the large subsite +1 accepts the small aglycone nonspecifically. C, PD cannot hold monosaccharide β-glucoside with no interaction in subsite −2 nor Trp residue of subsite +1.
As observed in the ambiguous electron density of phenyl group of PhPA, aglycone binding in subsite +1 is loose. PD recognizes β-primeveroside by subsites −2 and −1, with the binding of the β-1,6-Xyl-β-Glc moiety being sufficiently stable to fix the position of the substrate to the catalytic position even in the absence of specific interactions with aglycone (Fig. 4B). The β-1,6-Xyl-β-Glc of PhPA and BsPA had exactly the same structure despite varied aglycone structures. On the other hand, aglycones should associate nonspecifically with hydrophobic amino acids and the hydrophobic side of β-1,6-Xyl in subsite +1. The nonspecific association could enhance the specific polar interaction of β-1,6-Xyl in subsite −2, consistent with the rather potent inhibition (Ki = 26 μm) of BsPA filling subsite +1 with the large aglycone. The fact that the hydrophobic side of β-1,6-Xyl faces the aglycone suggests that subsites −2 and +1 could bind substrate in a concerted manner. The homology modeling of a diglycosdase cloned from Dalbergia nigrescens expected that the 6-O-linked sugar would be faced against the aglycone (37).
Monosaccharide β-glucosides are poor substrates for PD for two reasons: the absence of a β-1,6-Xyl moiety, and loose aglycone recognition in subsite +1 (Fig. 4C). Although PD conserves binding residues for β-Glc in subsite −1, it probably requires an additional interaction to fix the position of β-glucosides for catalysis. The aglycone binding in subsite +1 contributes to glycone binding in β-glucoside hydrolysis of GH1 β-glucosidases. Site-directed mutagenesis of GH1 β-glucosidases (38, 39) resulted in mutations of Trp in subsite +1 that markedly reduced activities and modified glycone specificity. Trp is missing in subsite +1 of PD, and therefore PD barely hydrolyzes β-glucosides, whereas the alternative small residue (Gly, Ala, and Leu) forms a large hydrophobic cavity to accept various aglycones and β-1,6-Xyl moiety. In the absence of β-1,6-Xyl, subsite +1 would lose the hydrophobic wall composed of β-1,6-Xyl, and the resultant large cavity would disfavor the binding of β-glucosides by diminishing the binding stability.
The glycone-specific and aglycone broadly specific activity of PD allows selective and multiple activation of temporally stored aglycones of β-primeveroside. The loose recognition of aglycone in subsite +1 can activate and release all four volatile compounds. This multiple emission can be more effective than single emission in defensive roles because aglycones have different antimicrobial spectra (9–14). Little or no activity of PD for β-glucosides is also important because phytohormones and toxic compounds are stored as glucose conjugates in the cell, and their activation is regulated by specific β-glucosidases (40–44). PD would barely hydrolyze these glucose conjugates even in the mixture of compounds in broken cells of wounded leaves, and avoids unnecessary activation of phytohormones. The system with β-primeveroside and PD would be efficient for controlling defensive volatile emission because PD can activate multiple volatile compounds by one-step specific hydrolysis of the β-glycosidic bond of β-primeverosides.
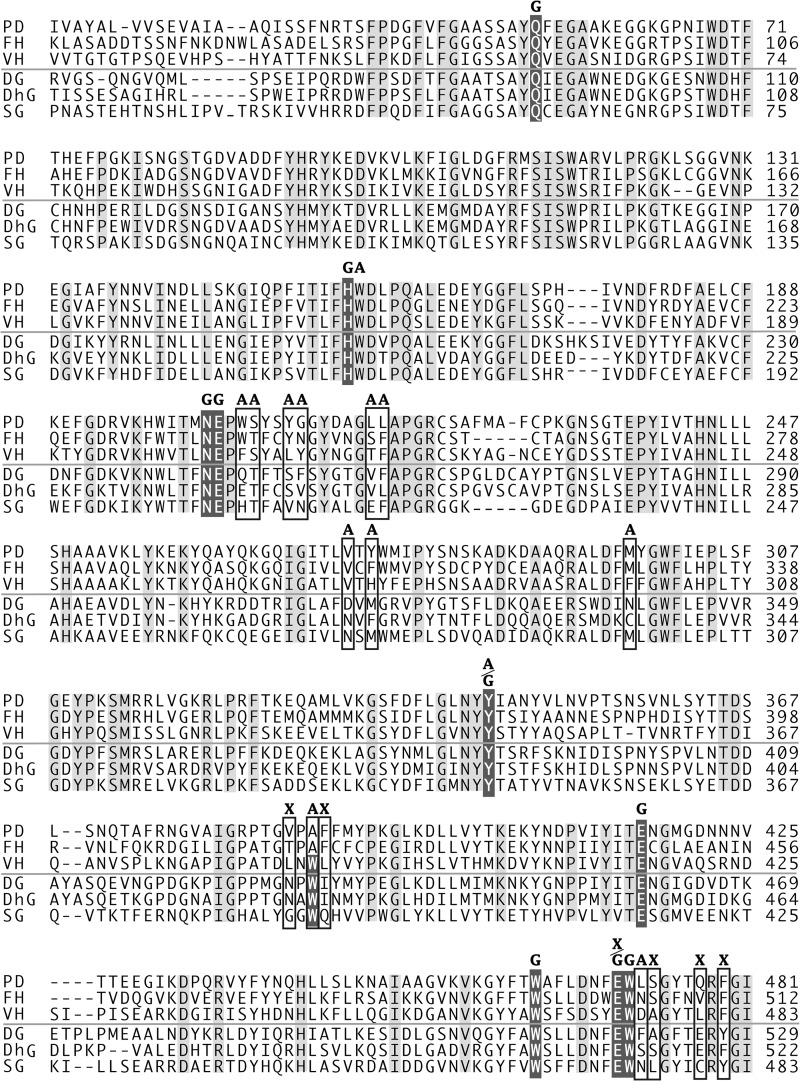
The substrate recognizing residues of subsite −2 and subsite +1 are not conserved in the other two disaccharide-specific glycosidases, furcatin hydrolase (FH) (45) and vicanin hydrolase (VH) (46). The sequence alignment of disaccharide-specific glycosidase (Fig. 5) indicates that subsites −2 and +1 are located at variable positions in GH1 sequences, consistent with their varied glycone/aglycone specificity. FH and VH are specific for both glycones and aglycones of their natural substrates, p-allylphenyl 6-O-β-d-apiofuranosyl-β-d-glucopyranosides (furcatin) and (R)-mandelonitrile 6-O-α-l-arabinopyranosyl-β-d-glucopyranoside (vicianin), respectively. The amino acid residues at the varied position probably form different architecture in the substrate-binding sites of FH and VH (47).
FIGURE 5.
Amino acid sequence alignment of disaccharide-specific glycosidases and aglycone-specific GH1 β-glucosidases from plants. The three sequences above the line are disaccharide-specific glycosidases, and the sequences below are GH1 β-glucosidases. PD, β-primeverosidase from C. sinensis; FH, furcatin hydrolase from Viburnum furcatum; VH, vicianin hydrolase from Vicia angustifolia; DG, DIMBOA β-glucosidase from Zea mays; DhG, dhurrinase, β-glucoside for (S)-4-hydroxymandelnitrile-β-d-glucopyranoside from Sorghum bicolor; SG, strictosidine β-glucosidase from Rauvolfia serpentina. The labels indicate amino acid residues of the substrate-binding site of PD with label X in subsite −2, G in subsite −1, and A in subsite +1, respectively. Boxes indicate varied residues in the substrate-binding site. The shaded white character indicates highly conserved glucose binding residues in GH1 β-glucosidase. The light shadow character indicates conserved residues in the aligned sequences. The region 15–481 including substrate-binding site is shown in 507 amino acids of PD.
Acknowledgment
We thank Prof. Masashi Miyano for critical reading of the manuscript and help with figure preparation.
This work was supported in part by Ministry of Education, Culture, Sports, Science, and Technology of Japan Grants-in-Aids 13460049 and 16380079 and the MEXT (Ministry of Education, Culture, Sports, Science and Technology)-supported Program for the Strategic Research Foundation at Private Universities (2013–2017).
The atomic coordinates and structure factors (codes 3WQ4, 3WQ5, and 3WQ6) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- PD
- β-primeverosidase
- FH
- furcatin hydrolase
- VH
- vicianin hydrolase
- DG
- (2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one)β-glucoside hydrolase
- Xyl
- d-xylopyranosyl
- Glc
- d-glucopyranosyl
- Ara
- l-arabinopyranosyl
- GH1
- glycoside hydrolase family 1
- DIMBOA
- 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one
- PhPA
- 2-phenyl-N-(6-O-β-d-xylopyranosyl-β-d-glucopyranosyl)ethylamidine
- BsPA
- 4-(N-benzylsuccinimide-3-sulfanyl)-N-(6-O-β-d-xylopyranosyl-β-d-glucopyranosyl)butylamidine.
REFERENCES
- 1. Sakata K., Mizutani M., Ma S. J., Hiratake J. (2003) Diglycoside-specific glycosidases. Methods Enzymol. 363, 444–459 [DOI] [PubMed] [Google Scholar]
- 2. Mizutani M., Nakanishi H., Ema J., Ma S. J., Noguchi E., Inohara-Ochiai M., Fukuchi-Mizutani M., Nakao M., Sakata K. (2002) Cloning of β-primeverosidase from tea leaves, a key enzyme in tea aroma formation. Plant Physiol. 130, 2164–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guo W., Hosoi R., Sakata K., Watanabe N., Yagi A., Ina K., Luo S. (1994) (S)-Linalyl, 2-phenylethyl, and benzyl disaccharide glycosides isolated as aroma precursors from oolong tea leaves. Biosci. Biotechnol. Biochem. 58, 1532–1534 [DOI] [PubMed] [Google Scholar]
- 4. Guo W., Sakata K., Watanabe N., Nakajima R., Yagi A., Ina K., Luo S. (1993) Geranyl 6-O-β-d-xylopyranosyl-β-d-glucopyranoside isolated as an aroma precursor from tea leaves for oolong tea. Phytochemistry 33, 1373–1375 [DOI] [PubMed] [Google Scholar]
- 5. Guo W., Yamauchi K., Watanabe N., Usui T., Luo S., Sakata K. (1995) A primeverosidase as a main glycosidase concerned with the alcoholic aroma formation in tea leaves. Biosci. Biotechnol. Biochem. 59, 962–964 [Google Scholar]
- 6. Gershenzon J., Dudareva N. (2007) The function of terpene natural products in the natural world. Nat. Chem. Biol. 3, 408–414 [DOI] [PubMed] [Google Scholar]
- 7. Dudareva N., Negre F., Nagegowda D. A., Orlova I. (2006) Plant volatiles: recent advances and future perspectives. Crit. Rev. Plant Sci. 25, 417–440 [Google Scholar]
- 8. Kessler A., Baldwin I. T. (2001) Defensive function of herbivore-induced plant volatile emissions in nature. Science 291, 2141–2144 [DOI] [PubMed] [Google Scholar]
- 9. Zhu Y. J., Zhou H. T., Hu Y. H., Tang J. Y., Su M. X., Guo Y. J., Chen Q. X., Liu B. (2011) Antityrosinase and antimicrobial activities of 2-phenylethanol. Food Chem. 124, 298–302 [Google Scholar]
- 10. Meyer B. K., Ni A., Hu B., Shi L. (2007) Antimicrobial preservative use in parenteral products: past and present. J. Pharm. Sci. 96, 3155–3167 [DOI] [PubMed] [Google Scholar]
- 11. Utama I. M., Wills R. B., Ben-Yehoshua S., Kuek C. (2002) In vitro efficacy of plant volatiles for inhibiting the growth of fruit and vegetable decay microorganisms. J. Agric. Food Chem. 50, 6371–6377 [DOI] [PubMed] [Google Scholar]
- 12. Griffin S. G., Wyllie S. G., Markham J. L., Leach D. N. (1999) The role of structure and molecular properties of terpenoids in determining their antimicrobial activity. Flavour Frag. J. 14, 322–332 [Google Scholar]
- 13. Kim J., Marshall M. R., Wei C. I. (1995) Antibacterial activity of some essential oil components against five foodborne pathogens. J. Agric. Food. Chem. 43, 2839–2845 [Google Scholar]
- 14. Berrah G., Konetzka W. A. (1962) Selective and reversible inhibition of the synthesis of bacterial deoxyribonucleic acid by phenethyl alcohol. J. Bacteriol. 83, 738–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cho J. Y., Mizutani M., Shimizu B., Kinoshita T., Ogura M., Tokoro K., Lin M. L., Sakata K. (2007) Chemical profiling and gene expression profiling during the manufacturing process of Taiwan oolong tea “oriental beauty”. Biosci. Biotechnol. Biochem. 71, 1476–1486 [DOI] [PubMed] [Google Scholar]
- 16. Ma S. J., Mizutani M., Hiratake J., Hayashi K., Yagi K., Watanabe N., Sakata K. (2001) Substrate specificity of β-primeverosidase, a key enzyme in aroma formation during oolong tea and black tea manufacturing. Biosci. Biotechnol. Biochem. 65, 2719–2729 [DOI] [PubMed] [Google Scholar]
- 17. Xia L., Ruppert M., Wang M., Panjikar S., Lin H., Rajendran C., Barleben L., Stöckigt J. (2012) Structures of alkaloid biosynthetic glucosidases decode substrate specificity. ACS Chem. Biol. 7, 226–234 [DOI] [PubMed] [Google Scholar]
- 18. Barleben L., Panjikar S., Ruppert M., Koepke J., Stöckigt J. (2007) Molecular architecture of strictosidine glucosidase: the gateway to the biosynthesis of the monoterpenoid indole alkaloid family. Plant Cell 19, 2886–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verdoucq L., Morinière J., Bevan D. R., Esen A., Vasella A., Henrissat B., Czjze M. (2004) Structural determinants of substrate specificity in family 1 β-glucosidases: novel insights from the crystal structure of sorghum dhurrinase-1, a plant β-glucosidase with strict specificity, in complex with its natural substrate. J. Biol. Chem. 279, 31796–31803 [DOI] [PubMed] [Google Scholar]
- 20. Saino H., Mizutani M., Hiratake J., Sakata K. (2008) Expression and biochemical characterization of β-primeverosidase and application of β-primeverosylamidine to affinity purification. Biosci. Biotechnol. Biochem. 72, 376–383 [DOI] [PubMed] [Google Scholar]
- 21. Inoue K., Hiratake J., Mizutani M., Takada M., Yamamoto M., Sakata K. (2003) β-Glycosylamidine as a ligand for affinity chromatography tailored to the glycon substrate specificity of β-glycosidases. Carbohydr. Res. 338, 1477–1490 [DOI] [PubMed] [Google Scholar]
- 22. Hiratake J., Sakata K. (2003) Glycosylamidines as potent selective and easily accessible glycosidase inhibitors and their application to affinity chromatography. Methods Enzymol. 363, 421–444 [DOI] [PubMed] [Google Scholar]
- 23. Guo W., Hiratake J., Ogawa K., Yamamoto M., Ma S. J., Sakata K. (2001) β-d-Glycosylamidines: potent, selective, and easily accessible β-glycosidase inhibitors. Bioorg. Med. Chem. Lett. 11, 467–470 [DOI] [PubMed] [Google Scholar]
- 24. Powell H. R. (1999) The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 55, 1690–1695 [DOI] [PubMed] [Google Scholar]
- 25. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vagin A., Teplyakov A. (1997) MOLREP: an Automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 27. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Automated macromolecular model building for X-ray crystallography using ARP/warp version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 32. DeLano W. L. (2002) PyMol: an open-source molecular graphics tool. CCP4. Newslett. Protein Crystallogr. 40, 44–53 [Google Scholar]
- 33. Altmann F., Staudacher E., Wilson I. B., März L. (1999) Insect cells as hosts for the expression of recombinant glycoproteins. Glycoconj. J. 16, 109–123 [DOI] [PubMed] [Google Scholar]
- 34. Czjzek M., Cicek M., Zamboni V., Bevan D. R., Henrissat B., Esen A. (2000) The mechanism of substrate (aglycone) specificity in β-glucosidases is revealed by crystal structures of mutant maize β-glucosidase-DIMBOA, -DIMBOAGlc, and -dhurrin complexes. Proc. Natl. Acad. Sci. U.S.A. 97, 13555–13560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ketudat Cairns J. R., Esen A. (2010) β-Glucosidases. Cell Mol. Life Sci. 67, 3389–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sue M., Nakamura C., Miyamoto T., Yajima S. (2011) Active-site architecture of benzoxazinone-glucoside β-d-glucosidases in Triticeae. Plant Sci. 180, 268–275 [DOI] [PubMed] [Google Scholar]
- 37. Chuankhayan P., Rimlumduan T., Tantanuch W., Mothong N., Kongsaeree P. T., Metheenukul P., Svasti J., Jensen O. N., Cairns J. R. (2007) Functional and structural differences between isoflavonoid β-glycosidases from Dalbergia sp. Arch. Biochem. Biophys. 468, 205–216 [DOI] [PubMed] [Google Scholar]
- 38. Tribolo S., Berrin J. G., Kroon P. A., Czjzek M., Juge N. (2007) The crystal structure of human cytosolic β-glucosidase unravels the substrate aglycone specificity of a family 1 glycoside hydrolase. J. Mol. Biol. 370, 964–975 [DOI] [PubMed] [Google Scholar]
- 39. Hancock S. M., Corbett K., Fordham-Skelton A. P., Gatehouse J. A., Davis B. G. (2005) Developing promiscuous glycosidases for glycoside synthesis: residues W433 and E432 in Sulfolobus solfataricus β-glycosidase are important glucoside- and galactoside-specificity determinants. ChemBioChem 6, 866–875 [DOI] [PubMed] [Google Scholar]
- 40. Hua Y., Sansenya S., Saetang C., Wakuta S., Ketudat Cairns J. R. (2013) Enzymatic and structural characterization of hydrolysis of gibberellin A4 glucosyl ester by a rice β-d-glucosidase. Arch. Biochem. Biophys. 537, 39–48 [DOI] [PubMed] [Google Scholar]
- 41. Himeno N., Saburi W., Wakuta S., Takeda R., Matsuura H., Nabeta K., Sansenya S., Ketudat Cairns J. R., Mori H., Imai R., Matsui H. (2013) Identification of rice β-glucosidase with high hydrolytic activity towards salicylic acid β-d-glucoside. Biosci. Biotechnol. Biochem. 77, 934–939 [DOI] [PubMed] [Google Scholar]
- 42. Lee K. H., Piao H. L., Kim H. Y., Choi S. M., Jiang F., Hartung W., Hwang I., Kwak J. M., Lee I. J., Hwang I. (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126, 1109–1120 [DOI] [PubMed] [Google Scholar]
- 43. Brzobohatý B., Moore I., Kristoffersen P., Bako L., Campos N., Schell J., Palme K. (1993) Release of active cytokinin by a β-glucosidase localized to the maize root meristem. Science 262, 1051–1054 [DOI] [PubMed] [Google Scholar]
- 44. Morant A. V., Jørgensen K., Jørgensen C., Paquette S. M., Sánchez-Pérez R., Møller B. L., Bak S. (2008) β-Glucosidases as detonators of plant chemical defense. Phytochemistry 69, 1795–1813 [DOI] [PubMed] [Google Scholar]
- 45. Ahn Y. O., Mizutani M., Saino H., Sakata K. (2004) Furcatin hydrolase from Viburnum furcatum Blume is a novel disaccharide-specific acuminosidase in glycosyl hydrolase family 1. J. Biol. Chem. 279, 23405–23414 [DOI] [PubMed] [Google Scholar]
- 46. Ahn Y. O., Saino H., Mizutani M., Shimizu B., Sakata K. (2007) Vicianin hydrolase is a novel cyanogenic β-glycosidase specific to β-vicianoside (6-O-α-l-arabinopyranosyl-β-d-glucopyranoside) in seeds of Vicia angustifolia. Plant Cell Physiol. 48, 938–947 [DOI] [PubMed] [Google Scholar]
- 47. Daiyasu H., Saino H., Tomoto H., Mizutani M., Sakata K., Toh H. (2008) Computational and experimental analyses of furcatin hydrolase for substrate specificity studies of disaccharide-specific glycosidases. J. Biochem. 144, 467–475 [DOI] [PubMed] [Google Scholar]