Background: Basal metabolic rate is regulated by thyroid hormone; the mechanism is unknown.
Results: NCOR1 and Rev-Erbα enrich at different sites from thyroid hormone receptor on the Tshb promoter.
Conclusion: NCOR1 and Rev-Erbα interact to regulate circadian expression of Tshb mRNA independent of thyroid hormone.
Significance: This novel role of Rev-Erbα in Tshb expression reveals new links between circadian rhythms and metabolism.
Keywords: Circadian Rhythm, Metabolism, Nuclear Receptor, Thyroid Hormone, Transcription Repressor, NCOR1, Rev-Erbα, Tshb
Abstract
Thyroid hormones (TH) are critical for development, growth, and metabolism. Circulating TH levels are tightly regulated by thyroid-stimulating hormone (TSH) secretion within the hypothalamic-pituitary-thyroid axis. Although circadian TSH secretion has been well documented, the mechanism of this observation remains unclear. Recently, the nuclear corepressor, NCOR1, has been postulated to regulate TSH expression, presumably by interacting with thyroid hormone receptors (THRs) bound to TSH subunit genes. We report herein the first in vitro study of NCOR1 regulation of TSH in a physiologically relevant cell system, the TαT1.1 mouse thyrotroph cell line. Knockdown of NCOR1 by shRNA adenovirus increased baseline Tshb mRNA levels compared with scrambled control, but surprisingly had no affect on the T3-mediated repression of this gene. Using ChIP, we show that NCOR1 enriches on the Tshb promoter at sites different from THR previously identified by our group. Furthermore, NCOR1 enrichment on Tshb is unaffected by T3 treatment. Given that NCOR1 does not target THR on Tshb, we hypothesized that NCOR1 targeted Rev-Erbα (NR1D1), an orphan nuclear receptor that is a potent repressor of gene transcription and regulator of metabolism and circadian rhythms. Using a serum shock technique, we synchronized TαT1.1 cells to study circadian gene expression. Post-synchronization, Tshb and Nr1d1 mRNA levels displayed oscillations that inversely correlated with each other. Furthermore, NR1D1 was enriched at the same locus as NCOR1 on Tshb. Therefore, we propose a model for Tshb regulation whereby NR1D1 and NCOR1 interact to regulate circadian expression of Tshb independent of TH negative regulation.
Introduction
Thyroid hormones (THs),3 T4 and T3, play an integral role in development, growth, and cellular metabolism (1–3), and circulating levels of THs are maintained within a narrow range by a finely tuned negative feedback system involving the hypothalamic-pituitary-thyroid (HPT) axis (4). Thyrotropin-releasing hormone (TRH) secreted from the hypothalamus stimulates pituitary thyrotrophs to produce biologically active thyroid stimulating hormone (TSH), which stimulates the thyroid gland to synthesize and secrete THs. Importantly, T3 exerts negative feedback at both the level of the pituitary and hypothalamus, thus completing the feedback loop (5). T3 action is classically thought to be mediated via THRs (6), which are members of the nuclear receptor superfamily and bind DNA both in the absence and presence of T3 (7–9). On positively regulated genes, the THR is thought to interact with corepressors, principally NCOR1, which is released and replaced by coactivators after T3 binding (10, 11). In contrast, less is known about mechanisms of genes repressed by T3. We are particularly interested in establishing mechanisms by which Tshb expression is regulated.
Circadian rhythms are fundamental phenomena in most living organisms whereby behavior and biological function are regulated through an autonomous clock. Control of this rhythm has been traced to a central clock in the suprachiasmatic nucleus of the hypothalamus (12). Disruptions of clock mechanisms are thought to be important in disorders of sleep, metabolism, and even cancer (13–15). The circadian cycle is triggered by a CLOCK/BMAL1 heterodimer that is regulated under a negative feedback loop mediated by the orphan nuclear receptor Rev-Erbα (NR1D1) (16–18). Feng et al. (19) mapped a NR1D1 cistrome in mouse liver, showing thousands of binding sites that have a rhythm, which correlate to the oscillating expression of NR1D1.
TSH and T3 secretion are also known to follow a photoperiodic circadian rhythm, with a nadir during the day and a peak secretory activity just before sleep (20). Interestingly, TSH may induce the expression of type 2 iodothyronine deiodinase in the hypothalamus, which enzymatically converts the prohormone T4 into its bioactive T3 product, providing an additional mechanism for controlling the HPT axis (21, 22). Given that this diurnal rhythm of TSH is disrupted in states as diverse as depression, poorly controlled diabetes, and mostly importantly, after pharmacologic T4 replacement to hypothyroid patients, further elucidation of this mechanism is warranted (23, 24).
Despite characterization of TSH diurnal rhythm in physiologic and pathologic states over many years, the upstream regulators are yet to be well characterized. We have established an appropriate and physiologically relevant mouse cell line model to study regulation of Tshb (25, 26). Using this model, our studies shed light on an unprecedented role of NCOR1 in repression of Tshb that is independent of THR action. We hypothesize that circadian regulation of TSH and T3 secretion is maintained by NCOR1 interaction with NR1D1, not THR, and that circadian changes in T3 levels may have a previously unrecognized role in controlling overall metabolism.
EXPERIMENTAL PROCEDURES
Cell Culture and Hormone Treatments
TαT1.1 cells were plated in DMEM (Corning Cellgro, Manassas, VA) containing 10% FBS (Invitrogen) and 1% penicillin/streptomycin (Invitrogen). Before the cells were seeded, the plates were coated with Matrigel (BD Biosciences) to facilitate adhesion. Matrigel was diluted 30-fold with Dulbecco's PBS (Invitrogen) before coating the plates. Cells were maintained at 37 °C in an environment of 5% CO2. Treatment of cells with either T3, TRH, or SR9011 (Sigma) was performed for the indicated durations after 24-h medium replacement with DMEM containing 10% FBS stripped of thyroid hormone by treatment with AG1X-8 resin (Bio-Rad) and charcoal (Sigma).
Adenoviral Transduction
Adenoviruses expressing nonspecific scrambled short hairpin RNA (shRNA) or shRNA against Ncor1 mRNA were generated using a BLOCK-iT adenoviral RNAi expression system following the manufacturer's instructions (Invitrogen). shNcor1 (3910–3930) targeted the sequence 5′-CATCCAAGGGCCATGTTATC-3′. TαT1.1 cells were transduced 24 h after the cells were seeded with adenoviruses to knock down the gene(s) of interest. The medium was changed the following day, and cells were harvested 72 h after infection. Hormone treatment with T3 was done using stripped serum 8 h before harvesting. The concentration of adenoviruses was determined and all solutions had equivalent titer. Scrambled adenovirus control was used in the same concentration as the virus of interest.
RNA Isolation and Quantitative PCR
Total RNA from TαT1.1 cells was extracted by standard methodology (TRIzol reagent; Invitrogen). One microgram of total RNA was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR analyses were performed in a fluorescent temperature cycler using SYBR Green reagent according to the recommendations of the manufacturer (Bio-Rad). Primers for Ncor1, Ncor2, Tshb, Cga, Gh, Nr1d1, and 36B4 are listed in supplemental Table S1. Cycle threshold values for 36B4 were used to normalize each sample. All results are expressed as a fraction of samples treated with vehicle only.
Western Blot Analysis
TαT1.1 cells were treated with experimental conditions, after which whole cell extracts were prepared in 1× radioimmune precipitation assay buffer (Sigma) containing protease and phosphatase inhibitors (Roche Applied Science). Extract (20 μg of total protein) was run on 10% SDS-PAGE gels and transferred to PVDF membranes (Millipore). Membrane was probed using antibodies for the following proteins from the indicated suppliers: NCOR1 (gift from A. Hollenberg, generated against the C-terminal portion of protein), SMRT (Affinity Bio), NR1D1 (Santa Cruz Biotechnology), and β-actin (Chemicon).
Chromatin Immunoprecipitation
ChIP assays were performed on TαT1.1 cells at ∼90% of confluence using ChIP-IT Express kit (Active Motif, Carlsbad, CA) following the manufacturer's instructions. Immunoprecipitation was performed at 4 °C overnight. Antibodies against the following proteins were used from the indicated suppliers: rabbit IgG (Santa Cruz Biotechnology), NCOR1 (Abcam), acetyl-H3 (Upstate Biotechnology), acetyl-H4 (Upstate), and Rev-Erbα (Abcam). Cross-links were reversed, and the DNA was purified using QIAquick PCR Purification Kit (Qiagen). The DNA recovered from the assay was subjected to qPCR using specific primers designed to detect enrichment in the proximal promoters of Tshb, Cga, and Gh (supplemental Table S2).
Cell Synchronization
TαT1.1 cells were seeded overnight in 6-cm dishes in DMEM supplemented with 10% FBS and antibiotics. Culture medium was replaced with serum-free DMEM for 4 h. Cells were then shocked with DMEM containing 50% horse serum for 2 h, after which medium was replaced with 10% stripped FBS (t = 0 h). Cells were harvested using TRIzol and stored at −80 °C until analyzed for RNA.
Statistical Analysis
Values are represented as mean ± S.E. of at least three independent experiments, unless otherwise noted. Statistical testing was performed using an unpaired two-tailed Student's t test. A p value of <0.05 was considered significant, and the respective levels of significance and group sizes are stated in respective figure legends.
RESULTS
Thyroid Hormone-responsive Genes Are Differentially Regulated by NCOR1
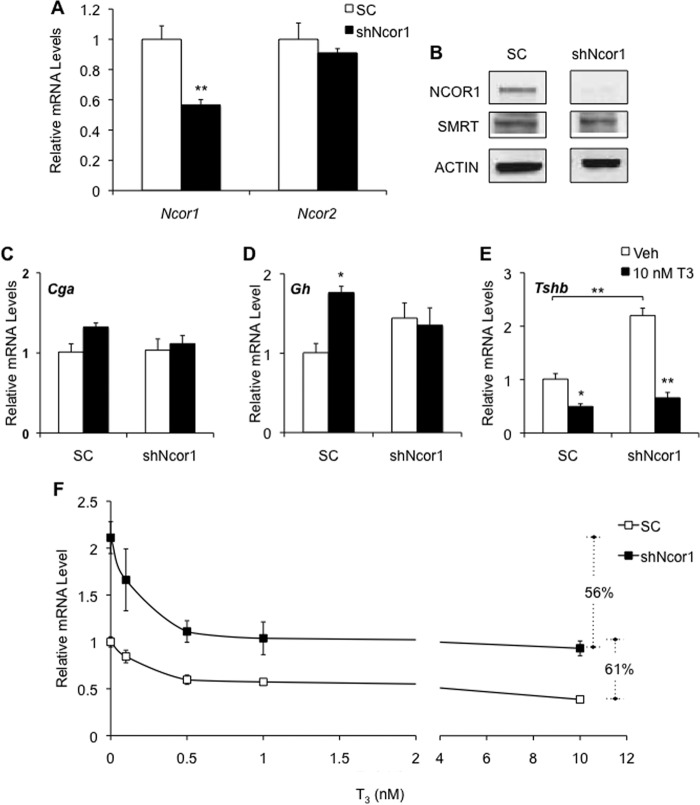
Studies have shown that T3 regulates thyrotopin subunit gene expression in the TαT1.1 thyrotroph cell line. Whereas T3 reduces Tshb mRNA levels, it has no effect on Cga mRNA levels. Negative regulation of transcription by nuclear receptors is often associated with recruitment of corepressors, and NCOR1 is thought to be the key corepressor that regulates the THR. To test the importance of NCOR1 on gene expression in TαT1.1 cells, we treated cells with adenovirus expressing NCOR1 shRNA (shNcor1). Ncor1 mRNA levels were reduced 45%, whereas Ncor2 mRNA levels that encode a closely related transcription factor, SMRTs (silencing mediator of retinoid or thyroid hormone receptors), were unaffected (Fig. 1A). Western blot analysis confirmed depletion of NCOR1 protein in these cells (Fig. 1B).
FIGURE 1.
Thyroid hormone-responsive genes are differentially regulated by NCOR1. TαT1.1 cells were transduced with adenovirus expressing scrambled shRNA (SC) or shRNA against NCOR1 (shNcor1). Cells were harvested for total RNA. A, RT-qPCR analysis for Ncor1 and Ncor2 (SMRT) mRNA was performed. B, Western blot analysis of NCOR1, SMRT, and actin (control) was performed. C–E, cells were treated with vehicle (Veh) or indicated concentrations of T3 for 24 h before harvesting. RT-qPCR analysis was performed for Cga (C), Gh (D), and Tshb (E and F) mRNA levels. F, total percent suppression after treatment with 10 nm T3 is indicated with dashed lines. Relative mRNA levels are expressed as fold change compared with vehicle-treated cells exposed to scrambled shRNA. *, p < 0.05 versus vehicle, SC treatment; **, p < 0.01.
Cga expression in TαT1.1 cells was not regulated by T3, and there was no change after treatment with shNcor1 (Fig. 1C). Throughout this work, we use Cga as a negative control. Gh, a gene up-regulated by T3 binding to THR, is generally repressed by NCOR1 (27). TαT1.1 cells express Gh at low levels such that this gene can be used as a positive control in our experiments. Knockdown of NCOR1 not only raised basal expression but also resulted in a reduction in T3-mediated activation of Gh (Fig. 1D). Although shNcor1 did not affect T3 repression of Tshb mRNA levels, the baseline level (vehicle treatment) did increase significantly compared with treatment with scrambled shRNA (Fig. 1E).
NCOR1 Regulation of Tshb Is Independent of T3
Given that knockdown of NCOR1 affects basal expression of Tshb, we next explored whether the T3 response is altered by determining a T3 concentration response after SC and shNcor1 treatment (Fig. 1F). Tshb mRNA levels were maximally repressed at 10 nm T3 after either SC treatment (71%) or shNcor1 treatment (66%). When the T3-treated Tshb mRNA levels for the shNcor1-treated cells were compared with shNcor1 vehicle, instead of scrambled shRNA vehicle, the dose response curves overlapped. The calculated EC50 for both treatments was 0.3 nm, indicating that NCOR1 does not mediate T3 regulation of Tshb.
Based on this novel finding that NCOR1 affects baseline Tshb mRNA expression, but not T3-mediated levels, we next determined whether NCOR1 interacts directly with Tshb. We performed ChIP-qPCR, scanning for NCOR1 enrichment around the transcription start site of Tshb (Fig. 2). Our laboratory has previously published that THRs enrich at sites −1861 and +18 bp relative to the transcription start site, regions containing several TRE half-sites (26). NCOR1, however, was found to be enriched at a different site, −2959 bp, and its occupancy was unaffected by T3 treatment (Fig. 2). On the Gh gene, THR is bound to a −220 bp site (28, 29). NCOR1 was enriched in this region and released upon T3 treatment, consistent with reports on Gh regulation (Fig. 2). We expected this response on Gh because NCOR1 recruitment is reported to be mediated through binding to THR-β (THRB). The contrasting action on Tshb lead us to hypothesize that NCOR1 may be bound by a different transcription factor than THRB.
FIGURE 2.
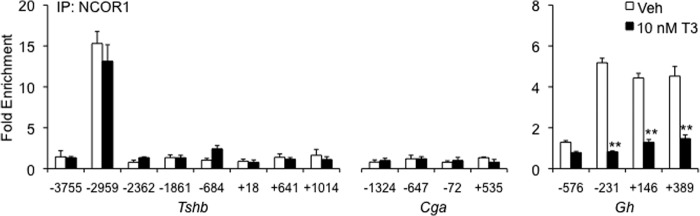
NCOR1 recruitment to Gh promoter, but not TSH subunit genes, is decreased with T3 treatment. ChIP-qPCR was performed to scan a region surrounding the Tshb, Gh, and Cga in TαT1.1 cells treated with vehicle (Veh) or 10 nm T3. The x axis shows the location of the forward primer relative to the transcription start site. Results are expressed as relative fold enrichment ± S.E. compared with background enrichment. **, p < 0.01 versus vehicle treatment.
TRH, but Not T3, Affects Acetylation of Histone H3 on the Tshb Promoter
Activation of gene expression is associated with histone acetylation, whereas repression is usually associated with deacetylation. Following this dogma, we assessed the enrichment of acetylated histones on the Tshb promoter. In Fig. 3A, a ChIP assay shows no significant changes in histone H3 acetylation on Tshb or Cga after treatment with 10 nm T3. TRH is known to stimulate Tshb expression, and this response was confirmed by increased acetyl-H3 after hormone treatment at sites centered on −684 and +18 bp (Fig. 3B). The latter region corresponds to a region also enriched with THRB (26). No changes in acetylated H4 were observed in Tshb or Cga after treatment with vehicle, T3, or TRH (Fig. 3, C and D). These findings indicate that the assembly of transcription factors that regulate Tshb is likely to contain both a T3-independent mechanism that uniquely involves NCOR1 and a TRH-dependent mechanism that is associated with histone acetylation. The interplay of these pathways was evidenced after knocking down NCOR1. Treating cells with shNcor1 resulted in a marked increase in acetyl-H3 at the +18 bp site (Fig. 3E). This study was performed without hormone treatment and could possibly be the explanation for the increase in baseline Tshb mRNA levels after shNcor1 transduction. For example, if more H3 is acetylated on Tshb, one would expect an increase in basal expression.
FIGURE 3.
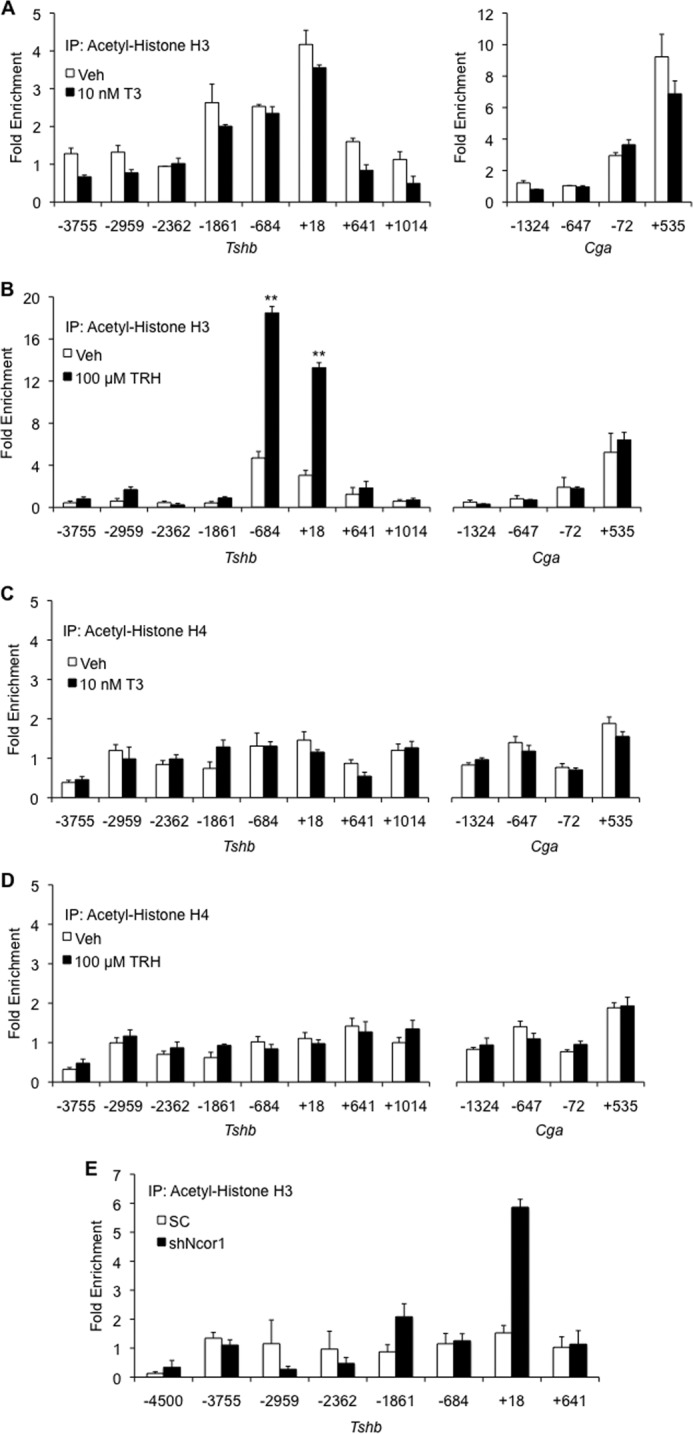
TRH, but not T3, affects histone acetylation of TSH subunit genes. ChIP-qPCR was performed to scan acetylation of histone H3 (A and B) or H4 (C and D) to a region surrounding the Tshb, Gh, and Cga promoters in TαT1.1 cells treated with vehicle (Veh), 10 nm T3 (A and C), or 100 μm TRH (B and D). E, cells were treated with scrambled shRNA (SC) or Ncor1 shRNA (shNcor1) and harvested 72 h after transduction for ChIP-qPCR analysis on the Tshb promoter. The x axis shows the location of the forward primer relative to the transcription start site. Results are expressed as relative fold enrichment ± S.E. compared with background enrichment. **, p < 0.01 versus vehicle.
Tshb mRNA Expression Oscillates in a Thyrotroph Cell Line
To define further the hormone-independent mechanism of Tshb expression, we studied gene expression over time in TαT1.1 cells. Although the cyclic secretion of TSH expression may be critical to cellular homeostasis, little is known about the overall impact of this physiologic event or the mechanism mediating the rhythm. By synchronizing TαT1.1 cells in culture, we were able to mimic Tshb mRNA cycling in this pituitary cell line. According to established protocols to study circadian regulation, we synchronized TαT1.1 cells by incubating them in serum-free medium, followed by a brief period of serum shock in 50% horse serum, and then harvesting mRNA at various time points for a qPCR analysis (Fig. 4). In this model, Tshb mRNA levels cycled with a 36-hour period, with peaks at 18 and 54 h, and nadirs at 0 and 36 h. Because expression amplitude dampened with each successive cycle, Tshb mRNA levels were difficult to measure beyond 72 h.
FIGURE 4.
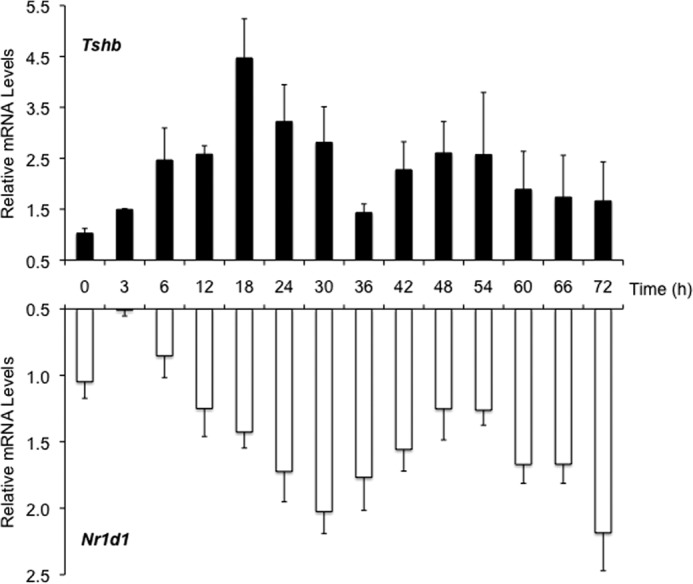
Tshb and Nr1d1 mRNA levels oscillate in TαT1.1 cells. TαT1.1 cells were synchronized by serum shock and harvested at the indicated times for total RNA. RT-qPCR was performed for Nr1d1 (Rev-Erbα) and Tshb mRNA (n = 4). Results are expressed as relative mRNA levels ± S.E. compared with initial levels at time = 0 h.
NR1D1, a heme receptor transcribed on the antisense strand of Thra, is a potent repressor of gene transcription and a key driver of circadian rhythms and metabolism. NR1D1 has also been associated with NCOR1-dependent repression of nuclear receptors, and direct binding between the two proteins has been established (11). We decided to investigate Nr1d1 expression in our synchronized TαT1.1 cells and also observed a significant oscillation in expression (Fig. 4). Initial Nr1d1 mRNA levels decreased before rising at 12 h, with peaks at 36 and 72h, and a nadir at 54 h. NR1D1 protein levels followed a similar pattern (data not shown). When comparing these mRNA levels, Tshb levels began to decline after the 18-h peak, whereas the Nr1d1 levels increased. From 30 to 54 h, Nr1d1 levels declined, and Tshb mRNA levels increased. The second decline in Tshb mRNA levels (54 to 72 h) was again marked with an increase in Nr1d1. The two mRNA oscillations appeared to oppose each other, suggesting that this hormone-independent circadian rhythm of Tshb may be controlled by Nr1d1.
NR1D1 and NCOR1 Co-localize to the Tshb Promoter
NR1D1 is an orphan nuclear receptor with a unique structure, in that it lacks the helix 12 motif that is typically required for coactivators to bind the receptor (30). Previously described as a DNA-binding protein that interacts with NCOR1, we hypothesize that NR1D1 regulates Tshb basal rhythm via recruitment of NCOR1. To study the interaction of these two proteins on DNA, we performed ChIP assays, scanning for NR1D1 and NCOR1 enrichment on the Tshb promoter (Fig. 5). When Nr1d1 expression is low at 4 h, it enriches at −2959 bp, but after 8 h, when NR1D1 levels are rising, additional enrichment at −2959 bp and a new enrichment at −2362 bp are noted (Fig. 5A). NCOR1 recruitment to the Tshb promoter showed a similar pattern (Fig. 5B). Previously, we only observed NCOR1 enrichment at the −2959 site, but 8 h post-synchronization, NCOR1 is additionally recruited to this new −2362-bp region.
FIGURE 5.
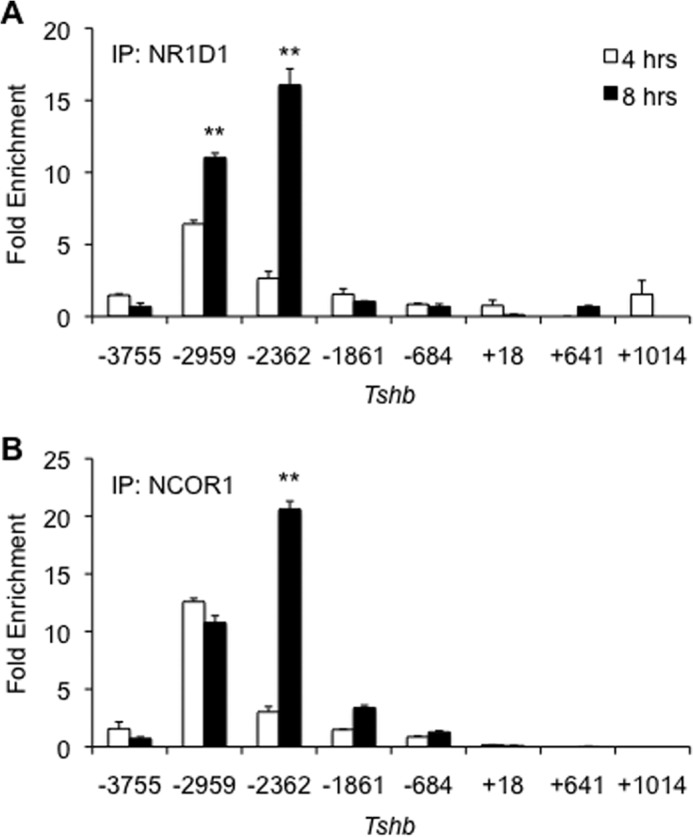
NR1D1 and NCOR1 are jointly recruited to additional sites on Tshb. TαT1.1 cells were synchronized and harvested 4 and 8 h after serum shock. ChIP-qPCR assays were performed with antibodies specific for NCOR1, NR1D1, or IgG (negative control). Results are expressed relative fold enrichment ± S.E. compared with background enrichment. **, p < 0.01 versus vehicle treatment.
An NR1D1 Agonist Promotes Tshb Repression
Our data show that NCOR1 binds directly to the proximal promoter of Tshb and represses its basal mRNA expression independent of thyroid hormone regulation. NR1D1 also influences basal regulation of Tshb mRNA levels, likely through its recruitment of NCOR1 to the promoter. To confirm that NCOR1-based repression of Tshb is dependent on NR1D1, an NR1D1 agonist was employed (SR9011). SR9011 is a potent agonist of NR1D1 and demonstrates minimal cross-reactivity with other nuclear receptors (31). TαT1.1 cells were treated with increasing concentrations of SR9011 (or vehicle) for 8 h, and mRNA was harvested for qPCR studies. SR9011 induced a concentration-dependent decrease in Tshb mRNA levels (Fig. 6, A and B). One nm T3 and 5 μm SR9011 treatments resulted in maximal inhibition of mRNA expression of 65 and 80%, respectively. High concentrations of either ligand had no significant effect on Cga mRNA levels in this cell line (data not shown). ChIP assays showed that addition of 5 μm SR9011 to TαT1.1 cells also promoted NCOR1 recruitment to the −2362-bp region of Tshb in addition to the −2959-bp site (Fig. 6C). In summary, these data confirm our hypothesis that NCOR1 is recruited to NR1D1, triggering repression of Tshb gene expression (Fig. 7).
FIGURE 6.
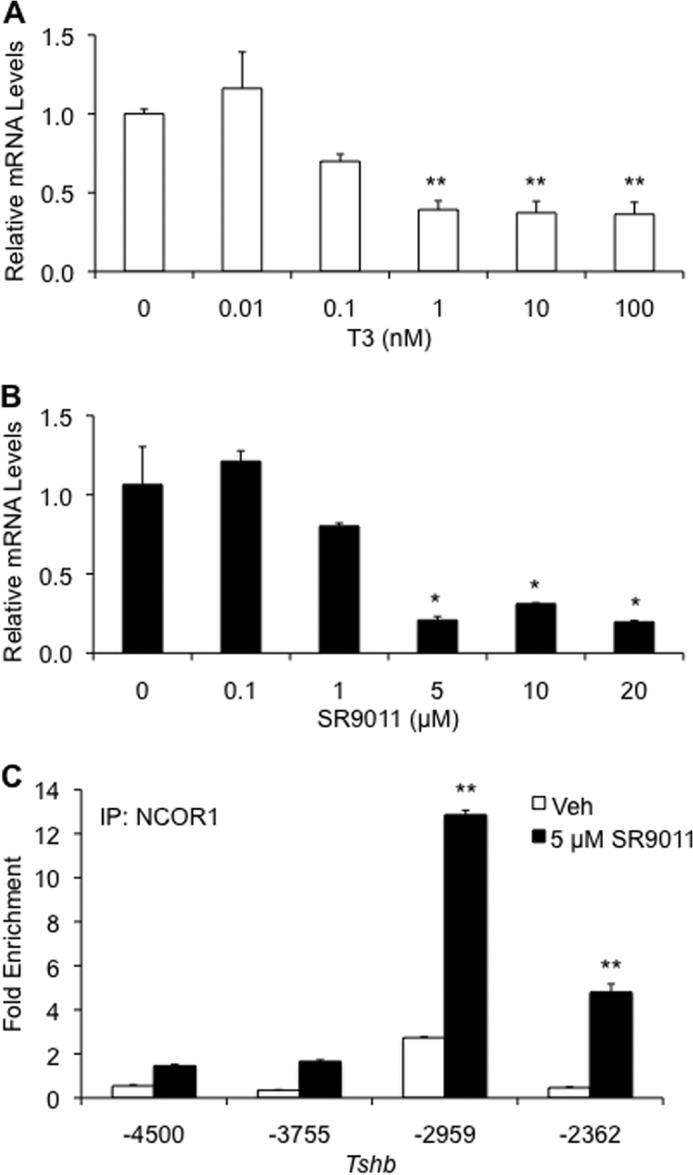
NR1D1 agonist causes a decrease in Tshb mRNA expression. TαT1.1 cells were treated with vehicle (Veh), T3 (A), or NR1D1 agonist SR9011 (B and C) and harvested after 8 h for total RNA. RT-qPCR was performed to assess Tshb and Cga mRNA expression. C, TαT1.1 cells were treated with vehicle or 5 μm SR9011 for 8 h. ChIP-qPCR was performed with antibody specific for NCOR1. Results are expressed as relative fold enrichment ± S.E. compared with background enrichment. *, p < 0.05 versus vehicle treatment; **, p < 0.01.
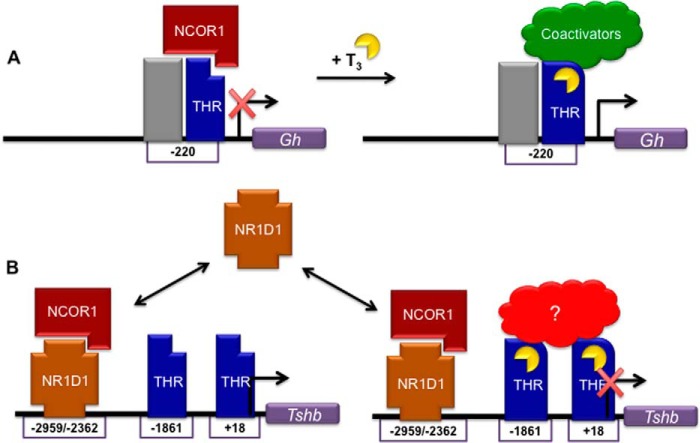
FIGURE 7.
Proposed model for NCOR1 regulation of gene expression. A, on a THR-mediated gene such as Gh, NCOR1 binds THR in the absence of T3 and represses transcriptional activity. Binding of T3 induces a conformation change of the receptor that releases NCOR1 and recruits coactivators to activate transcription. B, on Tshb, NR1D1 cycles on and off a primary site (−2959) and an inducible site (−2362) depending on protein levels. THR enriches at different downstream sites (−1861 and +18) mediate T3-dependent repression. The nature of the THR complex on Tshb has not been fully characterized.
DISCUSSION
For decades, researchers have studied the role of thyroid hormone in the regulation of metabolism. On positively regulated genes, T3 binds to THR, inducing a conformational change that favors coactivator over corepressor binding. The regulation of genes repressed by thyroid hormone, however, is not a simple reversal of this process. Thyroid hormone repression is critical for regulation of the HPT axis and hence thyroid hormone synthesis. TSH is central to regulation of the HPT axis, and both subunit genes that comprise TSH are down-regulated by T3 in vivo (29, 32). We have previously shown that T3-bound THRB represses Tshb subunit gene expression by recruiting cofactors to the ligand-binding domain (33). Our current findings show that there are two pathways for Tshb repression: one that involves T3 and THR and one that involves NCOR1 independent of T3. The latter pathway interested us in that it suggested a relationship between NCOR1 and NR1D1 on Tshb that has not been described previously.
Our first objective in establishing the role of NCOR1 on T3-responsive genes was to knock it down using an shRNA adenoviral construct. Knockdown of NCOR1 on the thyroid hormone positively regulated Gh gene demonstrated that NCOR1 was dismissed from the proximal Gh promoter as suggested previously. In the absence of T3, cells treated with shNcor1 displayed elevated basal Gh mRNA expression and eliminated any further increase after T3 treatment. On the negatively regulated Tshb, however, this increase in basal expression did not hinder T3 repression. In fact, comparing an extensive T3 concentration response between scrambled and shNcor1 treatments revealed absolutely no change in the relative T3 inhibition of Tshb mRNA levels.
In this regard, our findings are in agreement with in vivo models of NCOR1 action. Although global deletion of NCOR1 is embryonic lethal, mutating the inhibitory domains of NCOR1 revealed a hypersensitivity to TH in peripheral tissues that was not seen in the HPT axis (10, 34). This is likely due to changes in basal gene expression that we also find in the current investigation using a thyrotroph cell line. Because of its critical role in controlling T3-mediated development, metabolism, and other vital processes in the body, TSH secretion requires a highly conserved and tightly regulated system of control. The T3 negative feedback response seems to be protected at the level of the pituitary and is not affected by NCOR1. A T3-independent NCOR1 pathway was further confirmed when ChIP assays showed enrichment of NCOR1 binding to the Tshb promoter sites, which are >1000 bp upstream of reported THR binding sites (26). NCOR1 was neither released nor further enriched at these sites after treatment with T3.
Because acute repression of TSH by T3 was unaffected by NCOR1 depletion, we next evaluated whether baseline Tshb mRNA expression was affected by NCOR1 depletion. A well known characteristic of TSH and T3 secretion are their circadian secretory patterns. Metabolic processes can be affected by oscillations in hormone levels, light/dark periodicity, and feeding and are perpetuated by a core clock in the suprachiasmatic nucleus of the hypothalamus. BMAL1-CLOCK, the heterodimeric transcriptional controller of this central clock, is a direct target of NR1D1, and together, are required for the onset and continuation of each period in the cycle (16, 19). We explored the involvement of NR1D1 in our model because of its known role in circadian rhythm generation, as well as its direct interaction with NCOR1.
When the TαT1.1 thyrotroph cell line was synchronized, we were able to mimic a circadian cycle of Tshb mRNA levels in these cells. The mRNA expression of Tshb and Nr1d1 displayed opposing cycles, with peaks for one matching the nadir for the other, and vice versa. If NR1D1 were truly a mediator of NCOR1-driven Tshb repression, we would need to demonstrate its direct binding to Tshb, as well as interaction with NCOR1. NR1D1 did show enrichment to Tshb at the same locus as NCOR1. In addition, at a time when NR1D1 protein expression was increasing, it was enriched at a second site not described previously. Using the NR1D1 agonist SR9011, we showed that activation of NR1D1 was sufficient to elicit NCOR1 binding at the two sites we identified and lead to a reduction in Tshb mRNA levels.
Acute control of TSH secretion is tightly regulated through thyroid hormone negative feedback. We believe that basal regulation of Tshb is regulated by NR1D1, which recruits NCOR1 and leads to transcriptional repression. Recent studies have revealed a dynamic balance and highly protected role of NR1D1 in circadian and metabolic regulation (17). Conserving its activity on multiple levels demonstrates its vital importance and necessity in not just maintaining, but also driving, the core clock. Given that NR1D1 also drives circadian regulation of Tshb expression by binding to the promoter and recruiting NCOR1, it may provide a previously unknown way for metabolism to be regulated in a circadian rhythm by T3. Our work opens a new line of investigation linking NR1D1, circadian TSH and T3 secretion, and metabolism.
Supplementary Material
Acknowledgment
We thank Leticia A. Santiago for constructing the shRNA adenovirus against Ncor1.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DK49126. This work was also supported by The Johns Hopkins University-University of Maryland Diabetes Research Center Grant P60DK079637.

This article contains supplemental Tables S1 and S2.
- TH
- thyroid hormone
- T3
- 3,3′,5-triiodo-L-thyronine
- T4
- thyroxine
- NCOR1
- nuclear corepressor 1
- TSH
- thyroid stimulating hormone
- THR
- TH receptor
- TRH
- thyrotropin-releasing hormone
- qPCR
- quantitative PCR.
REFERENCES
- 1. Song Y., Yao X., Ying H. (2011) Thyroid hormone action in metabolic regulation. Protein Cell 2, 358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yen P. M., Ando S., Feng X., Liu Y., Maruvada P., Xia X. (2006) Thyroid hormone action at the cellular, genomic and target gene levels. Mol. Cell Endocrinol. 246, 121–127 [DOI] [PubMed] [Google Scholar]
- 3. Huang Y. Y., Gusdon A. M., Qu S. (2013) Cross-talk between the thyroid and liver: A new target for nonalcoholic fatty liver disease treatment. World J. Gastroenterol. 19, 8238–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Costa-e-Sousa R. H., Hollenberg A. N. (2012) Minireview: The neural regulation of the hypothalamic-pituitary-thyroid axis. Endocrinology 153, 4128–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yen P. M. (2001) Physiological and molecular basis of thyroid hormone action. Physiol. Rev. 81, 1097–1142 [DOI] [PubMed] [Google Scholar]
- 6. Richter C. P., Münscher A., Machado D. S., Wondisford F. E., Ortiga-Carvalho T. M. (2011) Complete activation of thyroid hormone receptor β by T3 is essential for normal cochlear function and morphology in mice. Cell Physiol. Biochem. 28, 997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Flamant F., Samarut J. (2003) Thyroid hormone receptors: lessons from knockout and knock-in mutant mice. Trends Endocrinol. Metab. 14, 85–90 [DOI] [PubMed] [Google Scholar]
- 8. Park E. A., Jerden D. C., Bahouth S. W. (1995) Regulation of phosphoenolpyruvate carboxykinase gene transcription by thyroid hormone involves two distinct binding sites in the promoter. Biochem. J. 309, 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oberste-Berghaus C., Zanger K., Hashimoto K., Cohen R. N., Hollenberg A. N., Wondisford F. E. (2000) Thyroid hormone-independent interaction between the thyroid hormone receptor β2 amino terminus and coactivators. J. Biol. Chem. 275, 1787–1792 [DOI] [PubMed] [Google Scholar]
- 10. Astapova I., Vella K. R., Ramadoss P., Holtz K. A., Rodwin B. A., Liao X. H., Weiss R. E., Rosenberg M. A., Rosenzweig A., Hollenberg A. N. (2011) The nuclear receptor corepressor (NCoR) controls thyroid hormone sensitivity and the set point of the hypothalamic-pituitary-thyroid axis. Mol. Endocrinol. 25, 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. You S. H., Liao X., Weiss R. E., Lazar M. A. (2010) The interaction between nuclear receptor corepressor and histone deacetylase 3 regulates both positive and negative thyroid hormone action in vivo. Mol. Endocrinol. 24, 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gachon F., Nagoshi E., Brown S. A., Ripperger J., Schibler U. (2004) The mammalian circadian timing system: from gene expression to physiology. Chromosoma 113, 103–112 [DOI] [PubMed] [Google Scholar]
- 13. Huang W., Ramsey K. M., Marcheva B., Bass J. (2011) Circadian rhythms, sleep, and metabolism. J. Clin. Invest. 121, 2133–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takahashi J. S., Hong H. K., Ko C. H., McDearmon E. L. (2008) The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat. Rev. Genet. 9, 764–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mannic T., Meyer P., Triponez F., Pusztaszeri M., Le Martelot G., Mariani O., Schmitter D., Sage D., Philippe J., Dibner C. (2013) Circadian clock characteristics are altered in human thyroid malignant nodules. J. Clin. Endocrinol. Metab. 98, 4446–4456 [DOI] [PubMed] [Google Scholar]
- 16. Lande-Diner L., Boyault C., Kim J. Y., Weitz C. J. (2013) A positive feedback loop links circadian clock factor CLOCK-BMAL1 to the basic transcriptional machinery. Proc. Natl. Acad. Sci. U.S.A. 110, 16021–16026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cho H., Zhao X., Hatori M., Yu R. T., Barish G. D., Lam M. T., Chong L. W., DiTacchio L., Atkins A. R., Glass C. K., Liddle C., Auwerx J., Downes M., Panda S., Evans R. M. (2012) Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bugge A., Feng D., Everett L. J., Briggs E. R., Mullican S. E., Wang F., Jager J., Lazar M. A. (2012) Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 26, 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feng D., Liu T., Sun Z., Bugge A., Mullican S. E., Alenghat T., Liu X. S., Lazar M. A. (2011) A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van Reeth O., Sturis J., Byrne M. M., Blackman J. D., L'Hermite-Balériaux M., Leproult R., Oliner C., Refetoff S., Turek F. W., Van Cauter E. (1994) Nocturnal exercise phase delays circadian rhythms of melatonin and thyrotropin secretion in normal men. Am. J. Physiol. 266, E964–E974 [DOI] [PubMed] [Google Scholar]
- 21. Ono H., Hoshino Y., Yasuo S., Watanabe M., Nakane Y., Murai A., Ebihara S., Korf H. W., Yoshimura T. (2008) Involvement of thyrotropin in photoperiodic signal transduction in mice. Proc. Natl. Acad. Sci. U.S.A. 105, 18238–18242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arrojo E Drigo R., Fonseca T. L., Werneck-de-Castro J. P.., Bianco A. C. (2013) Role of the type 2 iodothyronine deiodinase (D2) in the control of thyroid hormone signaling. Biochim. Biophys. Acta 1830, 3956–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fisher D. A. (1996) Physiological variations in thyroid hormones: physiological and pathophysiological considerations. Clin. Chem. 42, 135–139 [PubMed] [Google Scholar]
- 24. Sviridonova M. A., Fadeyev V. V., Sych Y. P., Melnichenko G. A. (2013) Clinical significance of TSH circadian variability in patients with hypothyroidism. Endocr. Res. 38, 24–31 [DOI] [PubMed] [Google Scholar]
- 25. Yusta B., Alarid E. T., Gordon D. F., Ridgway E. C., Mellon P. L. (1998) The thyrotropin β-subunit gene is repressed by thyroid hormone in a novel thyrotrope cell line, mouse TαT1 cells. Endocrinology 139, 4476–4482 [DOI] [PubMed] [Google Scholar]
- 26. Chiamolera M. I., Sidhaye A. R., Matsumoto S., He Q., Hashimoto K., Ortiga-Carvalho T. M., Wondisford F. E. (2012) Fundamentally distinct roles of thyroid hormone receptor isoforms in a thyrotroph cell line are due to differential DNA binding. Mol. Endocrinol. 26, 926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim S. W., Ahn I. M., Larsen P. R. (1996) In vivo genomic footprinting of thyroid hormone-responsive genes in pituitary tumor cell lines. Mol. Cell Biol. 16, 4465–4477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Flug F., Copp R. P., Casanova J., Horowitz Z. D., Janocko L., Plotnick M., Samuels H. H. (1987) Cis-acting elements of the rat growth hormone gene which mediate basal and regulated expression by thyroid hormone. J. Biol. Chem. 262, 6373–6382 [PubMed] [Google Scholar]
- 29. Machado D. S., Sabet A., Santiago L. A., Sidhaye A. R., Chiamolera M. I., Ortiga-Carvalho T. M., Wondisford F. E. (2009) A thyroid hormone receptor mutation that dissociates thyroid hormone regulation of gene expression in vivo. Proc. Natl. Acad. Sci. U.S.A. 106, 9441–9446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zamir I., Dawson J., Lavinsky R. M., Glass C. K., Rosenfeld M. G., Lazar M. A. (1997) Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Proc. Natl. Acad. Sci. U.S.A. 94, 14400–14405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Solt L. A., Wang Y., Banerjee S., Hughes T., Kojetin D. J., Lundasen T., Shin Y., Liu J., Cameron M. D., Noel R., Yoo S. H., Takahashi J. S., Butler A. A., Kamenecka T. M., Burris T. P. (2012) Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 485, 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abel E. D., Boers M. E., Pazos-Moura C., Moura E., Kaulbach H., Zakaria M., Lowell B., Radovick S., Liberman M. C., Wondisford F. (1999) Divergent roles for thyroid hormone receptor β isoforms in the endocrine axis and auditory system. J. Clin. Invest. 104, 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ortiga-Carvalho T. M., Shibusawa N., Nikrodhanond A., Oliveira K. J., Machado D. S., Liao X. H., Cohen R. N., Refetoff S., Wondisford F. E. (2005) Negative regulation by thyroid hormone receptor requires an intact coactivator-binding surface. J. Clin. Invest. 115, 2517–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Costa-e-Sousa R. H., Astapova I., Ye F., Wondisford F. E., Hollenberg A. N. (2012) The thyroid axis is regulated by NCoR1 via its actions in the pituitary. Endocrinology 153, 5049–5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.