Background: The acyltransferase Porcupine transfers a monounsaturated fatty acid, palmitoleate, to Wnt proteins.
Results: Structure-function analysis of Wnt3a and Porcupine identified key regions necessary for stability, activity, and signaling.
Conclusion: Residues in predicted transmembrane domain 9 within Porcupine and surrounding Ser-209 in Wnt3a are essential for palmitoleoylation.
Significance: This analysis provides insight into mechanisms operative within the membrane-bound O-acyltransferase family.
Keywords: Enzyme Mutation, Fatty Acid, Membrane Protein, Protein Palmitoylation, Wnt Signaling
Abstract
Wnts comprise a family of lipid-modified, secreted signaling proteins that control embryogenesis, as well as tissue homeostasis in adults. Post-translational attachment of palmitoleate (C16:1) to a conserved Ser in Wnt proteins is catalyzed by Porcupine (Porcn), a member of the membrane bound O-acyltransferase (MBOAT) family, and is required for Wnt secretion and signaling. Moreover, genetic alterations in the PORCN gene lead to focal dermal hypoplasia, an X-linked developmental disorder. Despite its physiological importance, the biochemical mechanism governing Wnt acylation by Porcn is poorly understood. Here, we use a cell-based fatty acylation assay that is a direct readout of Porcn acyltransferase activity to perform structure-function analysis of highly conserved residues in Porcn and Wnt3a. In total, 16-point mutations in Porcn and 13 mutations in Wnt3a were generated and analyzed. We identified key residues within Porcn required for enzymatic activity, stability, and Wnt3a binding and mapped these active site residues to predicted transmembrane domain 9. Analysis of focal dermal hypoplasia-associated mutations in Porcn revealed that loss of enzymatic activity arises from altered stability. A consensus sequence within Wnt3a was identified (CXCHGXSXXCXXKXC) that contains residues that mediate Porcn binding, fatty acid transfer, and Wnt signaling. We also showed that Ser or Thr, but not Cys, can serve as a fatty acylation site in Wnt, establishing Porcn as an O-acyltransferase. This analysis sheds light into the mechanism by which Porcn transfers fatty acids to Wnt proteins and provides insight into the mechanisms of fatty acid transfer by MBOAT family members.
Introduction
Post-translational modification of proteins with one or more fatty acids controls a dynamic range of cellular processes such as membrane trafficking and targeting, lipid raft targeting, and signaling activity (1–5). The most common form of fatty acylation is protein S-palmitoylation, which occurs by covalent attachment of the 16-carbon saturated fatty acid, palmitate (C16:0), to one or more cysteine residues through a thioester linkage. The mechanism of action of S-palmitoyl acyltransferases has been well documented and extensively reviewed (2). Recently, secreted signaling proteins (Hedgehog and Wnt) and hormones (Ghrelin) have been reported to contain fatty acids attached via amide (N)-linkage or oxyester (O)-linkage (3, 6, 7). The diverse mechanisms governing these reactions are currently being uncovered.
Wnts are secreted, lipid-modified signaling proteins that control embryonic development and self-renewal in adult tissue (8–12). Wnt proteins activate signaling cascades that result in the transcription of genes involved in proliferation, differentiation, and migration. Aberrant activation of the Wnt signaling pathway has been linked to a variety of human cancers, including melanoma, breast, colon, and head and neck (9, 13, 14). Because of its importance in diseases, there is significant interest in understanding the biochemistry of Wnt processing and secretion.
Production of an active Wnt signal is initiated by the acyltransferase Porcupine (Porcn),2 which catalyzes transfer of palmitoleate (C16:1), a 16-carbon monounsaturated fatty acid, to a highly conserved serine residue (Ser-209 in Wnt3a) (3, 15–17). Attachment of palmitoleate is required for Wnt function. Mutation of the conserved Ser to Ala in Wnt or pharmacological inhibition of Porcn blocks Wnt fatty acylation, intracellular trafficking and secretion, binding to its cell surface receptor, Frizzled, and signaling activity (3, 15, 17–19). Porcn is required for Wnt activity during development, because mutations in the PORCN gene cause focal dermal hypoplasia (FDH), also known as Goltz syndrome, an X-linked multisystem disorder characterized by developmental malformations (20–22). In adults, Porcn is a therapeutic target for blockade of Wnt pathway activation in Wnt-driven disorders (23, 24). Porcn inhibitors reduce tumor growth in Wnt-driven mammary cancers in mice and in head and neck cancer cell lines (25, 26).
Porcn is an endoplasmic reticulum (ER)-resident integral membrane protein. It has 11 predicted transmembrane domains (TMDs) and belongs to the membrane-bound O-acyltransferase (MBOAT) family (27). In mammals, 16 MBOAT members encoded by 11 genes have been identified. Biochemical analysis of this family has been hampered by the extreme hydrophobicity of the proteins because of the presence of multiple TMDs. Most MBOAT family members are involved in neutral lipid biosynthesis (ACAT1/2 and DGAT1/2) and phospholipid remodeling (LPEAT1–4 and GUP1). Only three members, Porcn, Hedgehog acyltransferase (Hhat), and ghrelin O-acyltransferase (GOAT), have been shown to have protein substrates. Hhat catalyzes attachment of palmitate to the N-terminal Cys of Hedgehog proteins (28), and GOAT mediates transfer of octanoate to Ser-3 of proghrelin, an appetite-stimulating hormone (29). All family members share a conserved MBOAT homology domain that is thought to be necessary for enzymatic activity. This region harbors an invariant His residue positioned within a stretch of highly conserved hydrophobic amino acids and a well conserved Asp/Asn surrounded by hydrophilic residues (27). Mutation of the conserved His reduces the catalytic activity of ACAT1/2, DGAT1, GOAT, Hhat, and Porcn, placing this residue as part of the putative active site (17, 29–32). Mutational analyses of other conserved residues in ACAT, DGAT, and Hhat have identified several residues within the MBOAT homology domain that are required for catalysis. However, no unifying consensus model for the MBOAT active site has been described.
In this study, we present a structure-function analysis of Porcn acyltransferase activity. Rather than relying on reporters of Wnt signaling activity as an indirect readout for Porcn activity (33), we directly monitored monounsaturated fatty acid transfer to Wnt by Porcn. We exploited a cell-based fatty acylation assay that uses [125I]iodo-pentadecenoic acid (125I-IC15:1), a radioiodinated palmitoleate analog, as a substrate for Porcn. 125I-IC15:1 is sterically similar to palmitoleate (C16:1) and provides a robust, accurate, and direct readout of Porcn acyltransferase activity in the cell (17). To determine the functional significance of highly conserved residues, we generated a series of truncation and point mutants within Porcn that targeted the MBOAT homology domain as well as residues that are mutated in FDH. We identified residues that are important for Wnt binding, acyltransferase activity, and stability of Porcn. In addition, the positional requirement of highly conserved residues surrounding the Wnt3a palmitoleoylation site was analyzed. Taken together, this structure-function analysis has enabled us to generate an initial working map of the active site of Porcn and to define a consensus sequence for Wnt palmitoleoylation.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
The following antibodies were purchased as follows: rabbit polyclonal anti-Wnt3a (Cell Signaling Technology), mouse monoclonal anti-FLAG M2, rabbit polyclonal anti-FLAG (Sigma-Aldrich), mouse monoclonal anti-Myc 9E10 (Monoclonal Antibody Core Facility, MSKCC), HRP-conjugated secondary antibodies (Thermo Scientific), goat anti-rabbit Alexa Fluor 594, and goat anti-mouse Alexa Fluor 488 (Invitrogen). Protein A/G plus agarose was purchased from Alpha Diagnostics Intl., Inc.
Plasmids, Cell Culture, and Transfection
Cloning of Myc-tagged Wnt3a into pcDNA3.1 has been previously described (28). N-terminal FLAG-tagged Porcn cDNA was generated from a murine Porcupine clone in pcDNA3.1 (a gift from Dr. Joseph Goldstein (UT Southwestern)). WT Porcn corresponds to NCBI accession number NP_076127.1. PCR fragments encoding C-terminally FLAG-tagged Porcn constructs Δ1–55 and Δ1–94 and N-terminally FLAG-tagged Porcn Δ416–613 and Δ371–461 were amplified by PCR and ligated into the NheI/KpnI restriction sites of pcDNA3.1. All Wnt3a and Porcn point mutants were generated by site-directed mutagenesis using the QuikChange mutagenesis kit (Agilent Technologies). COS-1 cells were obtained from ATCC and grown in DMEM containing 10% FBS. Super TOPFlash/FOPFlash and pRL-TK plasmids were a kind gift from Dr. Anthony M. C. Brown (Weill Cornell Medical College). Super TOPFlash (STF) has 8× T cell factor binding sites upstream of a thymidine kinase promoter and firefly luciferase ORF. Super FOPFlash contains 8× mutated T cell factor-binding sites. Transfections were performed with Lipofectamine 2000 (Invitrogen).
Synthesis of 125I-Iodo-Fatty Acids
Iodo cis-9-pentadecenoic acid (IC15:1) was synthesized by the Sloan-Kettering Organic Synthesis Core Lab. Conversion into 125I-iodo fatty acids was performed using 125I-NaI (PerkinElmer Life Sciences) as previously described (34, 35).
Metabolic Labeling of Cells with Radioiodinated Fatty Acid Analogs
COS-1 cells transfected with plasmids encoding Wnt3a-myc and FLAG-Porcn were incubated in DMEM containing 2% dialyzed FBS for 2 h. Cells were labeled with 30 μCi of 125I-IC15:1 for 5 h at 37 °C. Cells were lysed in 500 μl of radioimmunoprecipitation assay buffer, and cleared total cell lysates were immunoprecipitated with 5 μl of the indicated antibody and 60 μl of protein A/G-agarose beads for 16 h at 4 °C. Immunoprecipitates were eluted in sample buffer containing 60 mm DTT and electrophoresed on 12.5% SDS-PAGE gels. Radiolabel incorporation into Wnt3a was detected on a Fuji FLA-700 phosphorimager and quantified using ImageGauge software. Protein expression was detected by Western blotting using enhanced chemiluminescence (Thermo Scientific) and quantified with Quantity One software (Bio-Rad) on a GS-800 calibrated densitometer.
Immunofluorescence and Confocal Microscopy
Forty-eight hours after transfection, COS-1 cells were washed twice with PBS and fixed with 4% (v/v) paraformaldehyde in PBS for 20 min at room temperature. The cells were washed with PBS and permeabilized with 0. 2% Triton X-100 in PBS for 5 min at room temperature and then washed twice with PBS. The cells were stained with anti-FLAG antibody at 1:250 dilution in 3% BSA and incubated for 1 h. The cells were placed in PBS for 20 min, followed by a 45-min incubation with secondary antibodies and 1:5000 dilution of Hoechst dye in PBS. Coverslips were mounted on slides using ProLong Gold mounting solution (Invitrogen). The images were acquired on a Leica inverted SP5 confocal microscope using a 63× oil immersion objective.
Wnt Signaling Activity Assays
HEK293T cells in 100-mm plates were transfected with 3 μg of the indicated Wnt3a-Myc construct, 3 μg of STF or Super FOPFlash, and 0.3 μg pRL-TK. Twenty-four hours post-transfection, cells were seeded in 6-well plates at an initial density of 0.6 × 106 cells/well and grown for 24 h. Cells were lysed according to manufacturer specifications, and Wnt3a pathway activity was detected using the Dual-Luciferase® reporter assay (Promega). Firefly luciferase and Renilla luciferase (RL) activity were recorded as relative luciferase units using a VeritasTM microplate luminometer (Promega). Relative Wnt signaling activity measurements were calculated by normalizing firefly luciferase to RL, followed by normalization of TOPFlash to FOPFlash.
Protein Stability Assay
Porcn-transfected COS-1 cells were split into 60-mm dishes and cultured for 24 h at 37 °C. The cells were placed in DMEM containing 10% FBS, 100 μg/ml of cycloheximide, and 40 μg/ml of chloramphenicol and incubated for 0, 5, 10, and 24 h. At each time point, the cells were lysed in 500 μl of 1× radioimmunoprecipitation assay buffer, and Porcn was immunoprecipitated from the total cell lysate with anti-FLAG antibody and 60 μl of protein A/G-agarose beads for 16 h at 4 °C. Samples were eluted in sample buffer containing 60 mm DTT, electrophoresed on 12.5% SDS-PAGE gels, transferred onto PVDF membranes, and probed with anti-FLAG antibody. Protein expression was quantified as described above.
Co-immunoprecipitation Assay
COS-1 cells were co-transfected with 3 μg of Wnt3a-Myc and 4 μg of FLAG-Porcn cDNAs. Forty-eight hours after transfection, cells were rinsed with STE and lysed with 500 μl of modified radioimmunoprecipitation assay for 15 min (50 mm Tris, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 50 mm NaCl, 1 mm EDTA). Total cell lysates were immunoprecipitated with 5 μl of the indicated antibody and 60 μl of protein A/G-agarose beads for 16 h at 4 °C and eluted in sample buffer containing 60 mm DTT. Samples were electrophoresed on 12.5% SDS-PAGE gels, transferred onto PVDF membranes, and probed with anti-Wnt3a or anti-Myc or anti-FLAG antibodies, as indicated.
Bioinformatics
MBOAT family sequences (Mus musculus) were identified by querying the NCBI-conserved domain database against pfam03062. Multiple sequence alignment of murine MBOAT and Wnt family members was carried out using the ClustalW2 multiple sequence alignment server. Porcn membrane topology prediction was performed using the MEMSAT-SVM server (36), and a graphical model was generated by Protter Server (37).
RESULTS
N- and C-terminal Mutants of Porcn Lack Acyltransferase Activity and Aggregate in the ER
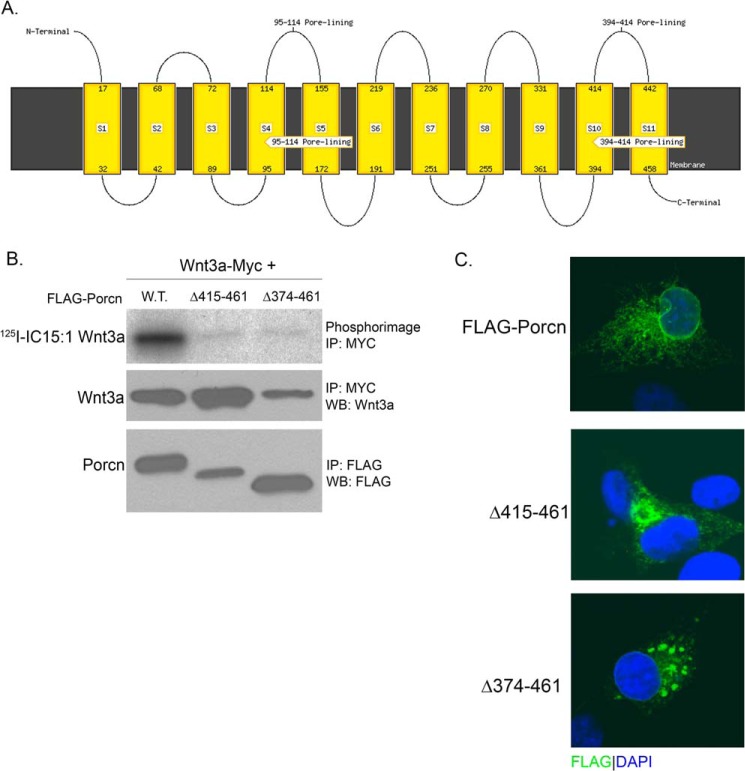
To identify a minimal region of Porcn retaining acyltransferase activity, we generated N- and C-terminal truncations. A web-based topology prediction program, MEMSAT-SVM (38), was utilized to aid in the design and placement of truncation points to avoid membrane-spanning regions. We used the MEMSAT-SVM topology prediction server for its consistency in predicting the topology of Porcn across species and its accuracy in predicting GOAT topology (39). MEMSAT-SVM predicted 11 TMDs with invariant His-341 embedded in TMD 9 (Fig. 1A). Based on this model, we engineered N- and C-terminal truncations, missing one or more predicted transmembrane helices, each carrying a FLAG tag at the opposite end of the truncation site. The enzymatic activity of these mutants was tested in a cell-based Wnt palmitoylation assay (17). COS-1 cells were co-transfected with cDNAs encoding Myc-tagged Wnt3a and either WT or truncated Porcn and labeled with 125I-IC15:1. Wnt3a was immunoprecipitated from cell lysates, and the amount of 125I-IC15:1 incorporated into Wnt3a was determined by phosphorimaging analysis after SDS-PAGE. Two C-terminal truncations (Δ415–461 and Δ374–461) expressed truncated Porcn proteins, but these mutants failed to promote Wnt3a acylation (Fig. 1B). We tested whether the enzymatic defects observed were due to altered intracellular localization. Analysis by indirect immunofluorescence and confocal imaging revealed punctate immunostaining, a pattern characteristic of aggregated, and misfolded protein (Fig. 1C). Expression of the N-terminal deletion mutants was not detected by Western blot, but immunofluorescent confocal imaging analysis again revealed aggregated protein (data not shown). Thus, these N- and C-terminal deletion mutants are misfolded and devoid of protein acyltransferase (PAT) activity.
FIGURE 1.
Porcn truncation mutants lack acyltransferase activity and aggregate in the ER. A, predicted transmembrane topology map of murine Porcn protein (NP_076127.1) generated using the MEMSAT-SVM server. The rectangles represent predicted transmembrane helices, and numbers indicate boundaries. B, COS-1 cells co-expressing Wnt3a-Myc and FLAG-Porcn, either WT or truncated, were labeled with 125I-IC15:1 for 5 h. Cell lysates were immunoprecipitated (IP) with anti-Myc or anti-FLAG antibodies and analyzed by SDS-PAGE and phosphorimaging (top panel) or Western blotting (WB, middle and bottom panels) with anti-Wnt3a or anti-FLAG antibodies. C, COS-1 cells expressing WT or truncated FLAG-tagged Porcn were fixed, stained with DAPI (blue), and analyzed by indirect immunofluorescence with anti-FLAG antibody (green) and confocal microscopy.
Identification of Key Residues in the MBOAT Homology Domain of Porcn
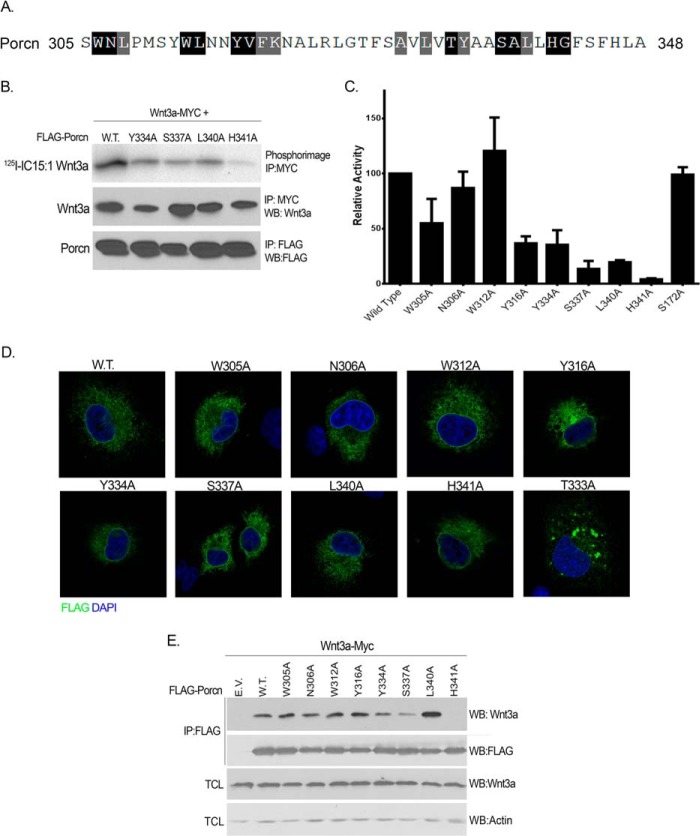
The MBOAT homology domain is the most conserved region of the MBOAT family, with residues sharing 50% or more conservation (Fig. 2A). Mutational analysis identified several residues in this region in ACAT1/2, DGAT, and Hhat that are critical for catalytic activity (30–32), but the requirement of these amino acids for Porcn PAT activity has not been explored. In murine Porcn, the MBOAT homology domain spans residues 305–342 and harbors putative catalytic sites Asn-306 and His-341. To determine the functional significance of this region in Porcn, nine conserved residues within the MBOAT homology domain were individually mutated to Ala. Based on their effect on Porcn PAT activity, these mutants could be classified into three categories. Two mutations did not significantly alter Porcn activity (N306A and W312A), indicating that these residues are not required for acyltransferase activity (Fig. 2C). Three other mutants (W305A, Y316A, and Y334) exhibited moderate defects in activity (30–50% of WT) (Fig. 2C). Three mutants (S337A, L340A, and H341A) had little to no activity (<20% of WT), indicating that these residues are critical for acyltransferase activity (Fig. 2, B and C). S172A was used as a control, because it does not localize to the MBOAT homology domain, and mutating this residue had no effect on Porcn activity. T333A was the only point mutant whose expression was not detected by Western blotting. Immunofluorescent confocal imaging analysis detected low expression levels, altered intracellular localization, and protein aggregation in the ER. Thus, a complete assessment of the effect of this mutant on Porcn activity and/or biochemical properties could not be established and was not included in further characterization (Fig. 2D). Taken together, this mutational analysis identified six residues within the MBOAT homology domain of Porcn that are essential for enzymatic activity.
FIGURE 2.
Mutational analysis of conserved residues within the MBOAT homology domain of Porcn. A, residues that are conserved (black boxes) or similar (gray boxes) in the MBOAT homology domain in 50% or more of the 16 MBOAT family members in the mouse (M. musculus) are highlighted. B, COS-1 cells co-expressing Wnt3a-Myc and either WT or mutant FLAG-Porcn were labeled and analyzed as in Fig. 1b. The experiment was performed three times in duplicate; a representative image is shown. C, quantification of experiments in B. Relative acyltransferase activity was determined by calculating the level of 125I-IC15:1 incorporation per amount of immunoprecipitated (IP) Wnt3a for each mutant Porcn construct and then normalizing to WT Porcn, which was set to 100%. Each bar represents the average of three experiments and is expressed as the percentage of WT Porcn activity; error bars indicate S.D. D, the intracellular localization of WT and mutant Porcn was analyzed as in Fig. 1C. E, lysates from COS-1 cells co-expressing Wnt3a and the indicated Porcn constructs were immunoprecipitated with anti-FLAG antibody followed by Western blotting (WB) with anti-Wnt3a or anti-FLAG antibody (top two panels). Total cell lysates (TCL) were analyzed directly by Western blotting with anti-Wnt3a and anti-actin antibodies.
We next tested the effect of the MBOAT homology domain mutations on protein stability. Porcn transfected COS-1 cells were treated with cycloheximide to block new protein synthesis, and levels of WT and mutant proteins were monitored as a function of time. WT Porcn was very stable, with 60% still present after 24 h of cycloheximide blockade. All mutants, except Y316A, exhibited stability comparable with WT, ranging from 48 to 98% remaining at 24 h (Table 1). Y316A was much less stable than WT, with 24% remaining after 24 h (Table 1). With the exception of T333A, proper intracellular localization of all point mutants to the ER was confirmed by indirect immunofluorescence and confocal microscopy, indicating that defects in enzymatic activity were not due to mislocalization or misfolding (Fig. 2D).
TABLE 1.
Relative stability of Porcn point mutants
| Porcn | Remaining at 24 ha | S.E. |
|---|---|---|
| % | ||
| MBOAT homology domain mutants | ||
| Wild type | 60.78 | 3.27 |
| S172A | 61.47 | 10.76 |
| W305A | 68.89 | 2.46 |
| N306A | 76.33 | 3.64 |
| W312A | 48.35 | 4.32 |
| Y316A | 24.74 | 10.03 |
| Y334A | 70.53 | 4.11 |
| S337A | 58.47 | 8.56 |
| L340A | 59.00 | 0.14 |
| H341A | 98.99 | 1.01 |
| FDH-associated mutants | ||
| S136F | 21.93 | 13.95 |
| R228C | 82.86 | 5.84 |
| L331R | 34.06 | 8.18 |
| R365Q | 30.28 | 19.31 |
a Relative stability of Porcn point mutants after 24 h of incubation with cycloheximide and chloramphenicol. The data are expressed as percentages of 0 h control (n = 2).
The ability of the Porcn mutants to bind to the Wnt3a protein substrate was assessed in a co-immunoprecipitation assay using COS-1 cells co-expressing Wnt3a and Porcn. Three Porcn mutants with severe enzymatic activity defects (Y334A, S337A, and H341A) exhibited impaired ability to co-immunoprecipitate with Wnt3a; all other mutants associated with Wnt3a to the same extent as WT (Fig. 2E). Interestingly, the catalytically dead mutant L340A was still able to bind Wnt3a, suggesting that this residue might be involved in facilitating fatty acid recognition rather than Wnt protein substrate binding.
FDH-associated Mutations Are Detrimental to Porcn Stability
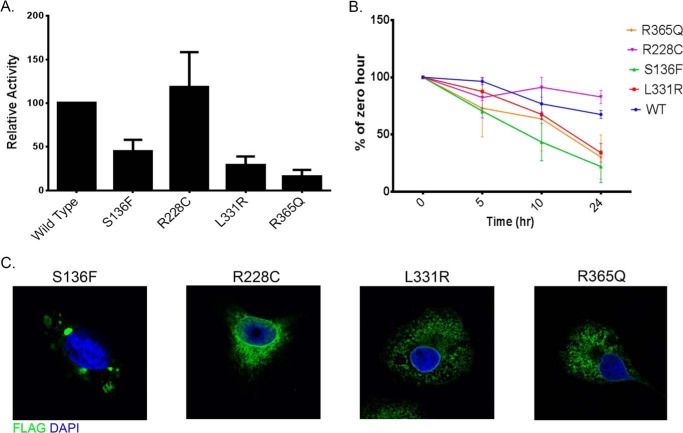
Over 70 mutations in the PORCN gene have been identified in FDH patients, including single nucleotide changes, microdeletions, insertions, and others (40–42). Approximately 26% of FDH-causing mutations are missense mutations in the PORCN coding region, but how these mutants affect Porcn function is not clear. To understand the biochemical mechanism by which missense mutations alter Porcn activity, the FDH-associated Porcn mutants L331R, R365Q, R228C, and S136F were generated and tested for PAT activity, stability and intracellular localization. In contrast to MBOAT homology domain mutants, all FDH-associated mutations except R228C exhibited moderate to severe enzymatic defects (Fig. 3A) and compromised protein stability (Fig. 3B). The R228C mutation was described in an FDH patient with two heterozygous mutations (43). Because Arg-228 is not conserved across species, and the patient's mother, who is clinically unaffected, has the same heterozygous mutation, the pathological significance of the R228C mutation was uncertain. Our finding that Arg-228 Porcn is essentially indistinguishable from WT Porcn suggests that this mutation does not contribute to the disease state. In addition, indirect immunofluorescence and confocal imaging analysis of S136F revealed protein aggregation, probably caused by misfolding (Fig. 3C). These findings suggest that altered stability and/or folding could be the underlying cause for enzymatic dysfunction in at least some of the FDH mutants described to date.
FIGURE 3.
FDH-associated mutations alter stability and intracellular localization of Porcn. A, COS-1 cells co-expressing Wnt3a-myc and the indicated Porcn constructs were labeled and analyzed as in Fig. 1B. Each bar represents the average of three experiments and is expressed as the percentage of WT Porcn activity (mean ± S.D.). B, COS-1 cells expressing the indicated Porcn constructs were treated with 100 μg/ml of cycloheximide and 40 μg/ml of chloramphenicol for 0, 5, 10, or 24 h. At each time point, lysates were immunoprecipitated with anti-FLAG antibodies and analyzed by Western blotting after SDS-PAGE. The amount of FLAG signal at each time point was determined by densitometry; each point indicates the mean ± S.D. (n = 2). The values for the percentage of Porcn protein remaining at 24 h were: 60.78% for WT, 21.93% for S136F, 82.86% for R228C, 34.06% for L331R, and 30% for R365Q. C, the intracellular localization of WT and mutant Porcn constructs was analyzed as in Fig. 1C.
Identification of Conserved Residues in Wnt Required for Porcn-dependent Fatty Acylation
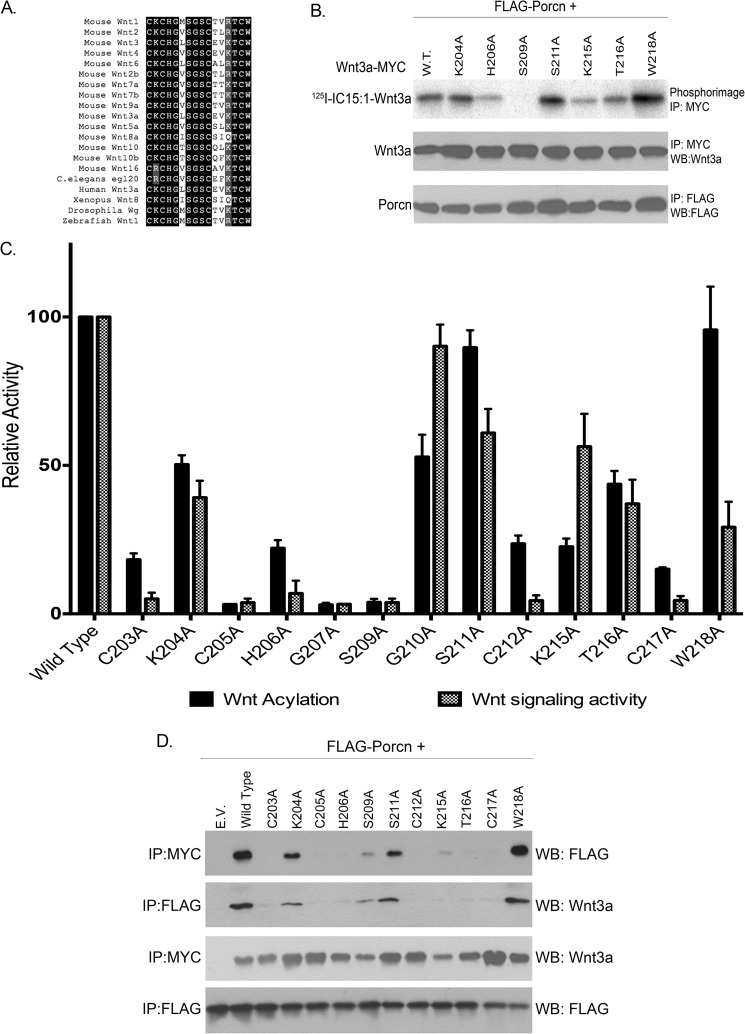
Although Wnt3a was originally reported to be palmitoylated on Cys-77 (44), further studies revealed that this residue is instead engaged in a disulfide bond and that the authentic fatty acylation site is Ser-209 (3, 15, 17). The residues surrounding Ser-209 in Wnt3a are highly conserved across species, from hydra to human Wnt proteins (Fig. 4A), but the importance of these amino acids for Porcn-mediated fatty acylation has not been explored. Alanine-scanning mutagenesis was used to alter conserved residues surrounding Ser-209 in Wnt3a. The ability of each mutant protein to incorporate 125I-IC15:1 was tested. Five of the Wnt3a mutants were acylated to the same extent (S211A and W218A) or to levels 40–50% (K204A, G210A, and T216A) of WT Wnt3a (Fig. 4, B and C). The other mutants exhibited severe acylation defects (<20% of WT) (Fig. 4C). Of note, the presence of each of the four Cys that form disulfide bonds in Wnt proteins (15, 45) was required for fatty acylation. We then tested the ability of the Wnt3a mutants to bind Porcn in a co-immunoprecipitation assay (Fig. 4D). S211A and W218A, which are acylated to WT levels, associated with Porcn, as did K204A, which has moderately decreased fatty acylation. None of the acylation-deficient mutants were able to co-immunoprecipitate with Porcn.
FIGURE 4.
Mutational analysis of conserved residues surrounding Ser-209 in Wnt3a. A, multiple sequence alignment of a highly conserved region of Wnt family members from diverse species. Black boxes denote residues that are conserved in 80% or more sequences. B, COS-1 cells transfected with FLAG-Porcn and the indicated Wnt3a constructs were labeled with 125I-IC15:1 for 5 h. Cell lysates were immunoprecipitated (IP) with anti-Myc or anti-FLAG antibodies and analyzed by SDS-PAGE and phosphorimaging (top panel) or Western blotting (WB, middle and bottom panels) with anti-Wnt3a or anti-FLAG antibodies. The experiment was repeated three times in duplicate; a representative image is shown. C, black bars, quantification of experiments in B. Phosphorimaging signals were normalized to Wnt3a protein levels and then expressed as a percentage of WT Wnt3a (set at 100%). Each bar represents the mean ± S.D. (n = 3–6). Gray patterned bars, HEK293T cells were transfected with STF or FOPFlash, RL, and either WT or mutant Wnt3a. 48 h after transfection, STF luciferase activity was measured, normalized to RL and FOPFlash activity, and expressed as a percentage of WT; bars represent means ± S.E. (n = 3). D, lysates from COS-1 cells co-expressing WT Porcn and the indicated Wnt3a constructs were immunoprecipitated with anti-Myc or anti-FLAG antibody followed by Western blotting with anti-Wnt3a or anti-FLAG antibody. E. V., empty vector.
The S209A mutant of Wnt3a has been shown to have defective signaling activity, and this has led to the conclusion that Wnt palmiteoylation is required for signal transduction (3, 17, 33). To examine the correlation between protein fatty acylation levels and Wnt signaling, we utilized a luciferase-based reporter of Wnt activity, STF. HEK293T cells were transfected with STF, RL, and Wnt3a, and luciferase activity was measured. All of the Wnt3a point mutants exhibited reduced signaling activity compared with WT Wnt3a (Fig. 4C). These reductions in signaling output, in general, correlated with reduced levels of fatty acylation. However, the W218A construct exhibited near WT levels of 125I-IC15:1 incorporation but displayed signaling defects, arguing that this residue might be involved in mediating events downstream of Porcn activity. In summary, these results indicate that the highly conserved region surrounding Ser-209 plays a critical role in mediating the interaction between Porcupine and Wnt and that disruption of this region results in acylation defects, thus affecting downstream signaling activity.
Porcupine Can Acylate Ser and Thr but Not Cys Residues
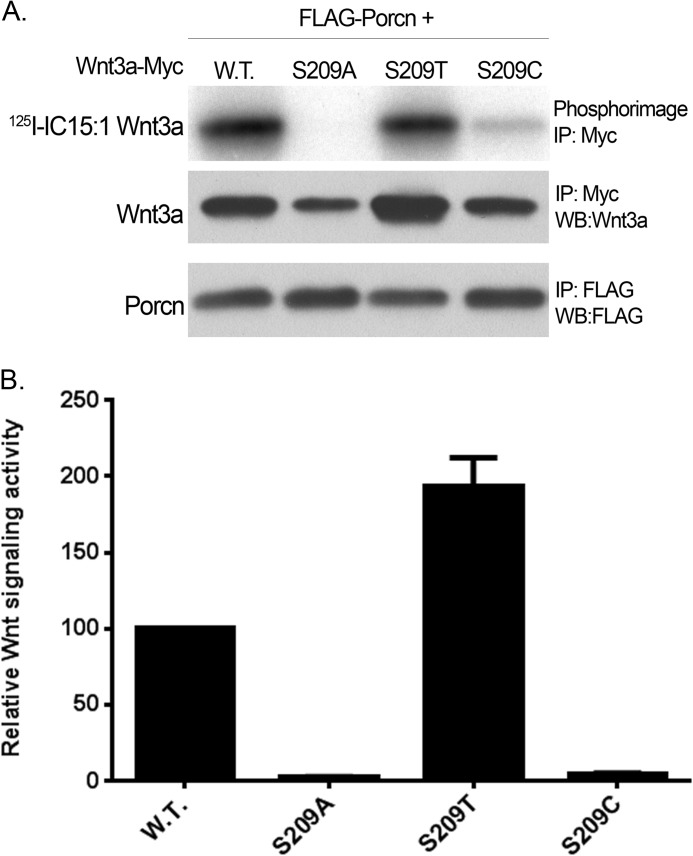
With the exception of Hhat, all MBOAT family members with known substrates are O-acyltransferases, transferring fatty acids to hydroxyl groups (-OH) of proteins or lipids. For Porcn and GOAT, the modified residue on the protein substrate is Ser. We tested whether Thr or Cys in position 209 of Wnt3a would substitute for Ser as fatty acid acceptor for Porcn. As depicted in Fig. 5A, 125I-IC15:1 was transferred to Wnt3a containing S209T but not to S209C. The S209T construct exhibited signaling activity even greater than that of WT Wnt3a, whereas S209A and S209C mutants were inactive (Fig. 5B). Thus, Porcn strictly functions as an O-acyltransferase and can form oxyester linkages to either Ser or Thr.
FIGURE 5.
Porcn can transfer a fatty acid to either Ser or Thr at position 209 in Wnt3a. A, COS-1 cells expressing WT Porcn and the indicated Wnt3a constructs were labeled and analyzed as in Fig. 1b. The experiment was repeated two times in duplicate; a representative image is shown. B, relative luciferase activity for the Wnt3a constructs in A was determined as in Fig. 4C. IP, immunoprecipitation; WB, Western blot.
DISCUSSION
A Map of the Active Site of Porcn
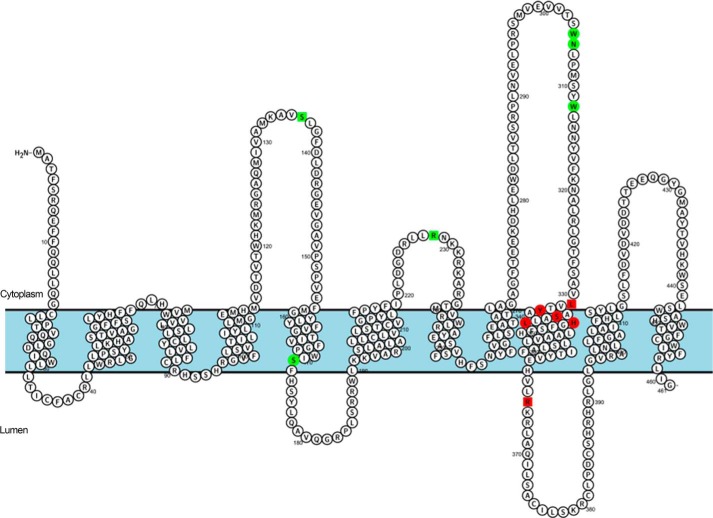
Porcn-mediated attachment of palmitoleate to Wnt proteins ensures Wnt passage through the secretory pathway and proper signaling activity. At the biochemical level, this reaction has been difficult to study in a quantitative manner, partly because of the hydrophobic nature of Porcn and the lack of tools to accurately detect acylated Wnt. In this report, we undertook a systematic approach to dissect the complexities of the Porcn-mediated fatty acylation reaction. A cell-based fatty acylation assay that provides an accurate and direct measurement of Porcn acyltransferase activity (17) was combined with alanine-scanning mutagenesis of highly conserved residues and regions within Porcn and Wnt3a. Here we identify key residues that are crucial for the intracellular localization, stability, and activity of these proteins and provide molecular level information regarding interactions between Wnt and Porcn. In total, 16 Porcn mutants (2 truncation mutants, 9 MBOAT homology domain mutants, 1 random site mutant, and 4 FDH-associated mutants) and 13 Wnt point mutants were analyzed. When mapped within a predicted topology model of Porcn, a striking pattern was observed for the Porcn mutants. Nearly all of the inactive Porcn mutants localize to TMD 9, whereas all of the constructs retaining catalytic activity localize to predicted cytoplasmic or luminal loops (Fig. 6). This analysis defines a critical active site region within predicted TMD 9 essential for catalysis by Porcn.
FIGURE 6.
Predicted transmembrane topology map of the active site of Porcn. Graphical representation of the predicted topology of Porcn as generated by Protter Server (37). MBOAT homology domain (circles) and FDH-associated mutants (squares) are highlighted according to their effect on Porcn PAT activity. Red boxes represent residues that are essential for enzymatic activity (inactive mutants), and green boxes indicate residues that are not required for PAT activity (active mutants).
A recent study examined the effect of multiple Porcn mutations that occur in FDH on Wnt signaling activity (33). Several of the mutants overlap with those tested in this study. Three of the four FDH mutants, we analyzed (S136F, L331R, and R365Q) showed decreased stability and mislocalization, which is likely the cause of the decreased fatty acyltransferase activity that we observed (Fig. 3) and could potentially explain the disease phenotype. Proffit et al. (33) also noted decreased expression levels for R365Q Porcn but obtained the opposite result for S136F and L331R Porcn, which exhibited nearly normal activity when assayed in a β-catenin signaling assay. The reason for this difference may be related to the use of an assay that is distal and not a direct readout of the initial biochemical event, Porcn-mediated acylation of Wnt.
We observed that N- and C-terminal truncation mutants of Porcn were unstable and inactive. These residues are likely needed to maintain enzyme integrity and when deleted cause the protein to become misfolded and more susceptible to degradation. However, this was not the case for the MBOAT homology domain mutants which, except for Y316A, had stabilities close to that of WT Porcn. Despite its lower stability, Y316A retained a moderate level of acyltransferase activity. With the exception of T333A, none of the MBOAT homology domain mutants that we examined exhibited observable alterations in subcellular localization. Thus, residues in the inactive mutants are likely involved in mediating the acyltransferase reaction, by either binding to the protein or the fatty acid substrate or directly catalyzing fatty acid transfer.
Invariant His and Asn/Asp residues are highly conserved and regarded as putative catalytic sites for all MBOATs. However, several differences regarding the requirement for these residues have been observed. Mutation of the invariant histidine residues abrogates activity in all family members tested but only reduces activity by 50% in Hhat. The residue immediately adjacent to the conserved His is required for catalytic activity in Porcn and Hhat, but not in ACAT1 (30, 32). Of note, the L340A Porcn mutant was catalytically inactive but could still bind to Wnt3a. This suggests that Leu-340 might be involved in binding to palmitoleolate, consistent with its localization within a TMD (Fig. 6). Conversely, the conserved Asn/Asp is not required for Porcn acyltransferase activity (Asn-306), but it is essential for Hhat and GOAT activity (29, 32). Mutational analyses of other conserved residues within the MBOAT homology domain have been performed for ACAT1, ACAT2, DGAT1, and DGAT2, based on sequence conservation across species, and in Hhat, based on sequence homology among MBOATs with protein acyltransferase activity. Similarities and differences among the overlapping mutants are summarized in Table 2. Mutation of the Ser-337 equivalent results in complete inactivity or severe enzymatic defects in ACAT1 and ACAT2 (46). We observed a similar result for Porcn, whereas another group reported that the S337A Porcn mutation was able to fully restore Wnt signaling activity to Porcn null cells (33). In Porcn, Y334A exhibited moderate defects in activity, whereas mutation of this residue in Hhat has no detectable effect (32). Mutations of Trp-305 in Porcn or Phe-338 in Hhat results in partial loss of acyltransferase activity (32). Considering that Porcn, Hhat, and GOAT catalyze distinct biochemical reactions with different fatty acid and protein substrates, it is perhaps not surprising that the active sites of these enzymes are not identical.
TABLE 2.
Relative acyltransferase activity of other MBOAT family member mutants
Percentages indicate relative enzymatic activity compared with wild type controls.
| Residue in Porcn | PORCN | HHAT | GOAT | ACAT1 | ACAT2 |
|---|---|---|---|---|---|
| % | % | % | % | % | |
| Trp-305 | 50 | 32 | |||
| Asn-306 | 80 | 10 | 0 | ||
| Tyr-334 | 40 | 100 | |||
| Ser-337 | 13 | 0 | 27 | ||
| Leu-340 | 20 | 30 | 70a | ||
| H341A | 4 | 50 | 0 | 0 | 0 |
| References | (32) | (29) | (46) | (48) | |
a This residue in ACAT1 was mutated to a Cys. All others represent Ala mutations.
Identification of a Consensus Sequence within Wnt Necessary for Porcn-mediated Fatty Acylation
Site-directed mutagenesis of the sequences immediately upstream and downstream of the conserved Ser acylation site led to identification of residues required for fatty acylation of Wnt3a. Based on this analysis, we propose the following consensus sequence for Porcn-mediated acylation: CXCHGXSXXCXXKXC. This sequence overlaps significantly with the Wnt family sequence conservation surrounding Ser-209 (Fig. 4A) but identifies multiple conserved residues (Lys-204, Gly-210, Ser-211, Thr-216, and Trp-218) that are not required for Wnt acylation. The products of a blastp interrogation of the human proteome are nearly all Wnt proteins. However, we found two proteins that retain six of the residues surrounding Ser-209: β-defensin and netrin G1, both of which are secreted proteins that could be potential Porcn substrates. Further experiments will be required to explore this possibility.
A close correlation was observed between 125I-IC15:1 incorporation and signaling activity of the Wnt mutants (Fig. 4C). However, W218A Wnt3a was acylated to levels near WT but displayed signaling defects. This suggests that this residue might be involved in mediating distal events downstream of Porcn, such as the interaction between Wnt and its cell surface receptor, Frizzled. In fact, the equivalent Trp-218 residue in Xenopus Wnt8 (Trp-196) is engaged in van der Waals interactions with an Asn residue in the cysteine-rich domain of Frizzled (15). The K215A construct was a poor substrate for Porcn-mediated acylation but exhibited nearly 50% of the signaling activity of WT Wnt3a. One could speculate that the Lys to Ala mutation induces a conformational change that is conducive to Wntless binding and Wnt pathway activation, thereby bypassing the acylation step.
Both GOAT and Porcn transfer a fatty acid to a Ser residue via oxyester linkage. Although hedgehog proteins, which are the only known substrates for mammalian Hhat, are palmitoylated on an N-terminal Cys, we have shown that Hhat can also attach palmitate to Ser (47). This could potentially occur through initial attachment via an oxyester link, followed by intramolecular rearrangement to an amide bond. Here we show that Porcn can also recognize a Thr residue placed within the consensus sequence for Wnt3a palmitoleoylation, a property shared by GOAT (29). This finding supports the designation of Porcn as an O-acyltransferase that attaches fatty acids via oxyester linkage. Because all Wnt proteins described to date contain Ser and not Thr at this site, it is likely that Ser is required for downstream functions of Wnt that occur after acylation.
This work was supported, in whole or in part, by National Institutes of Health Grants GM57966 (to M. D. R.) and F31GM100691 (to J. R. E.).
- Porcn
- Porcupine
- MBOAT
- membrane-bound O-acyl transferase
- FDH
- focal dermal hypoplasia
- PAT
- palmitoyl acyltransferase
- Hhat
- Hedgehog acyltransferase
- GOAT
- ghrelin O-acyltransferase
- ACAT
- acyl CoA cholesterol acyltransferase
- DGAT
- diacylglycerol acyltransferase
- ER
- endoplasmic reticulum
- TMD
- transmembrane domain
- 125I-IC15:1
- [125I]iodo-pentadecenoic acid
- STF
- Super TOPFlash
- RL
- Renilla luciferase.
REFERENCES
- 1. Rocks O., Peyker A., Kahms M., Verveer P. J., Koerner C., Lumbierres M., Kuhlmann J., Waldmann H., Wittinghofer A., Bastiaens P. I. (2005) An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307, 1746–1752 [DOI] [PubMed] [Google Scholar]
- 2. Smotrys J. E., Linder M. E. (2004) Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 73, 559–587 [DOI] [PubMed] [Google Scholar]
- 3. Takada R., Satomi Y., Kurata T., Ueno N., Norioka S., Kondoh H., Takao T., Takada S. (2006) Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev. Cell 11, 791–801 [DOI] [PubMed] [Google Scholar]
- 4. Moffett S., Brown D. A., Linder M. E. (2000) Lipid-dependent targeting of G proteins into rafts. J. Biol. Chem. 275, 2191–2198 [DOI] [PubMed] [Google Scholar]
- 5. Resh M. D. (2006) Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2, 584–590 [DOI] [PubMed] [Google Scholar]
- 6. Kojima M., Hosoda H., Date Y., Nakazato M., Matsuo H., Kangawa K. (1999) Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature 402, 656–660 [DOI] [PubMed] [Google Scholar]
- 7. Pepinsky R. B., Zeng C., Wen D., Rayhorn P., Baker D. P., Williams K. P., Bixler S. A., Ambrose C. M., Garber E. A., Miatkowski K., Taylor F. R., Wang E. A., Galdes A. (1998) Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem. 273, 14037–14045 [DOI] [PubMed] [Google Scholar]
- 8. Lim X., Tan S. H., Koh W. L., Chau R. M., Yan K. S., Kuo C. J., van Amerongen R., Klein A. M., Nusse R. (2013) Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science 342, 1226–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lim X., Nusse R. (2013) Wnt signaling in skin development, homeostasis, and disease. Cold Spring Harbor Perspectives Biol. 5, pii: a008029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clevers H., Nusse R. (2012) Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 [DOI] [PubMed] [Google Scholar]
- 11. Blauwkamp T. A., Nigam S., Ardehali R., Weissman I. L., Nusse R. (2012) Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat. Commun. 3, 1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reya T., Duncan A. W., Ailles L., Domen J., Scherer D. C., Willert K., Hintz L., Nusse R., Weissman I. L. (2003) A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature 423, 409–414 [DOI] [PubMed] [Google Scholar]
- 13. Green J. L., La J., Yum K. W., Desai P., Rodewald L. W., Zhang X., Leblanc M., Nusse R., Lewis M. T., Wahl G. M. (2013) Paracrine Wnt signaling both promotes and inhibits human breast tumor growth. Proc. Natl. Acad. Sci. U.S.A. 110, 6991–6996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bafico A., Liu G., Goldin L., Harris V., Aaronson S. A. (2004) An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell 6, 497–506 [DOI] [PubMed] [Google Scholar]
- 15. Janda C. Y., Waghray D., Levin A. M., Thomas C., Garcia K. C. (2012) Structural basis of Wnt recognition by Frizzled. Science 337, 59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhai L., Chaturvedi D., Cumberledge S. (2004) Drosophila wnt-1 undergoes a hydrophobic modification and is targeted to lipid rafts, a process that requires porcupine. J. Biol. Chem. 279, 33220–33227 [DOI] [PubMed] [Google Scholar]
- 17. Rios-Esteves J., Resh M. D. (2013) Stearoyl CoA desaturase is required to produce active, lipid-modified Wnt proteins. Cell Rep. 4, 1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Herr P., Basler K. (2012) Porcupine-mediated lipidation is required for Wnt recognition by Wls. Dev. Biol. 361, 392–402 [DOI] [PubMed] [Google Scholar]
- 19. Coombs G. S., Yu J., Canning C. A., Veltri C. A., Covey T. M., Cheong J. K., Utomo V., Banerjee N., Zhang Z. H., Jadulco R. C., Concepcion G. P., Bugni T. S., Harper M. K., Mihalek I., Jones C. M., Ireland C. M., Virshup D. M. (2010) WLS-dependent secretion of WNT3A requires Ser209 acylation and vacuolar acidification. J. Cell Sci. 123, 3357–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X., Reid Sutton V., Omar Peraza-Llanes J., Yu Z., Rosetta R., Kou Y. C., Eble T. N., Patel A., Thaller C., Fang P., Van den Veyver I. B. (2007) Mutations in X-linked PORCN, a putative regulator of Wnt signaling, cause focal dermal hypoplasia. Nat. Genet. 39, 836–838 [DOI] [PubMed] [Google Scholar]
- 21. Grzeschik K. H., Bornholdt D., Oeffner F., König A., del Carmen Boente M., Enders H., Fritz B., Hertl M., Grasshoff U., Höfling K., Oji V., Paradisi M., Schuchardt C., Szalai Z., Tadini G., Traupe H., Happle R. (2007) Deficiency of PORCN, a regulator of Wnt signaling, is associated with focal dermal hypoplasia. Nat. Genet. 39, 833–835 [DOI] [PubMed] [Google Scholar]
- 22. Barrott J. J., Cash G. M., Smith A. P., Barrow J. R., Murtaugh L. C. (2011) Deletion of mouse Porcn blocks Wnt ligand secretion and reveals an ectodermal etiology of human focal dermal hypoplasia/Goltz syndrome. Proc. Natl. Acad. Sci. U.S.A. 108, 12752–12757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen B., Dodge M. E., Tang W., Lu J., Ma Z., Fan C. W., Wei S., Hao W., Kilgore J., Williams N. S., Roth M. G., Amatruda J. F., Chen C., Lum L. (2009) Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dodge M. E., Moon J., Tuladhar R., Lu J., Jacob L. S., Zhang L. S., Shi H., Wang X., Moro E., Mongera A., Argenton F., Karner C. M., Carroll T. J., Chen C., Amatruda J. F., Lum L. (2012) Diverse chemical scaffolds support direct inhibition of the membrane-bound O-acyltransferase porcupine. J. Biol. Chem. 287, 23246–23254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Proffitt K. D., Madan B., Ke Z., Pendharkar V., Ding L., Lee M. A., Hannoush R. N., Virshup D. M. (2013) Pharmacological inhibition of the Wnt acyltransferase PORCN prevents growth of WNT-driven mammary cancer. Cancer Res. 73, 502–507 [DOI] [PubMed] [Google Scholar]
- 26. Liu J., Pan S., Hsieh M. H., Ng N., Sun F., Wang T., Kasibhatla S., Schuller A. G., Li A. G., Cheng D., Li J., Tompkins C., Pferdekamper A., Steffy A., Cheng J., Kowal C., Phung V., Guo G., Wang Y., Graham M. P., Flynn S., Brenner J. C., Li C., Villarroel M. C., Schultz P. G., Wu X., McNamara P., Sellers W. R., Petruzzelli L., Boral A. L., Seidel H. M., McLaughlin M. E., Che J., Carey T. E., Vanasse G., Harris J. L. (2013) Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc. Natl. Acad. Sci. U.S.A. 110, 20224–20229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hofmann K. (2000) A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem. Sci. 25, 111–112 [DOI] [PubMed] [Google Scholar]
- 28. Buglino J. A., Resh M. D. (2008) Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. J. Biol. Chem. 283, 22076–22088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang J., Brown M. S., Liang G., Grishin N. V., Goldstein J. L. (2008) Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell 132, 387–396 [DOI] [PubMed] [Google Scholar]
- 30. Guo Z. Y., Lin S., Heinen J. A., Chang C. C., Chang T. Y. (2005) The active site His-460 of human acyl-coenzyme A:cholesterol acyltransferase 1 resides in a hitherto undisclosed transmembrane domain. J. Biol. Chem. 280, 37814–37826 [DOI] [PubMed] [Google Scholar]
- 31. McFie P. J., Stone S. L., Banman S. L., Stone S. J. (2010) Topological orientation of acyl-CoA:diacylglycerol acyltransferase-1 (DGAT1) and identification of a putative active site histidine and the role of the N terminus in dimer/tetramer formation. J. Biol. Chem. 285, 37377–37387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Buglino J. A., Resh M. D. (2010) Identification of conserved regions and residues within Hedgehog acyltransferase critical for palmitoylation of Sonic Hedgehog. PLoS One 5, e11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Proffitt K. D., Virshup D. M. (2012) Precise regulation of porcupine activity is required for physiological Wnt signaling. J. Biol. Chem. 287, 34167–34178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alland L., Peseckis S. M., Atherton R. E., Berthiaume L., Resh M. D. (1994) Dual myristylation and palmitylation of Src family member p59fyn affects subcellular localization. J. Biol. Chem. 269, 16701–16705 [PubMed] [Google Scholar]
- 35. Peseckis S. M., Deichaite I., Resh M. D. (1993) Iodinated fatty acids as probes for myristate processing and function. Incorporation into pp60v-src. J. Biol. Chem. 268, 5107–5114 [PubMed] [Google Scholar]
- 36. Nugent T., Jones D. T. (2009) Transmembrane protein topology prediction using support vector machines. BMC Bioinformatics 10, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Omasits U., Ahrens C. H., Müller S., Wollscheid B. (2014) Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886 [DOI] [PubMed] [Google Scholar]
- 38. Nugent T., Jones D. T. (2012) Detecting pore-lining regions in transmembrane protein sequences. BMC Bioinformatics 13, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taylor M. S., Ruch T. R., Hsiao P. Y., Hwang Y., Zhang P., Dai L., Huang C. R., Berndsen C. E., Kim M. S., Pandey A., Wolberger C., Marmorstein R., Machamer C., Boeke J. D., Cole P. A. (2013) Architectural organization of the metabolic regulatory enzyme ghrelin O-acyltransferase. J. Biol. Chem. 288, 32211–32228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Froyen G., Govaerts K., Van Esch H., Verbeeck J., Tuomi M. L., Heikkilä H., Torniainen S., Devriendt K., Fryns J. P., Marynen P., Järvelä I., Ala-Mello S. (2009) Novel PORCN mutations in focal dermal hypoplasia. Clin. Genet. 76, 535–543 [DOI] [PubMed] [Google Scholar]
- 41. Lombardi M. P., Bulk S., Celli J., Lampe A., Gabbett M. T., Ousager L. B., van der Smagt J. J., Soller M., Stattin E. L., Mannens M. A., Smigiel R., Hennekam R. C. (2011) Mutation update for the PORCN gene. Hum. Mutat. 32, 723–728 [DOI] [PubMed] [Google Scholar]
- 42. Nakanishi G., Hasegawa K., Oono T., Koshida S., Fujimoto N., Iwatsuki K., Tanaka H., Tanaka T. (2013) Novel and recurrent PORCN gene mutations in almost unilateral and typical focal dermal hypoplasia patients. Eur. J. Dermatol. 23, 64–67 [DOI] [PubMed] [Google Scholar]
- 43. Leoyklang P., Suphapeetiporn K., Wananukul S., Shotelersuk V. (2008) Three novel mutations in the PORCN gene underlying focal dermal hypoplasia. Clin. Genet. 73, 373–379 [DOI] [PubMed] [Google Scholar]
- 44. Willert K., Brown J. D., Danenberg E., Duncan A. W., Weissman I. L., Reya T., Yates J. R., 3rd, Nusse R. (2003) Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423, 448–452 [DOI] [PubMed] [Google Scholar]
- 45. Chu M. L., Ahn V. E., Choi H. J., Daniels D. L., Nusse R., Weis W. I. (2013) structural Studies of Wnts and identification of an LRP6 binding site. Structure 21, 1235–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Das A., Davis M. A., Rudel L. L. (2008) Identification of putative active site residues of ACAT enzymes. J. Lipid Res. 49, 1770–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hardy R. Y., Resh M. D. (2012) Identification of N-terminal residues of Sonic Hedgehog important for palmitoylation by Hedgehog acyltransferase. J. Biol. Chem. 287, 42881–42889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lin S., Lu X., Chang C. C., Chang T. Y. (2003) Human acyl-coenzyme A:cholesterol acyltransferase expressed in chinese hamster ovary cells: membrane topology and active site location. Mol. Biol. Cell 14, 2447–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]