Background: NOS enzymes are large, dimeric complexes essential in mammalian physiology.
Results: EM structural analysis and three-dimensional models reveal nNOS reductase-oxygenase arrangements and a CaM-dependent rotation of the FMN domain.
Conclusion: Coordinated conformational changes act to reposition the FMN domain for electron transfer.
Significance: This work captures structural states of the NOS holoenzyme that drive the NO synthesis cycle.
Keywords: Calmodulin, Electron Microscopy (EM), Flavoproteins, Nitric-oxide Synthase, Oxidation-Reduction
Abstract
Nitric-oxide synthase (NOS) is required in mammals to generate NO for regulating blood pressure, synaptic response, and immune defense. NOS is a large homodimer with well characterized reductase and oxygenase domains that coordinate a multistep, interdomain electron transfer mechanism to oxidize l-arginine and generate NO. Ca2+-calmodulin (CaM) binds between the reductase and oxygenase domains to activate NO synthesis. Although NOS has long been proposed to adopt distinct conformations that alternate between interflavin and FMN-heme electron transfer steps, structures of the holoenzyme have remained elusive and the CaM-bound arrangement is unknown. Here we have applied single particle electron microscopy (EM) methods to characterize the full-length of the neuronal isoform (nNOS) complex and determine the structural mechanism of CaM activation. We have identified that nNOS adopts an ensemble of open and closed conformational states and that CaM binding induces a dramatic rearrangement of the reductase domain. Our three-dimensional reconstruction of the intact nNOS-CaM complex reveals a closed conformation and a cross-monomer arrangement with the FMN domain rotated away from the NADPH-FAD center, toward the oxygenase dimer. This work captures, for the first time, the reductase-oxygenase structural arrangement and the CaM-dependent release of the FMN domain that coordinates to drive electron transfer across the domains during catalysis.
Introduction
Nitric oxide (NO) serves fundamental roles in neurotransmission, cardiovascular function, and cellular defense (1, 2). The three major NOS isoforms in mammals, neuronal (nNOS),2 endothelial (eNOS), and inducible (iNOS), produce NO in diverse signaling pathways critical to physiology. Calmodulin (CaM) is an essential activator of NOS, binding to nNOS and eNOS in a Ca2+ concentration-dependent manner, but it binds iNOS at all intracellular Ca2+ concentrations (3). Dysfunctional NOS produces reactive oxygen species and lowers NO, contributing to oxidative stress in stroke, diabetes, and neurodegeneration (4).
NOS is a large (260–330-kDa) homodimer distinguished by a C-terminal reductase and N-terminal oxygenase domains that together catalyze the conversion of l-Arg and molecular oxygen to citrulline and NO. Crystal structures of the oxygenase domain identified substrate, heme, and BH4 binding sites that are required for oxidizing Arg during catalysis and stabilizing the dimer interface in the holoenzyme complex (5, 6). NOS is a diflavin oxidoreductase, homologous to cytochrome P450, that contains distinct FAD and FMN domains that are connected by a hinge that facilitates stepwise NADPH-FAD-FMN-heme electron transfer (7). The final FMN-heme electron transfer step is rate-limiting and proposed to involve a trans-monomer mechanism (8, 9). CaM interacts with a recognition sequence in a flexible region between the reductase and oxygenase domains and accelerates flavin reduction and FMN-heme electron transfer steps (10, 11).
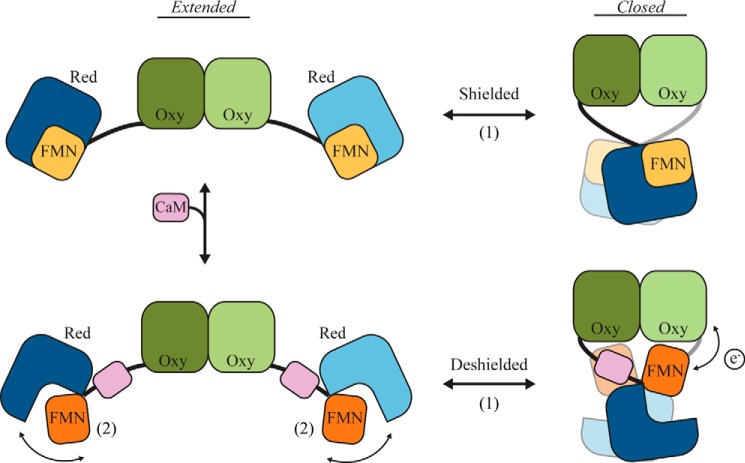
In the nNOS reductase domain structure, the FMN cofactor is buried and positioned adjacent to the NADPH and FAD sites to give a 5 Å separation that would facilitate FAD-FMN electron transfer (12). However, this orientation is not compatible for subsequent electron transfer to the heme. Kinetic and biochemical experiments have led to models proposing that the FMN subdomain rotates about the hinge in a conformational switch from a “shielded” to “deshielded” state where the FMN becomes accessible and transfers electrons to the oxygenase domain of the adjacent monomer (13, 14). Although structures of cytochrome P450 have identified flexibility about this hinge, the large change in the position of the FMN domain proposed by kinetic experiments and other models has never been observed structurally (15). CaM interactions increase FMN-heme electron transfer, possibly by destabilizing the shielded state through bridging contacts with the FMN or interactions with the reductase connecting domain (CD) (16–20). Electrostatic interactions via structural control elements in the reductase domain, including the autoinhibitory loop, C-terminal tail, and connecting domain, favor the shielded state and are proposed to become disrupted for precise release of the FMN domain during the catalytic cycle (21, 22).
The dual function of CaM in promoting both intradomain and interdomain electron transfer in nNOS indicates that the CaM-bound complex is likely structurally dynamic and can support conformational states that coordinate FAD-FMN and FMN-heme electron transfer (23, 24). Recent hydrogen-deuterium exchange experiments of isolated iNOS domains identify interaction surfaces between the oxygenase, FMN domain, and CaM, supporting a CaM-dependent arrangement for electron transfer to the heme (25). However, structures of the full-length complex have not been solved, and thus the arrangement of the reductase and oxygenase domains and the CaM-bound conformation of the holoenzyme remain fundamental questions in understanding the NOS catalytic cycle.
Here we have characterized the structure of the nNOS holoenzyme complex by electron microscopy (EM) and identified distinct domain arrangements of the dimer and a CaM-dependent reorganization of the reductase domain that together explain the conformational changes required for FMN-heme electron transfer. Two-dimensional classification and analysis of nNOS reveal an ensemble of extended, V-shaped, and closed conformations, as well as a dramatic opening of the reductase domain following CaM binding. By chemical cross-linking, we stabilized nNOS-CaM in the closed conformation and performed a three-dimensional reconstruction. In the complete three-dimensional model, the reductase domains are positioned adjacent to the oxygenase dimer, whereas the FMN domain is rotated away from the NADPH-FAD center toward the oxygenase domain of the adjacent monomer. From this work, we propose that the NO synthesis cycle involves large conformational changes that transiently position the reductase domains across the oxygenase dimer, whereas CaM-specific activation triggers release and rotation of the FMN subdomain to expose the flavin for electron transfer to the heme.
EXPERIMENTAL PROCEDURES
Expression and Purification of nNOS and CaM
The cDNA for rat nNOS was kindly provided by Dr. S. Snyder (Johns Hopkins University). BH4 was purchased from Schircks Laboratories (Jona, Switzerland). Heme, ATP, creatine phosphokinase, l-arginine, NG-nitro-l-arginine, and NG-nitro-d-arginine were purchased from Sigma, and creatine phosphate was from Fluka (St. Louis, MO). Untagged nNOS dimer protein was expressed using a recombinant baculovirus/Sf9 insect cell system and purified by 2′,5′-ADP-Sepharose and gel-filtration chromatography as described (26). Heme was added as an albumin conjugate during the expression to convert all of the nNOS to the holo-nNOS dimer. The human CaM plasmid, pACYC/trc-hCaM, was a gift from the R. Neubig laboratory (University of Michigan), and expression and purification were performed essentially as described (27).
NOS Activity assay and SEC-MALS Analysis
NO synthesis activity was determined by measuring the conversion of oxyhemoglobin to methemoglobin, as described (28). Samples were incubated at 37 °C in 100 μm CaCl2, 100 μm l-arginine, 100 μm BH4, 100 units/ml catalase, 5 μg/ml CaM, 25 μm oxyhemoglobin, and an NADPH-regenerating system consisting of 400 mm NADP+, 10 mm glucose 6-phosphate, and 1 unit/ml glucose-6-phosphate dehydrogenase, expressed as final concentrations, in a total volume of 180 μl of 50 mm potassium phosphate, pH 7.4. The rate of oxyhemoglobin oxidation was monitored by UV absorbance (λ = 401–411 nm) with a microplate reader. The average molecular weight (Mw) and hydrodynamic radius (RH) of nNOS and nNOS-CaM were determined by separation using a WTC-050S5 SEC column (Wyatt Technology Corp.) with an ÄKTAmicro (GE Healthcare) and analysis with a DAWN HELEOS II MALS detector equipped with a WyattQELS dynamic light scattering detector, and Optilab rEX differential refractive index detector using ASTRA VI software (Wyatt Technology). The Mw was determined from the Raleigh ratio calculated by measuring the static light scattering and corresponding protein concentration of a selected peak. Bovine serum albumin served as a calibration standard. For size exclusion chromatography and multiangle light scattering (SEC-MALS), 30 μm nNOS was incubated with or without 60 μm CaM for 20 min on ice in the running buffer, containing 20 mm HEPES, pH 7.5, 100 mm KCl, 1 mm DTT, 2 mm CaCl2, and 15 μm BH4.
EM Sample Preparation and Data Collection
Peak fractions collected following SEC-MALS were diluted 1/20 in buffer and negatively stained with 0.75% uranyl formate (pH 5.5–6.0) on thin carbon-layered 400-mesh copper grids (Pelco) as described (29). Chemical cross-linking was performed following SEC-MALS by incubating nNOS-CaM with 0.01% glutaraldehyde for 20 min and then quenching with 20 mm Tris, pH 7.5, as described previously (30). Cross-linked nNOS-CaM was then repurified by SEC using a Superdex 200 precision column (GE Healthcare), and EM grids were prepared as above. Samples were imaged under low dose conditions using a G2 Spirit transmission electron microscope (FEI) operated at 120 keV. Micrographs were taken at 52,000× magnification with 2.16 Å per pixel using a 4k x 4k CCD camera (Gatan). Single particles of nNOS and nNOS-CaM were manually selected using E2boxer (EMAN2) (31) and totaled 12,207 and 13,340, respectively. Data for random conical tilt (RCT) reconstructions were collected by imaging at 0° and 60° and selecting 12,323 single particle tilt pairs using e2RCTboxer (EMAN2).
EM Data Processing and Molecular Modeling
Reference-free classification of uncross-linked nNOS and nNOS-CaM was performed using the Iterative Stable Alignment and Clustering (ISAC) method (32). The total particle sets were processed through 20 initial iterations of classification and then five iterations of two-way matching, generating 124 classes from 7,326 particles for nNOS and 139 classes from 8,818 particles for nNOS-CaM. For the RCT reconstructions, reference-free two-dimensional classification of the untilted data was performed, and initial three-dimensional maps were calculated from the single classes using the corresponding tilted data with SPIDER (33, 34). Three-dimensional refinement using the untilted data was then performed with RELION (35) where the RCT model was used as an initial model and low pass-filtered to 60 Å, and two-fold symmetry was imposed. The 3D classification procedure was used with two classes and 20 rounds of refinement. The map that agreed best with the oxygenase dimer crystal structure was from 5,570 particles and selected for an additional six rounds of refinement using 3D auto-refine. Crystal structures were docked in the final map using Chimera, with the Fit in Map command for the oxygenase dimer and manually for the reductase domain (36). The deshielded arrangement was achieved by manually rotating the FMN domain around a pivot point between residues 944 and 968. The structure of the CaM-FMN-CaM binding helix complex was aligned to the FMN domain, and the arrangement was symmetrized for the final molecular model. The degree of rotation between the FMN positions was determined from the angle between two planes drawn through the center of the domains using Chimera.
RESULTS
Solution Conformation, Activity, and Negative Stain EM of nNOS and nNOS-CaM
Current models indicate that NO synthesis by NOS likely involves large conformational changes and CaM-dependent rearrangements to bring the reductase and oxygenase domains together (15). However, the architecture of the full-length NOS holoenzyme complex is unknown. To begin we examined the overall size and shape of purified, recombinant rat nNOS with and without CaM by SEC-MALS. In the absence of CaM, nNOS elutes as a single peak at 7.9 ml by SEC with an Mw of 330,000 (Fig. 1A). This indicates that purified nNOS alone forms a stable dimer complex when compared with the 164,000 Mw of the monomer calculated from the amino acid sequence (Mwaa). Following incubation with excess CaM, a single peak elutes earlier, at 7.7 ml with a Mw of 360,000 determined by MALS, indicating complete binding of CaM at a 2:2 ratio of nNOS to CaM when compared with the Mwaa of 361,000 for the holoenzyme complex. To characterize the overall shape of nNOS in solution, the hydrodynamic radius (RH) was determined by dynamic light scattering. For nNOS alone and nNOS-CaM, an RH of 85 and 86 Å was determined, respectively (Fig. 1A). Based on comparison with a known globular protein of similar size, mammalian phosphofructokinase tetramer (345 kDa), which has an RH of 63 Å (37, 38), nNOS is predicted to be in an elongated arrangement in solution, and CaM binding results in no significant change in the overall RH.
FIGURE 1.
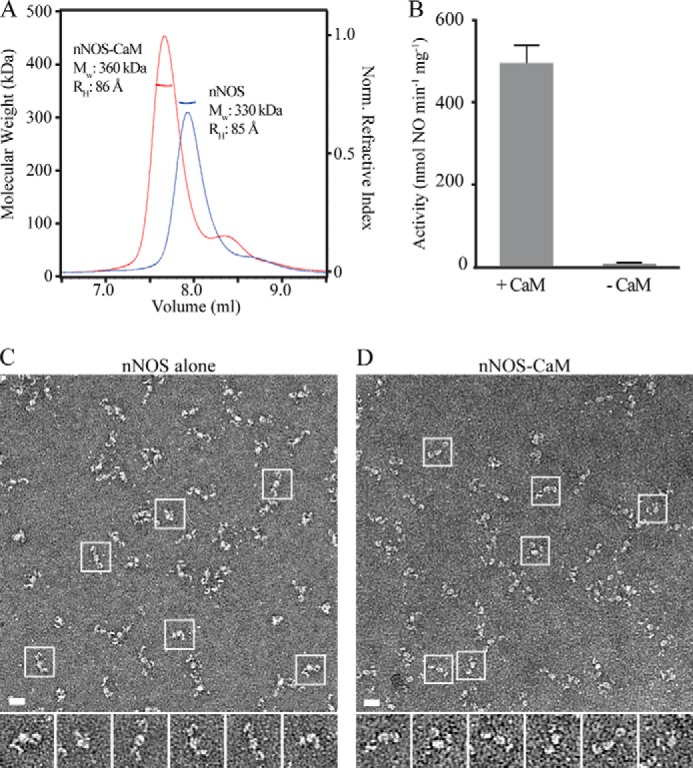
Solution analysis, activity, and EM images of nNOS and nNOS-CaM. A, SEC-MALS analysis of the nNOS dimer (blue), showing a 7.9-ml elution volume, a 330,000 Mw (Mwaa = 164,000 for the monomer), and an 85 Å RH. Following incubation with CaM, the nNOS-CaM complex (red) elutes at 7.7 ml with a 360,000 Mw (Mwaa = 361,000 for a 2:2 tetramer) and an 86 Å RH. B, NO synthesis activity for nNOS incubated with CaM and nNOS alone, measured by the oxyhemoglobin assay. Error bars indicate mean ± S.D calculated from six experiments. C and D, representative micrograph images and boxed single particles of negatively stained nNOS (C) and nNOS-CaM isolated by SEC-MALS (D) (scale bars = 200 Å).
NO synthesis activity was investigated by an oxyhemoglobin activity assay for these nNOS and nNOS-CaM preparations, and the rates were determined to be 6 and 492 nmol of NO/min/mg, respectively (Fig. 1B). This significant 80-fold increase in activity indicates the critical dependence on CaM binding for the catalytic activation of nNOS. Based on the unchanged elongated state of nNOS-CaM in solution, it remained unclear how electron transfer could occur across the domains. We hypothesized that although the average shape of nNOS-CaM may be extended, additional conformations that support NO synthesis may exist in equilibrium.
To explore the conformation and structure of nNOS and nNOS-CaM, we used single particle EM. Because of the expected flexibility of the nNOS complexes, high contrast negative-stain methods were used. Following SEC-MALS analysis, samples were collected and immediately prepared for EM. In micrographs and single particle images, nNOS (Fig. 1C) and nNOS-CaM (Fig. 1D) were confirmed to be in an overall elongated conformation with an approximate length of 250 Å. Additional conformations were also prevalent including V-shaped structures with a variable open angle and a more compact, closed structure. This demonstrates that indeed nNOS exists in an ensemble of conformational states. The presence of saturating amounts of substrate (Arg) and active site inhibitors (NG-hydroxy-l-Arg and 7-nitroindazole) was also tested with and without CaM; however, no differences in nNOS were observed (data not shown).
Distinct Conformational States of nNOS and nNOS-CaM by Two-dimensional EM Analysis
To further characterize the conformational states of NOS and the consequence of CaM binding, we collected single particle data sets of nNOS alone and nNOS-CaM and performed two-dimensional reference-free averaging and analysis using the recently developed ISAC method (32). Three lobes of density are well defined in the majority of two-dimensional averages of nNOS alone, enabling domain localization based on comparisons with crystal structures (Fig. 2A). The oxygenase dimer forms a central, oval-shaped structure, whereas reductase domains are distal with little density connecting the domains. Extended, V-shaped, and closed conformations are clearly apparent in the two-dimensional averages. Notably, the V-shaped structures have a symmetrical arrangement of the reductase domains, indicating that the extended-to-closed conformational change is potentially coordinated across the monomers.
FIGURE 2.
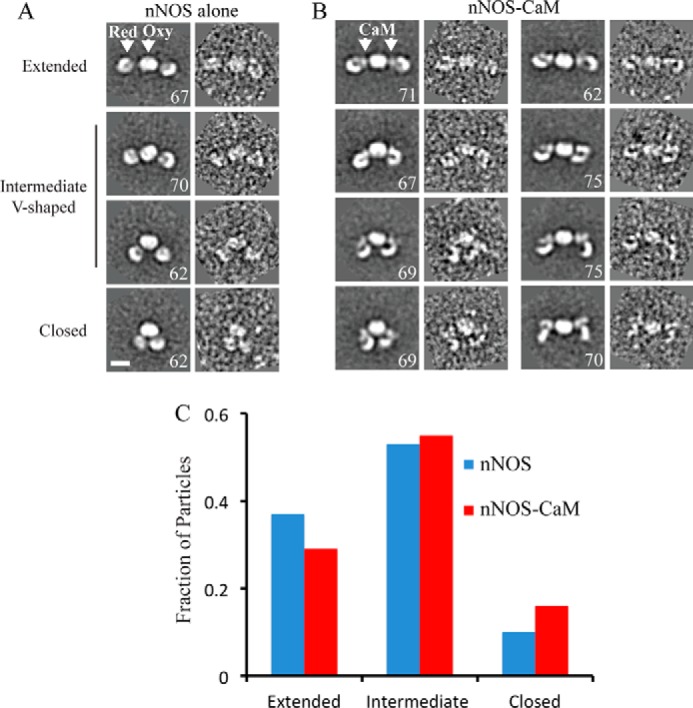
Comparison of the conformational states of nNOS and nNOS-CaM by EM. A and B, representative averages and single particle images from each class, highlighting an extended-to-closed conformational change for the nNOS dimer (A) and nNOS-CaM (B, left panel), with the indicated location of the reductase domain (Red), oxygenase dimer (Oxy), and CaM. B, right panel, representative averages and single particles of nNOS-CaM are shown highlighting conformational changes in the reductase domain. The number of single particle images for each average is shown (scale bar = 100 Å). C, bar graph showing the fraction of nNOS and nNOS-CaM single particles that adopt extended, intermediate V-shaped, or closed conformations, based on the arrangements identified in the two-dimensional averages.
In the two-dimensional averages of the nNOS-CaM complex, both reductase domains are well defined, and similar extended, V-shaped, and closed conformations are observed (Fig. 2B, left panel). However, two significant distinctions are apparent when compared with nNOS alone. First, additional density is observed between the reductase and oxygenase domains. Although small, this density is observed on both sides of the dimer and appears to increase the connectivity between the domains when compared with NOS alone. This likely corresponds to CaM based on the known binding site between the domains. Difference images were generated between nNOS and nNOS-CaM in an attempt to better visualize the density, but additional conformational changes prevented precise alignment (data not shown).
Secondly, in the CaM-bound complex, the reductase domains are in an alternate conformational state, forming a clamp-like structure with a subdomain portion that separates and rotates away from the complex in several distinct arrangements (Fig. 2B, right panel). This large rearrangement of the reductase domain is not observed in the averages of nNOS alone.
The overall conformational equilibrium was characterized for nNOS and nNOS-CaM by comparing the fraction of particles that adopt extended, intermediate, and closed conformations, based on the two-dimensional averages (Fig. 2C). For nNOS alone, about 40% are extended, whereas 10% are in the closed conformation when compared with 30% extended and 15% closed for nNOS-CaM. This indicates a slight shift to the closed conformation for nNOS-CaM; however, both complexes are primarily in an intermediate arrangement (53 and 55%, respectively). From this we conclude that CaM binding does not significantly alter the overall conformational equilibrium of the dimer, supporting our SEC-MALS analysis.
For complete characterization of the changes in the reductase domain resulting from CaM binding, the entire set of class averages of nNOS alone and nNOS-CaM is shown (Fig. 3). The reductase domains remain globular for nearly all averages of nNOS alone, but clearly adopt a variety of different structural states in the averages of the nNOS-CaM complex. Although this conformational change appears symmetrical in some averages, many are asymmetric with each reductase domain adopting a different arrangement. Furthermore, these reductase conformations are present in combination with the different extended-closed states of the dimer. Overall these results demonstrate that nNOS and nNOS-CaM undergo a large extended-to-closed conformational change that brings the reductase domains together. CaM is localized between the domains and induces a significant rearrangement of the reductase domain appearing as an opening clamp that separates the reductase domain into two distinct subdomains.
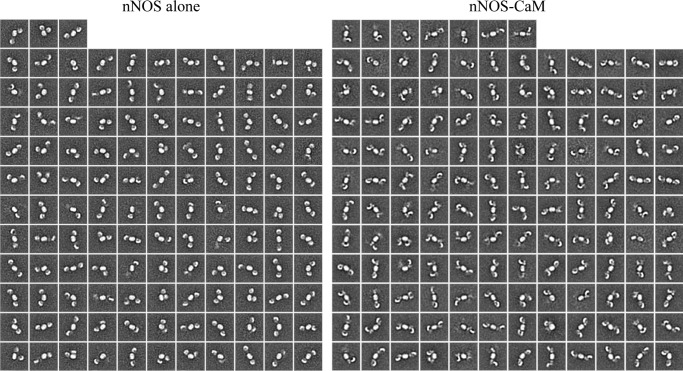
FIGURE 3.
The complete set of reference-free two-dimensional class averages of nNOS and nNOS-CaM. A total of 124 averages were determined from 7,326 particles for nNOS alone (left panel), and 139 classes were determined from 8,818 particles for nNOS-CaM (right panel) using the ISAC image clustering method (32).
Chemical Cross-linking Stabilizes nNOS-CaM in a Closed State
Although our two-dimensional EM analysis points to significant structural changes that result from CaM binding, a three-dimensional reconstruction was required to determine the architecture of this CaM-stabilized arrangement. However, the ensemble of different conformational states we identified presented significant challenges for determining a three-dimensional reconstruction. Chemical cross-linking, typically with glutaraldehyde, is an established method we and others have used previously to stabilize a single conformation and improve homogeneity (39, 40). Therefore, we tested a glutaraldehyde cross-linking method where nNOS-CaM, isolated by SEC-MALS, was incubated with low concentrations (0.01%) of cross-linker for short time points and then quenched. Uniform cross-linking and the absence of aggregation were established by additional SEC separation of a single species (data not shown). In negative-stain micrographs and single particle images, glutaraldehyde-cross-linked nNOS-CaM appeared remarkably homogeneous and in a closed conformational state (Fig. 4A). An initial single particle data set was collected and analyzed by reference-free two-dimensional averaging methods. In the projection averages, the reductase domains are clearly identifiable in a closed arrangement adjacent to the oxygenase dimer (Fig. 4B). This closed arrangement is similar to the small population of closed particles observed with nNOS-CaM (Fig. 4C). For the cross-linked complex, the domains appear better defined with significant additional views. Cross-linked nNOS dimer alone was also tested and appeared to form a similar closed state (data not shown). Because nNOS is inactive in the absence of CaM and the reductase domain rearrangement we observe was only found in the presence of CaM, additional structural characterization of the nNOS dimer alone was not performed. In previous work, cross-linking by glutaraldehyde was shown to stabilize nucleotide-dependent structural states of the heat shock protein 90 that are normally sampled during the conformational cycle (30). Our data here demonstrate that cross-linking nNOS-CaM similarly captures a functional arrangement we observe in the uncross-linked sample. Based on the shorter distance between the reductase and oxygenase domains when compared with the other states, this closed conformation is the most probable arrangement for supporting a cross-monomer FMN-heme electron transfer mechanism.
FIGURE 4.
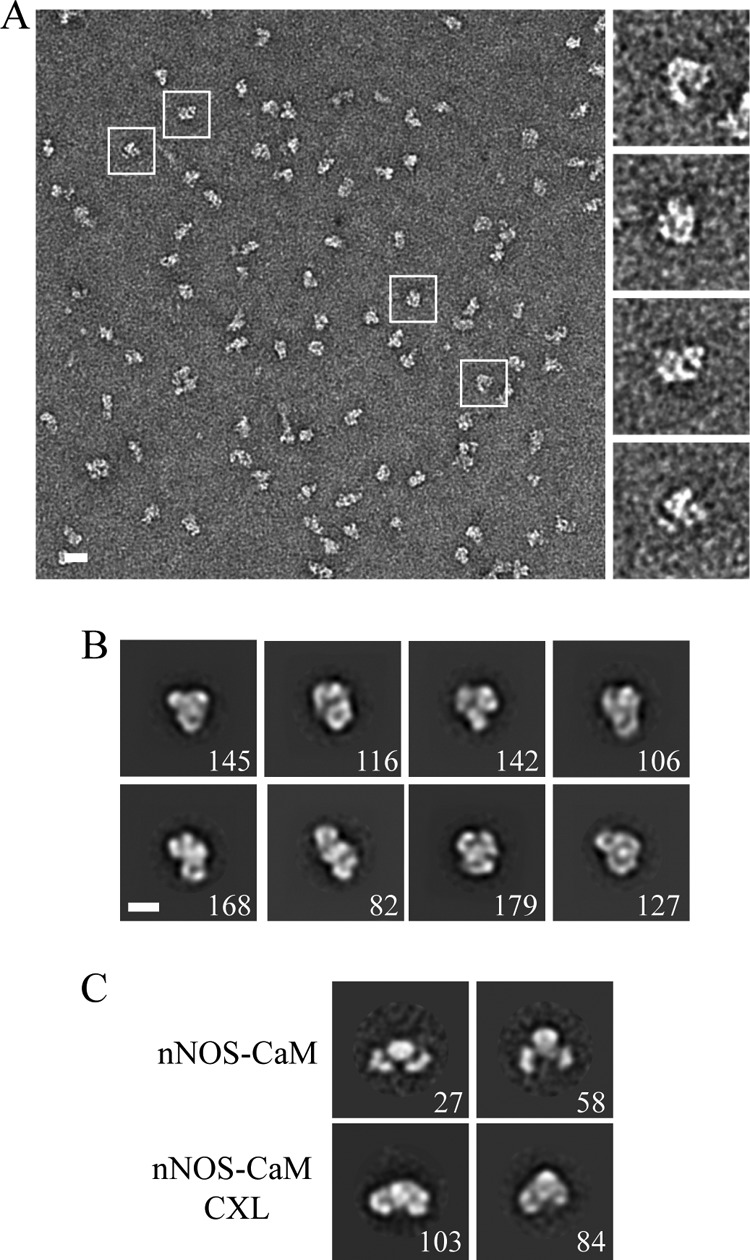
Two-dimensional EM analysis of nNOS-CaM cross-linked with glutaraldehyde. A, nNOS-CaM cross-linked (CXL) with 0.01% glutaraldehyde is shown in a representative micrograph image and selected particles (scale bar = 200 Å). B, two-dimensional reference-free averages of nNOS-CaM CXL with the number of single particles per average (scale bar = 100 Å). C, comparison of two-dimensional averages of uncross-linked nNOS-CaM in the closed conformation and nNOS-CaM CXL, following two-dimensional alignment.
Three-dimensional EM Reconstruction of nNOS-CaM Identifies a Closed, Cross-monomer Arrangement
From our two-dimensional analysis, the cross-linked nNOS-CaM complex appeared homogenous and adopted multiple orientations with clearly defined features, making it suitable for three-dimensional reconstruction methods. To achieve an accurate three-dimensional initial model, we relied on the RCT method, where a three-dimensional reconstruction from a single two-dimensional class is determined from data tilted to 60° (33). We utilized this method to compare several reconstructions from different classes without imposing symmetry to generate unbiased initial models. The RCT models were then used as starting models in three-dimensional refinements carried out with the entire untilted dataset. As with our analysis above, untilted projection averages showed that the particles adopt multiple orientations (Fig. 5A). Following three-dimensional back projection of the tilted data from well populated classes (>200 particles), it was clear that particles that orient with “side” views with three distinct domains provided the most consistent RCT reconstruction (Fig. 5B). Similar RCT reconstructions were achieved from several different classes, identifying a two-fold symmetric arrangement of the complex. The model from class 13 was used for the refinements with the full data set, but all models were tested and converged similarly (data not shown). Three-dimensional refinements were then carried out using the RELION method (35). Projections of the final model match the reference-free averages generated independently, and there is good particle distribution among the projection angles, although some views were preferred (Fig. 5, C and D).
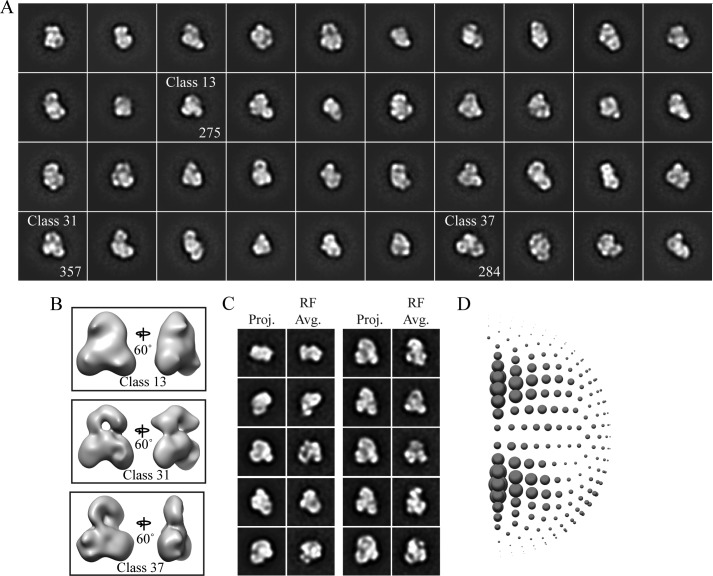
FIGURE 5.
RCT initial model generation and analysis of the final reconstruction. A, two-dimensional reference-free class averages of cross-linked nNOS-CaM from untilted images with the indicated classes used in the RCT reconstructions. B, three-dimensional RCT reconstructions, generated from tilted data corresponding to the indicated two-dimensional class. The reconstruction from class 13 was used as the starting model for additional three-dimensional refinement. C, two-dimensional projections (Proj.) of the final three-dimensional reconstruction aligned to reference-free class averages (RF Av.) of cross-linked nNOS-CaM. D, angle distribution image from the final round of refinement with a 7.5° increment, generated with XMIPP (48).
The final model was calculated to be at 23 Å resolution using the gold standard Fourier shell correlation procedure (Fig. 6A). The model appears similar to the RCT models but with improved definition of the three lobes of density, identifying a cross-monomer arrangement (Fig. 6B). In the structure, a large “head” domain is connected to two globular base domains by an “arm” that extends across the structure, with a central space between the head and base domains. The crystal structure of the nNOS oxygenase dimer fits well into the head region of the structure with the C terminus and α1-α2 helices aligning to where the arms project downward (Fig. 6C). Agreement with the crystal structure is further confirmed in a cross-section side view that reveals a cavity that matches the solvent-accessible site of the heme binding pocket (Fig. 6D). Overall no significant conformational change is observed in the oxygenase dimer when compared with the crystal structure. In this structural arrangement, CaM is expected to be located in the connecting arms across the complex, whereas the reductase domains are positioned adjacent the oxygenase domain of the alternate monomer.
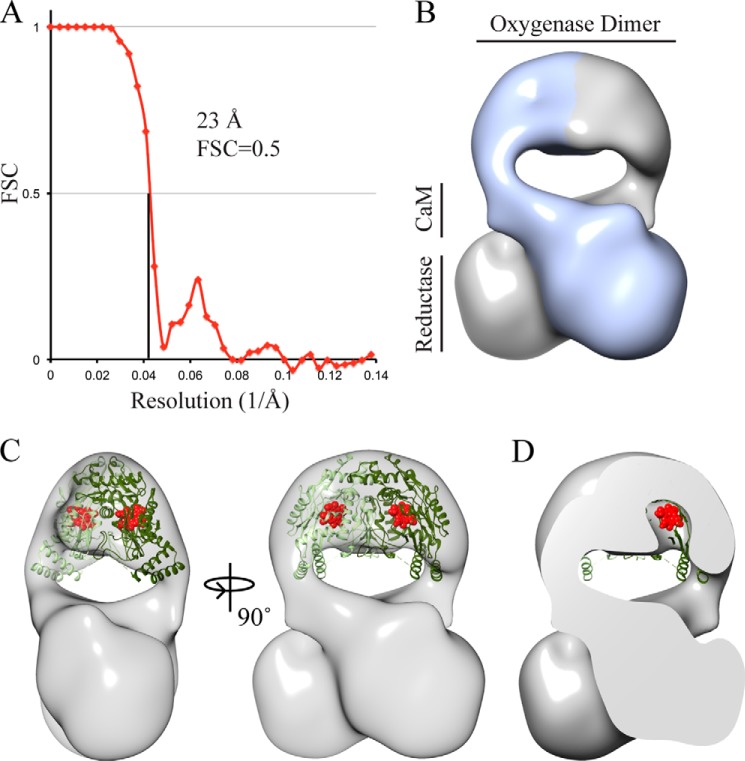
FIGURE 6.
Final three-dimensional model of the cross-linked nNOS-CaM complex. A, gold standard Fourier shell correlation (FSC) curve of the final round of refinement showing a 23 Å resolution at a Fourier shell correlation = 0.5 criterion. B, three-dimensional model of nNOS-CaM, colored along the two-fold axis to show the monomer arrangement. C, the oxygenase dimer (Protein Data Bank (PDB) ID: 1RS9) docked into the EM map and locally optimized using Chimera. The heme cofactors are shown in red. D, cross-section view showing a pocket in the EM model that matches the location of the heme cofactor.
Molecular Model of the nNOS-CaM Complex Reveals an FMN Rotation to a Deshielded State
Next we worked to build a complete molecular model of the nNOS-CaM complex to determine whether nNOS is in a conformation that could support electron transfer across the domains. When the crystal structure of the nNOS reductase domain was docked into the EM map, the NADPH and FAD domains aligned well to the curved globular base and the FMN domain was positioned above, near where the arms connect across to the oxygenase domain and where CaM is expected to be localized (Fig. 7A) (12). The CaM binding helix was included in the fit and oriented based on the crystal structure of the FMN domain-CaM complex (19). In this arrangement, the CaM binding helix projects away from the oxygenase domain, across the dimer in an orientation that is incompatible with the EM model. Furthermore, the FMN cofactor is positioned in the center of the reductase domain in a shielded conformation, nearly 100 Å away from the heme pocket; thus a different arrangement of the FMN domain would be required for electron transfer to the oxygenase domain.
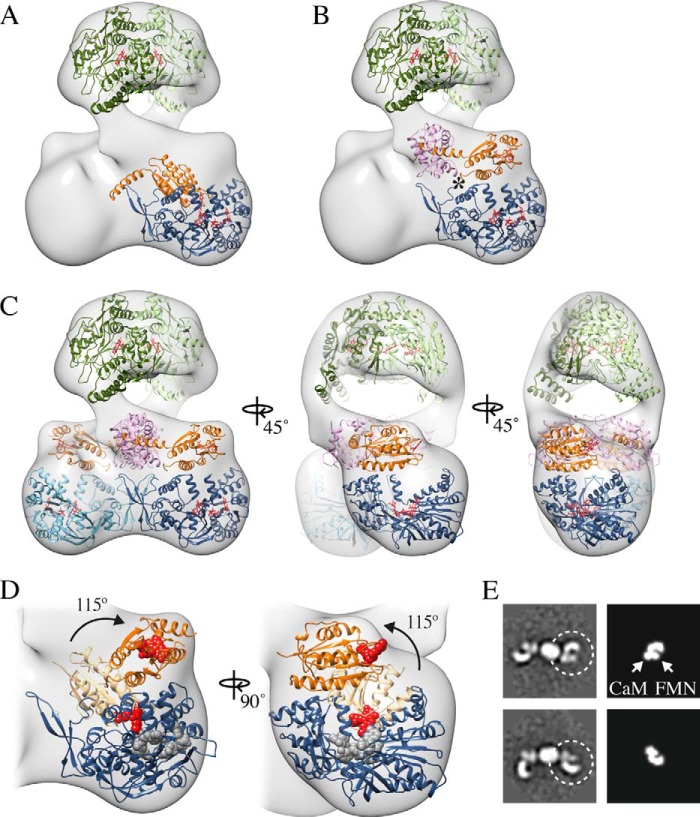
FIGURE 7.
Molecular model of the nNOS-CaM holoenzyme identifying a rotation of the FMN domain to a deshielded state. A, fit of the reductase domain (PDB ID: 1TLL) showing the FMN domain (orange) in the shielded arrangement with the aligned CaM binding helix (PDB ID: 3HR4) and NADPH-FAD domains (blue). B, fit of the reductase domain with CaM (pink) and showing the rotation of the FMN domain around the flexible hinge (*) to a proposed FMN-released, deshielded state. Cofactors are shown in red. C, complete molecular model the nNOS-CaM complex in the deshielded state. D, close-up view of the reductase domain with the FMN in the shielded (light orange) and deshielded (dark orange) states indicating a 115° rotation. E, comparison of two-dimensional averages of uncross-linked nNOS-CaM (left panels) and two-dimensional projections of the proposed reductase-CaM deshielded arrangement (right panels).
To achieve a more accurate model, the FMN domain, including the CaM binding helix, was rotated to fit in the density that extends toward the oxygenase domain (Fig. 7B). This was achieved by treating residues 511–951 as a rigid body and rotating around a pivot point set in the flexible “hinge” region (residues 944–968). Notably, there are two forms of the reductase domain in the crystal structure that are related by a minor rotation about this hinge, supporting the basis for the rotation tested here (12). The autoinhibitory helix was excluded from the fit because of its known flexibility. In this proposed deshielded state, the FMN domain fits more completely into the density and the cofactor is now exposed and projects upward toward the oxygenase domain of the adjacent monomer (Fig. 7B and supplemental Movie S1). In addition, CaM fits into the connecting arm with the CaM binding helix oriented toward the oxygenase domain. The 29 residues between the CaM binding helix and the oxygenase domain that are missing in existing crystal structures likely make up the remaining density in the connecting arm.
When this reductase-CaM conformation is symmetrized to form a complete holoenzyme complex, there are no clashes across the dimer, supporting the overall arrangement (Fig. 7C and supplemental Movie S1). The β-finger that includes the CD2A loop (residues 1060–1082) in the reductase domain is positioned along the two-fold axis in our model, making possible contacts across the dimer that could support this closed dimer conformation. This loop forms part of a dimer interface in the crystal form of the reductase structure and has functional roles in facilitating CaM binding and NO synthesis (12, 20). The shielded-deshielded conformational change we identify here involves a large, 115° rotation of the FMN domain that reorients the cofactor below the oxygenase domain of the adjacent monomer, at about 25 Å away (Fig. 7D). Importantly, this alternate reductase conformation highlights a rotation that would increase accessibility to the FMN cofactor and potentially favor electron transfer to the heme or other electron acceptors. To further verify the rotated position of the FMN domain, we compared two-dimensional projections of our reductase-CaM deshielded model, calculated from the crystal structures, with the two-dimensional averages of the uncross-linked nNOS-CaM complex (Fig. 7E). An open clamp structure is identified in the two-dimensional projections of the deshielded model, and the conformation and position of CaM and the FMN domain agree with the two-dimensional experimental class averages of nNOS-CaM. Although a more extended reductase conformation is observed in some two-dimensional averages, suggesting an increased rotation, the majority are similar to this open clamp structure, confirming that the proposed rotation of the FMN domain is the underlying CaM-dependent conformational change.
DISCUSSION
From the data presented here we propose a model where large conformational changes of the nNOS dimer are coupled to CaM-driven release of the FMN domain during the NO synthesis cycle (Fig. 8). In the absence of CaM, nNOS exists in an equilibrium of extended, intermediate, and closed states, defined by changes in the connecting arm that alter the relative reductase-oxygenase position. The FMN domain remains protected in the shielded conformation, and NO synthesis does not occur. CaM binding does not significantly change this equilibrium but induces flexibility in the reductase domain that causes the FMN domain to release and undergo a large rotation that exposes the FMN cofactor, enabling electron transfer across the domains.
FIGURE 8.
Proposed conformational model for NO synthesis by nNOS. NO synthesis involves two distinct changes in the NOS holoenzyme complex: 1) an extended-to-closed conformational equilibrium that brings the reductase domains together in a cross-monomer arrangement, and 2) release and rotation of the FMN domain triggered by CaM binding that positions the FMN cofactor for electron transfer across to the adjacent oxygenase domain in the closed state.
Our EM structure and pseudo-atomic model of nNOS captures, for the first time, this CaM-dependent conformational change in the FMN that has been proposed in previous kinetic studies (13, 14). Our two-dimensional averages indicate that the deshielded, FMN-released state occurs in extended, intermediate, and closed dimer arrangements. Because of the long distance between the reductase and oxygenase domains in the more extended states (potentially >100 Å), we argue FMN-heme electron transfer across the monomers would be optimal in the closed state. However, given the dynamic conformations of nNOS-CaM we have identified, certain asymmetric arrangements of the complex may also facilitate FMN-heme electron transfer.
Flanking hinge elements are thought to enable the FMN to shift between shielded and deshielded states to favor respective NADPH-FAD-FMN and FMN-heme electron transfer (22). Although the requirement of CaM binding is unique to NOS, flexibility of the FAD-FMN hinge has been observed in several cytochrome P450 structures and is known to be important to catalytic function (7, 41). Thus, the large rotation of the FMN we have identified here may be a conserved mechanism to control redox cycling by the oxidoreductase enzymes.
From our model the FMN domain does not directly contact the oxygenase domain, as proposed from previous work (21, 25). However, a minor rotation of the connecting arm would bring the domains closer and position the FMN adjacent the heme pocket. Therefore, the cross monomer arrangement and FMN domain rotation captured in the EM structure are arguably the primary conformational changes on-path for the final steps of NO synthesis. The closed conformation in the three-dimensional reconstruction appears to be supported by additional reductase-reductase interactions based on our molecular model. Although this supports a potential role for a reductase-reductase interaction in stabilizing the arrangement, previous experiments established that heterodimer complexes containing one reductase domain are catalytically active (42). Therefore this interaction would not be required for synthesis, but based on our model we argue the contacts, such as through the CD2A loop (12, 20), could provide additional catalytic or regulatory control. Additional experiments and higher resolution structures are required to better characterize this interaction and potential function.
From the two-dimensional classification of nNOS-CaM, the reductase domain adopts several conformations, suggesting that both shielded and deshielded states are accessible in the CaM-bound state. This fits with kinetic models where CaM remains bound throughout the catalytic cycle to accelerate both interflavin and FMN-heme electron transfer steps (43). The conformational equilibrium we observe indicates that the FMN domain would be closest to the oxygenase domain of the adjacent monomer in the closed state, supporting a cross-monomer electron transfer mechanism when this conformation is sampled. Although the V-shaped and closed conformations in the two-dimensional averages appear to be symmetric, indicating a coupling between the monomers, the CaM-dependent opening of the reductase domains is largely asymmetric. Thus, the final electron transfer step may occur independently or alternate between the monomers.
A conserved reductase domain Arg residue (Arg-753 in nNOS), identified to form a salt bridge with CaM in the iNOS FMN domain-CaM structure, was shown in recent studies to be important for enhancing CaM-dependent catalytic activity (19, 21, 44). Our results provide additional structural framework for this interaction and support a model whereby this salt bridge and other interactions that stabilize the FMN domain-CaM arrangement function to drive the release of the FMN domain from the shielded state and stabilize the open-clamp conformation we have identified. We argue that these changes function together with the inherent extended-to-closed conformational equilibrium to transiently position the FMN adjacent to the oxygenase domain.
Finally, although the core mechanism of electron transfer is highly conserved, the NO synthesis rates vary significantly among the isoforms and have been attributed to differences in sequence elements between the domains (45, 46). In particular, the 40-residue autoinhibitory loop found in nNOS and eNOS affects the Ca2+ response in CaM binding, whereas the C-terminal tail segment has been shown to regulate multiple steps in electron transfer (47). Structurally how these elements might control the catalytic cycle is unknown, but this could occur by altering the open-closed conformational equilibrium or FMN-release state observed with nNOS, and thus provide a unique means for regulatory control of NO synthesis for the different, isoform-specific aspects of cellular function.
Supplementary Material
Acknowledgment
We thank Y. Skiniotis for many helpful comments and suggestions.
This work was supported by an American Heart Association Scientist Development Grant, National Institutes of Health Grant GM077430 (to Y. O.), and the University of Michigan Protein Folding Diseases Initiative.
This article was selected as a Paper of the Week.

This article contains supplemental Movie S1.
The three-dimensional map (code 5940) has been deposited in the Electron Microscopy Data Bank.
- nNOS
- neuronal nitric-oxide synthase
- eNOS
- endothelial nitric-oxide synthase
- iNOS
- inducible nitric-oxide synthase
- CaM
- calmodulin
- BH4
- (6R)-5,6,7,8-tetrahydro-l-biopterin
- ISAC
- Iterative Stable Alignment and Clustering
- SEC-MALS
- size exclusion chromatography and multiangle light scattering
- RCT
- random conical tilt
- Mw
- average molecular weight
- Mwaa
- Mw of the monomer calculated from the amino acid sequence.
REFERENCES
- 1. Bogdan C. (2001) Nitric oxide and the immune response. Nat. Immunol. 2, 907–916 [DOI] [PubMed] [Google Scholar]
- 2. Bredt D. S., Snyder S. H. (1990) Isolation of nitric oxide synthetase, a calmodulin-requiring enzyme. Proc. Natl. Acad. Sci. U.S.A. 87, 682–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marletta M. A. (1994) Nitric oxide synthase: aspects concerning structure and catalysis. Cell 78, 927–930 [DOI] [PubMed] [Google Scholar]
- 4. Förstermann U., Sessa W. C.(2012) Nitric oxide synthases: regulation and function. Eur. Heart J. 33, 829–837, 837a–837d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Crane B. R., Arvai A. S., Ghosh D. K., Wu C., Getzoff E. D., Stuehr D. J., Tainer J. A. (1998) Structure of nitric oxide synthase oxygenase dimer with pterin and substrate. Science 279, 2121–2126 [DOI] [PubMed] [Google Scholar]
- 6. Ghosh S., Wolan D., Adak S., Crane B. R., Kwon N. S., Tainer J. A., Getzoff E. D., Stuehr D. J. (1999) Mutational analysis of the tetrahydrobiopterin-binding site in inducible nitric-oxide synthase. J. Biol. Chem. 274, 24100–24112 [DOI] [PubMed] [Google Scholar]
- 7. Iyanagi T., Xia C., Kim J. J. (2012) NADPH-cytochrome P450 oxidoreductase: prototypic member of the diflavin reductase family. Arch. Biochem. Biophys. 528, 72–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adak S., Aulak K. S., Stuehr D. J. (2001) Chimeras of nitric-oxide synthase types I and III establish fundamental correlates between heme reduction, heme-NO complex formation, and catalytic activity. J. Biol. Chem. 276, 23246–23252 [DOI] [PubMed] [Google Scholar]
- 9. Miller R. T., Martásek P., Omura T., Siler Masters B. S. (1999) Rapid kinetic studies of electron transfer in the three isoforms of nitric oxide synthase. Biochem. Biophys. Res. Commun. 265, 184–188 [DOI] [PubMed] [Google Scholar]
- 10. Gachhui R., Presta A., Bentley D. F., Abu-Soud H. M., McArthur R., Brudvig G., Ghosh D. K., Stuehr D. J. (1996) Characterization of the reductase domain of rat neuronal nitric oxide synthase generated in the methylotrophic yeast Pichia pastoris: calmodulin response is complete within the reductase domain itself. J. Biol. Chem. 271, 20594–20602 [DOI] [PubMed] [Google Scholar]
- 11. Craig D. H., Chapman S. K., Daff S. (2002) Calmodulin activates electron transfer through neuronal nitric-oxide synthase reductase domain by releasing an NADPH-dependent conformational lock. J. Biol. Chem. 277, 33987–33994 [DOI] [PubMed] [Google Scholar]
- 12. Garcin E. D., Bruns C. M., Lloyd S. J., Hosfield D. J., Tiso M., Gachhui R., Stuehr D. J., Tainer J. A., Getzoff E. D. (2004) Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase. J. Biol. Chem. 279, 37918–37927 [DOI] [PubMed] [Google Scholar]
- 13. Welland A., Garnaud P. E., Kitamura M., Miles C. S., Daff S. (2008) Importance of the domain-domain interface to the catalytic action of the NO synthase reductase domain. Biochemistry 47, 9771–9780 [DOI] [PubMed] [Google Scholar]
- 14. Ilagan R. P., Tiso M., Konas D. W., Hemann C., Durra D., Hille R., Stuehr D. J. (2008) Differences in a conformational equilibrium distinguish catalysis by the endothelial and neuronal nitric-oxide synthase flavoproteins. J. Biol. Chem. 283, 19603–19615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Daff S. (2010) NO synthase: structures and mechanisms. Nitric Oxide 23, 1–11 [DOI] [PubMed] [Google Scholar]
- 16. Roman L. J., Martásek P., Miller R. T., Harris D. E., de La Garza M. A., Shea T. M., Kim J. J., Masters B. S. (2000) The C termini of constitutive nitric-oxide synthases control electron flow through the flavin and heme domains and affect modulation by calmodulin. J. Biol. Chem. 275, 29225–29232 [DOI] [PubMed] [Google Scholar]
- 17. Adak S., Santolini J., Tikunova S., Wang Q., Johnson J. D., Stuehr D. J. (2001) Neuronal nitric-oxide synthase mutant (Ser-1412 → Asp) demonstrates surprising connections between heme reduction, NO complex formation, and catalysis. J. Biol. Chem. 276, 1244–1252 [DOI] [PubMed] [Google Scholar]
- 18. Abu-Soud H. M., Stuehr D. J. (1993) Nitric oxide synthases reveal a role for calmodulin in controlling electron transfer. Proc. Natl. Acad. Sci. U.S.A. 90, 10769–10772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xia C., Misra I., Iyanagi T., Kim J. J. (2009) Regulation of interdomain interactions by calmodulin in inducible nitric-oxide synthase. J. Biol. Chem. 284, 30708–30717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Knudsen G. M., Nishida C. R., Mooney S. D., Ortiz de Montellano P. R. (2003) Nitric-oxide synthase (NOS) reductase domain models suggest a new control element in endothelial NOS that attenuates calmodulin-dependent activity. J. Biol. Chem. 278, 31814–31824 [DOI] [PubMed] [Google Scholar]
- 21. Tejero J., Haque M. M., Durra D., Stuehr D. J. (2010) A bridging interaction allows calmodulin to activate NO synthase through a bi-modal mechanism. J. Biol. Chem. 285, 25941–25949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haque M. M., Fadlalla M. A., Aulak K. S., Ghosh A., Durra D., Stuehr D. J. (2012) Control of electron transfer and catalysis in neuronal nitric-oxide synthase (nNOS) by a hinge connecting its FMN and FAD-NADPH domains. J. Biol. Chem. 287, 30105–30116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Panda K., Ghosh S., Stuehr D. J. (2001) Calmodulin activates intersubunit electron transfer in the neuronal nitric-oxide synthase dimer. J. Biol. Chem. 276, 23349–23356 [DOI] [PubMed] [Google Scholar]
- 24. Rozhkova E. A., Fujimoto N., Sagami I., Daff S. N., Shimizu T. (2002) Interactions between the isolated oxygenase and reductase domains of neuronal nitric-oxide synthase: assessing the role of calmodulin. J. Biol. Chem. 277, 16888–16894 [DOI] [PubMed] [Google Scholar]
- 25. Smith B. C., Underbakke E. S., Kulp D. W., Schief W. R., Marletta M. A. (2013) Nitric oxide synthase domain interfaces regulate electron transfer and calmodulin activation. Proc. Natl. Acad. Sci. U.S.A. 110, E3577–E3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bender A. T., Silverstein A. M., Demady D. R., Kanelakis K. C., Noguchi S., Pratt W. B., Osawa Y. (1999) Neuronal nitric-oxide synthase is regulated by the Hsp90-based chaperone system in vivo. J. Biol. Chem. 274, 1472–1478 [DOI] [PubMed] [Google Scholar]
- 27. Li H., Das A., Sibhatu H., Jamal J., Sligar S. G., Poulos T. L. (2008) Exploring the electron transfer properties of neuronal nitric-oxide synthase by reversal of the FMN redox potential. J. Biol. Chem. 283, 34762–34772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bender A. T., Nakatsuka M., Osawa Y. (2000) Heme insertion, assembly, and activation of apo-neuronal nitric-oxide synthase in vitro. J. Biol. Chem. 275, 26018–26023 [DOI] [PubMed] [Google Scholar]
- 29. Ohi M., Li Y., Cheng Y., Walz T. (2004) Negative staining and image classification: powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Southworth D. R., Agard D. A. (2008) Species-dependent ensembles of conserved conformational states define the Hsp90 chaperone ATPase cycle. Mol. Cell 32, 631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J. (2007) EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 [DOI] [PubMed] [Google Scholar]
- 32. Yang Z., Fang J., Chittuluru J., Asturias F. J., Penczek P. A. (2012) Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure 20, 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Radermacher M., Wagenknecht T., Verschoor A., Frank J. (1987) Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J. Microsc. 146, 113–136 [DOI] [PubMed] [Google Scholar]
- 34. Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 [DOI] [PubMed] [Google Scholar]
- 35. Scheres S. H. (2012) RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 37. Erickson H. P. (2009) Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online 11, 32–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paradies H. H. (1979) Structure of cross-linked rabbit muscle phosphofructokinase in solution. J. Biol. Chem. 254, 7495–7504 [PubMed] [Google Scholar]
- 39. Southworth D. R., Agard D. A. (2011) Client-loading conformation of the Hsp90 molecular chaperone revealed in the cryo-EM structure of the human Hsp90:Hop complex. Mol. Cell 42, 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stark H. (2010) GraFix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 481, 109–126 [DOI] [PubMed] [Google Scholar]
- 41. Hamdane D., Xia C., Im S. C., Zhang H., Kim J. J., Waskell L. (2009) Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J. Biol. Chem. 284, 11374–11384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Siddhanta U., Wu C., Abu-Soud H. M., Zhang J., Ghosh D. K., Stuehr D. J. (1996) Heme iron reduction and catalysis by a nitric oxide synthase heterodimer containing one reductase and two oxygenase domains. J. Biol. Chem. 271, 7309–7312 [DOI] [PubMed] [Google Scholar]
- 43. Abu-Soud H. M., Yoho L. L., Stuehr D. J. (1994) Calmodulin controls neuronal nitric-oxide synthase by a dual mechanism: activation of intra- and interdomain electron transfer. J. Biol. Chem. 269, 32047–32050 [PubMed] [Google Scholar]
- 44. Panda S. P., Polusani S. R., Kellogg D. L., 3rd, Venkatakrishnan P., Roman M. G., Demeler B., Masters B. S., Roman L. J. (2013) Intra- and inter-molecular effects of a conserved arginine residue of neuronal and inducible nitric oxide synthases on FMN and calmodulin binding. Arch. Biochem. Biophys. 533, 88–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ilagan R. P., Tejero J., Aulak K. S., Ray S. S., Hemann C., Wang Z. Q., Gangoda M., Zweier J. L., Stuehr D. J. (2009) Regulation of FMN subdomain interactions and function in neuronal nitric oxide synthase. Biochemistry 48, 3864–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roman L. J., Masters B. S. (2006) Electron transfer by neuronal nitric-oxide synthase is regulated by concerted interaction of calmodulin and two intrinsic regulatory elements. J. Biol. Chem. 281, 23111–23118 [DOI] [PubMed] [Google Scholar]
- 47. Tiso M., Tejero J., Panda K., Aulak K. S., Stuehr D. J. (2007) Versatile regulation of neuronal nitric oxide synthase by specific regions of its C-terminal tail. Biochemistry 46, 14418–14428 [DOI] [PubMed] [Google Scholar]
- 48. de la Rosa-Trevín J. M., Otón J., Marabini R., Zaldívar A., Vargas J., Carazo J. M., Sorzano C. O. (2013) Xmipp 3.0: an improved software suite for image processing in electron microscopy. J. Struct. Biol. 184, 321–328 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.