Background: The contribution of autophagy to acquired cisplatin resistance in ovarian cancer has not been studied.
Results: Cisplatin treatment activates ERK and subsequently promotes autophagy and counteracts cisplatin-induced cell death. Inhibition of autophagy enhances cisplatin sensitivity.
Conclusion: Cisplatin-induced autophagy contributes to cisplatin resistance.
Significance: Targeting autophagy may overcome cisplatin resistance.
Keywords: Autophagy, Cancer Therapy, Drug Resistance, Extracellular Signal-regulated Kinase (ERK), Ovarian Cancer
Abstract
Cisplatin-based treatment is the first line chemotherapy for several cancers including ovarian cancer. The development of cisplatin resistance results in treatment failure, but the underlying mechanisms are not fully understood. Here we show that the induction of autophagy plays an important role in cisplatin resistance in ovarian cancer cells. Specifically, we show that cisplatin resistance is correlated with autophagy induction in a panel of ovarian cancer cells but not in immortalized human ovarian surface epithelial cells. Mechanistically, cisplatin treatment activates ERK and subsequently promotes autophagy. The inhibition of ERK activation with MEK inhibitors or knockdown of ERK expression with siRNA decreases cisplatin-induced autophagy and subsequently sensitizes ovarian cancer cells to cisplatin-induced apoptosis. In ovarian cancer cells that have developed acquired cisplatin resistance, both ERK activation and autophagy induction are increased. Importantly, knockdown of ERK or inhibition of autophagy promotes cisplatin-induced apoptosis in acquired cisplatin-resistant cells. Collectively, our data indicate that ERK-mediated autophagy can lead to cisplatin resistance and suggest that cisplatin resistance can be overcome by inhibition of autophagy in ovarian cancer cells.
Introduction
Ovarian cancer is the fifth leading cause of cancer-related death among women in the United States. 90% of primary ovarian tumors are epithelial cell in origin (1, 2). Ovarian cancer can be classified into serous, endometrioid, mucinous, and clear cell (1, 2), with a significant majority of ovarian carcinomas being the serous type (3). Currently available therapeutic options include tumor debulking surgery and chemotherapy. The standard first line chemotherapy includes platinum (cisplatin/carboplatin)-based therapy.
Cisplatin (cis-diamminedichloroplatinum) is a platinum compound that was discovered in the 1960s and has been an important chemotherapeutic drug for the treatment of many cancers, including ovarian, as a single agent or in combination with other anticancer agents (4–6). Mechanistic studies indicate that cisplatin covalently binds to the N-7 atoms of purines on DNA to form DNA adducts. DNA adducts distort DNA conformation and inhibit replication and transcription, leading to the activation of multiple signaling pathways to induce cell cycle arrest and apoptosis. The induction of apoptosis primarily accounts for cisplatin anticancer activity (7). Although cisplatin is the first line chemotherapeutic agent for several cancers, particularly for patients with testicular and ovarian cancers, it turns out that some cancer cells develop acquired cisplatin resistance whereas other tumor cells are intrinsically resistant. Cisplatin resistance can lead to cisplatin-based treatment failure. For example, 70% of ovarian cancer patients initially respond to cisplatin, but the majority of responsive patients relapse due to the development of resistance (3, 8). The mechanisms of cisplatin resistance are not fully understood, but are believed to be multifactorial in nature, which include insufficient DNA binding, increased detoxification, increased DNA repair, deregulated expression of transporters, and altered expression and activation of genes involved in cell death pathways (5, 9–15). Recent studies suggest that cancer drug resistance including cisplatin resistance can be mediated by autophagy (16–18).
Autophagy is a highly conserved process by which cytoplasmic components are sequestered in double membrane vesicles called autophagosomes and degraded upon fusion with lysosomal compartments (19). Autophagy plays key roles in the quality control of cellular components and supplying nutrients and materials for newly constructed structures in cells under metabolic stress. Autophagy can be induced by a number of stress conditions including starvation and drug treatment and is associated with both cell survival and cell death. The molecular mechanism of autophagy regulation is not fully understood. It is believed that a number of molecules are involved, including the target of rapamycin, 5′-AMP-activated protein kinase and the eukaryotic initiation factor 2α (elF2α) (19, 20). Target of rapamycin kinase is the major regulator that inhibits autophagy in the presence of growth factors and abundant nutrients whereas 5′-AMP-activated protein kinase and elF2α control autophagy in response to low energy and nutrient deprivation, respectively (19). Autophagy is carried by a number of autophagy-related genes (Atgs)2 (19). These Atgs are involved either in autophagosome formation, fusing autophagosome with lysosomes for degradation, or reuse of macromolecules (21). Recent studies indicated that acute cisplatin treatment activates an autophagic response that serves as a survival factor to counteract cisplatin-induced cell death (22, 23). In lung cancer cells, autophagy has been suggested to play a role in acquired cisplatin resistance (24, 25). However, the role of autophagy in cisplatin resistance in ovarian cancer cells has not been determined.
In this study, we show that induction of autophagy is correlated with cisplatin resistance in a panel of human ovarian cancer cell lines. We find that cisplatin induces extracellular signal-regulated kinase (ERK), which promotes autophagy induction. We also show that knockdown of ERK or inhibition of mitogen-activated protein (MAP) kinase (MAPK)/ERK (MEK) activity decreases autophagy induction while increasing cisplatin-induced cell death. Furthermore, we show that ovarian cancer cells that have developed cisplatin resistance have increased ERK activation and autophagy. Finally, we show that inhibition of autophagy sensitizes acquired cisplatin-resistant cells to cisplatin. Thus, these data suggest that targeting autophagy may be a vital strategy for overcoming cisplatin resistance.
EXPERIMENTAL PROCEDURES
Reagents
Cisplatin, 3-methyladenine (3-MA), bafilomycin A1 (Baf A1), and anti-actin antibody were purchased from Sigma. The MEK inhibitor U0126 and the p38 inhibitor SB203580 were purchased from Promega. The JNK inhibitor SP600125 was purchased from Calbiochem. Rabbit antibodies against P-ERK1/2, ERK1/2, P-p38, p38, P-JNK1/2, JNK1/2, P-CREB, CREB, P-c-Jun, c-Jun, LC3, Atg5, p62, and PARP were purchased from Cell Signaling Technology (Beverly, MA). pEGFP-c1-LC3 plasmid was provided by Dr. Hong-Gang Wang (Pennsylvania State University).
Cell Lines and Culture Conditions
Human ovarian cancer cell lines RMG-1, OV90, OV433, OVCA420, and CAOV3 were maintained as described previously (26). Immortalized human ovarian surface epithelial IOSE385 cells were obtained from Dr. Nelly Auersperg (University of British Columbia, Vancouver, Canada). CAOV3 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS). The rest of cell lines were maintained in MCDB105/M199 supplemented with 10% FBS and 1% l-Glu. All cells were maintained at 37 °C in a humidified atmosphere consisting of 5% CO2 and 95% air. The cisplatin-resistant ovarian cancer cell line OV433-CR was established by chronically exposing parental OV433 cells (OV433-P) to gradually increased concentrations of cisplatin starting from 0.1 μg/ml to 0.8 μg/ml for over 6 months, as described previously (27).
RNA Interference of ERK1/2 and Atg5
On-TARGETplus SMARTpool siRNAs for ERK1/ERK2 and Atg5 and corresponding control nontarget siRNA were purchased from Dharmacon Research (Lafayette, CO). Transfections were performed as suggested by the manufacturer's instruction with slight modifications as described previously (26). Briefly, cells were seeded in 6-well plates (5 × 105 cells/well). The next day, cells were transfected with siRNA oligonucleotides using Lipofectamine RNAiMAX (Invitrogen). After 3 days, transfected cells were left untreated or treated with cisplatin and then harvested for examining protein expression by Western blot analysis. For cisplatin sensitivity experiments, transfected cells were placed at 8000 cells/well in 96-well plates and then left untreated or treated with cisplatin, and cell proliferation was determined by MTT assays.
MTT Assays
MTT assays were performed as described previously (28). Briefly, cells were left untreated or pretreated with U0126 (10 μm), SB203580 (10 μm), SP600125 (10 μm), or 3-MA (1 mm) for 30 min and then left untreated or treated with cisplatin. After incubation with MTT solution for 4 h, isopropyl alcohol was added to dissolve the formazan crystals. OD was measured using a Vmax Microplate Reader (Molecular Devices, Sunnyvale, CA) at 570 nm. The cell proliferation was calculated from the mean of pooled data from three separate experiments with five wells each.
Western Blot Analysis
Cell lysates were prepared as described previously (28), and protein concentration was determined using the Protein Assay Kit (Bio-Rad). Cell lysates were electrophoresed through denaturing polyacrylamide gels and transferred to a PVDF membrane (Millipore). The blots were probed or re-probed with the antibodies and detected using Enhanced Chemiluminescence (ECL) or Odyssey Infrared Imaging System according to the manufacturer's protocol.
Quantification of Western Blot and Statistical Analysis
Quantification of Western blots was performed using ImageJ, and statistical analyses were performed using Student's t test. The data were presented as the mean ± S.D., and p value < 0.001 was considered significant.
RESULTS
Elevation of the LC3-II Level Is Correlated with Cisplatin Resistance in a Panel of Human Ovarian Cancer Cell Lines
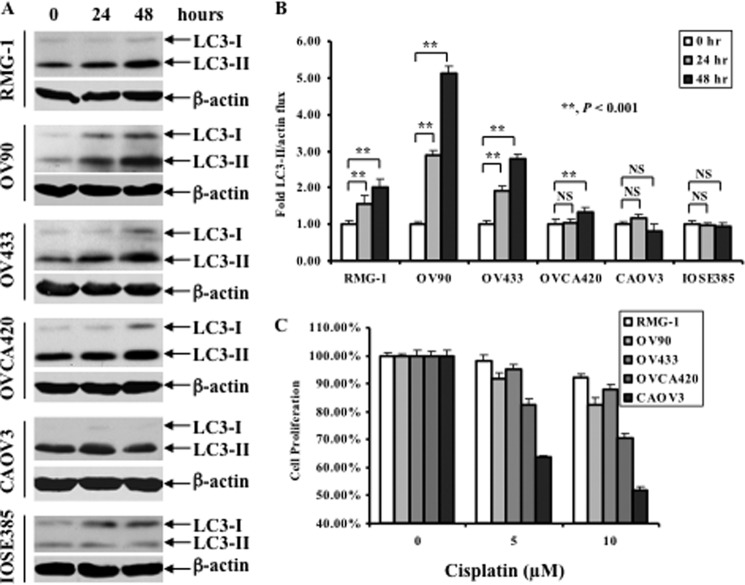
Accumulating evidence suggests that autophagy plays an important role in chemoresistance (24, 25), yet, its involvement in cisplatin resistance in ovarian cancer cells has not been tested. In this regard, a panel of human ovarian cancer cell lines including RMG-1, OV433, OV90, OVCA420, and CAOV3 was treated with 10 or 20 μm cisplatin for 24 and 48 h, and changes in LC3-II levels were assessed by Western blot analysis. LC3 is a microtubule-associated structural protein and a mammalian homologue of the yeast gene Atg8, which is required for autophagy (29). Shortly after translation, LC3 is cleaved to yield LC3-I (29). When autophagy occurs, LC3-I is covalently linked to phosphatidylethanolamine to yield LC3-II, which associates with the autophagosome. Because LC3-II migrates more rapidly than LC3-I on denaturing gels, the presence of LC3-II is a biochemical marker of autophagy. Fig. 1, A and B, shows that cisplatin treatment caused a significant increase in LC3-II in RMG-1, OV90, and OV433 cells, a slight increase in LC3-II in OVCA420 cells. In contrast, cisplatin treatment caused a slight decrease in CAOV3 cells at a 48-h treatment. Importantly, cisplatin treatment did not increase the levels of LC3-II in the immortalized human ovarian surface epithelial IOSE385 cells (a nontumorigenic cell line). In parallel, MTT assays were performed to evaluate their cisplatin sensitivities. Fig. 1C shows that all cancer cell lines exhibited the differential cisplatin sensitivity; RMG-1, OV90, and OV433 cells were resistant to cisplatin, and CAOV3 cells were sensitive to cisplatin whereas OVCA420 cells were in between (modest resistance). We found that IOSE358 was a cisplatin-sensitive cell line (data not shown). Further analysis revealed a correlation between an increase in the LC3-II level and cisplatin resistance; LC3-II was increased significantly in the resistant cell lines RMG-1, OV90, and OV433, but not in the sensitive CAOV3 and IOSE385 cells, and slightly in modest resistant OVCA420 cells. Thus, our data indicate that elevation of LC3-II levels may predict cisplatin resistance in ovarian cancer cells.
FIGURE 1.
Effect of cisplatin treatment on LC3 levels and growth inhibition in a panel of human ovarian cell lines. A, Western blot analyses of the levels of LC3. Ovarian cancer cell lines were left untreated or treated with 20 μm cisplatin (RMG-1, OV433, OV90, and OVCA420 cells) or 10 μm cisplatin (CAOV3 cells) for the indicated time periods. IOSE385 cells were treated with 3.33 μm cisplatin (at IC50) for the indicated time periods. β-Actin was used as a loading control. B, quantification of LC3-II. The level of LC3-II was quantified by densitometry. Data represent mean ± S.D. (error bars) of three independent experiments. **, p < 0.001, statistically significant; NS, not statistically significant. C, MTT assays of growth inhibition. Ovarian cancer cells in A were left untreated or treated with cisplatin with the indicated concentrations for 48 h.
Cisplatin Treatment Induces the Changes Associated with Autophagy
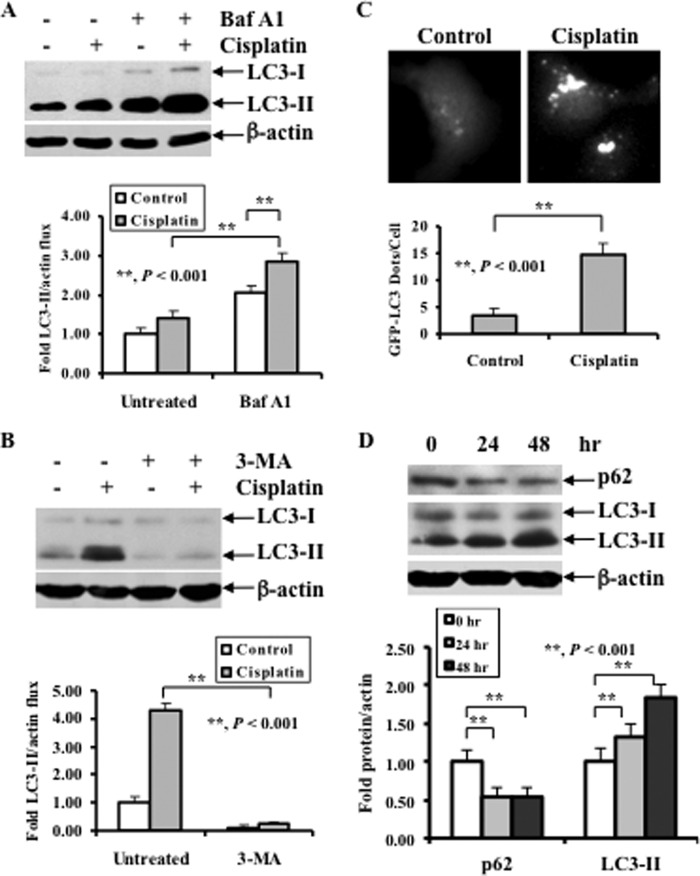
Although increased LC3-II levels indicate autophagy induction, it is not absolutely certain that these cells undergo autophagy. To characterize cisplatin-induced autophagy, we performed analyses of “autophagic flux” by employing Baf A1 to intentionally prevent autophagosome-lysosome fusion and degradation to better determine the extent to which the complete autophagic process occurred in OV433 cells. We chose OV433 cells because this cell line is a cisplatin-resistant line. Fig. 2A shows a greater accumulation of LC3-II in cisplatin-treated OV433 cells relative to untreated cells following Baf A1 treatment. This result indicates that cisplatin is able to cause autophagy in ovarian cancer cells. To determine whether cisplatin-induced LC3-II elevation can be blocked by autophagy inhibition, we treated OV433 cells with cisplatin in the absence or presence of the autophagy inhibitor 3-MA. Fig. 2B shows that 3-MA decreased cisplatin-induced LC3-II levels compared with cisplatin treatment alone. To further confirm the role of cisplatin in inducing autophagy, we used direct fluorescence to monitor LC3 punctate formation as an index for autophagosome accumulation in live cells. We stably transfected GFP-LC3 into OV433 cells in the presence and absence of cisplatin treatment. Fig. 2C shows that a punctuate pattern of LC3 was detected in cisplatin-treated but not in untreated cells. In addition, p62, another marker for autophagy, was decreased following cisplatin treatment, and this decrease inversely correlated with an increase in the levels of LC3-II (Fig. 2D). Taken together, these data indicate that cisplatin treatments activate an autophagic response in human ovarian cancer cells.
FIGURE 2.
Effect of Baf A1 and 3-MA on cisplatin-induced LC3-II, and the role for cisplatin treatment in p62 levels and LC3 punctate formation. A, Western blot analyses of the levels of LC3 in cells treated with Baf A1 (upper panel) and quantification of LC3-II (lower panel). OV433 cells were left untreated or treated with 20 μm cisplatin in the presence or absence of 2 nm Baf A1 for 24 h. B, Western blot analyses of LC3 in cells treated with 3-MA (upper panel) and quantification of LC3-II (lower panel). OV433 cells were left untreated or treated with 20 μm cisplatin in the presence or absence of 1 mm 3-MA for 24 h. C, representative fluorescent images of GFP-LC3 (upper panel) and quantification of GFP-LC3 dots (lower panel). OV433 cells transfected with GFP-LC3 were selected with G418 for stable GFP-LC3 and then subjected to cisplatin treatment (20 μm, 20 h) or left alone. Bright dots denote autophagosomes. Bars represent mean ± S.D. (error bars) of triplicate samples with >20 cells analyzed per sample. D, Western blot analyses of LC3 and p62 (upper panel) and quantification of LC3-II and p62 (lower panel). OV433 cells were left untreated or treated with 20 μm cisplatin for 24 h. In A, B, and D, data represent mean ± S.D. of three independent experiments. **, p < 0.001, statistically significant.
Cisplatin Treatment Activates ERK, which Promotes Autophagy
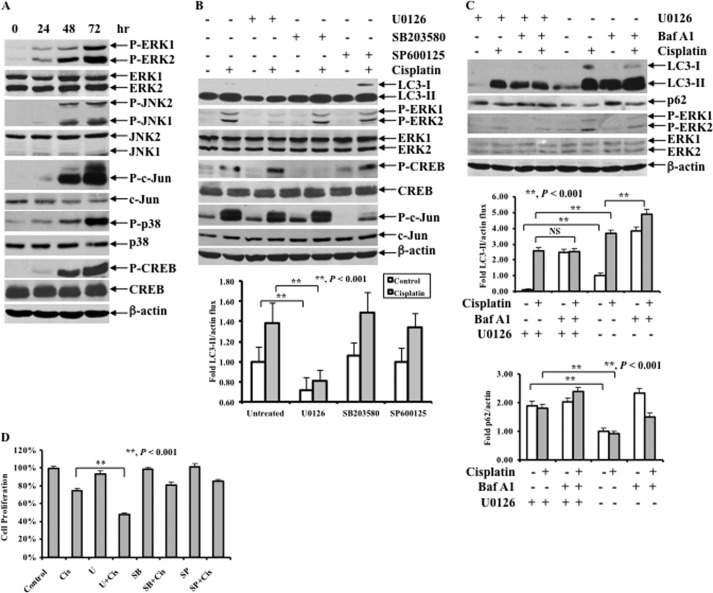
Emerging evidence suggests that all three MAPK subfamilies may regulate autophagy (30–35). To determine whether MAPKs are responsible for cisplatin-induced autophagy, we first tested the effect of cisplatin treatment on MAPK activation. OV433 cells were treated with cisplatin, and the activation of MAPK pathways was then determined. Fig. 3A shows that cisplatin treatment caused phosphorylation of ERK, p38, and c-Jun N-terminal kinases (JNK) and their downstream targets including CREB, and c-Jun, confirming our previous study showing that cisplatin activates all three major MAPK pathways (26). Next, we determined which MAPK is responsible for cisplatin-induced autophagy. OV433 cells were left untreated or treated with 20 μm cisplatin in the presence or absence of the MEK1/2 inhibitor U0126 (10 μm), the p38 inhibitor SB203580 (10 μm), or the JNK inhibitor SP600125 (10 μm) for 24 h, and the levels of LC3-II and the activation of MAPK pathways were examined. As shown in Fig. 3B, cisplatin treatment increased the level of LC3-II, which was inhibited by U0126 but not by SB203580 or SP600125. Furthermore, we performed analyses of autophagic flux by employing Baf A1 and showed that there was less LC3-II accumulation in cisplatin and Baf A1-treated OV433 cells in the presence of U0126 compared with the same treatments in the absence of U0126. By contrast, the p62 level was inversely correlated with the LC3-II level (Fig. 3C). In parallel, we showed that inhibition of ERK by U0126 but not inhibition of JNK by SP600125 or inhibition of p38 by SB203580 significantly increased cisplatin-induced cell death (Fig. 3D). Because autophagy can be a cell survival mechanism, these data indicate that ERK-mediated autophagy may promote cell survival.
FIGURE 3.
Effect of cisplatin treatment on MAPK pathways, LC3 levels, and cisplatin sensitivity following ERK inhibition. A, Western blot analyses of the activation of the MAPK pathways including P-ERK1/2, ERK1/2, P-JNK1/2, JNK1/2, P-c-Jun, c-Jun, P-p38, p38, P-CREB, and CREB. OV433 cells were left untreated or treated with 20 μm cisplatin for the indicated time periods. B, Western blot analyses of LC3, P-ERK1/2, ERK1/2, P-CREB, CREB, P-c-Jun, and c-Jun (upper panel) and quantification of LC3-II (lower panel). OV433 cells were left untreated or treated with 20 μm cisplatin in the presence or absence of 10 μm U0126 or 10 μm SB203580 or 10 μm SP600125 for 24 h. C, Western blot analyses of the levels of LC3 and p62 in untreated or 20 μm cisplatin-treated cells with or without 2 nm Baf A1 treatment in the presence or absence of 10 μm U0126 (upper panel) and quantification of LC3-II (middle panel) and p62 (lower panel) in OV433 cells. D, MTT analyses of growth inhibition. OV433 cells were left untreated or treated with 20 μm cisplatin (Cis) in the presence or absence of 10 μm MEK inhibitor U0126 (U), 10 μm p38 inhibitor SB203580 (SB), or 10 μm JNK inhibitor SP600125 (SP) for 24 h. Data represent mean ± S.D. (error bars) of three independent experiments. **, p < 0.001, statistically significant.
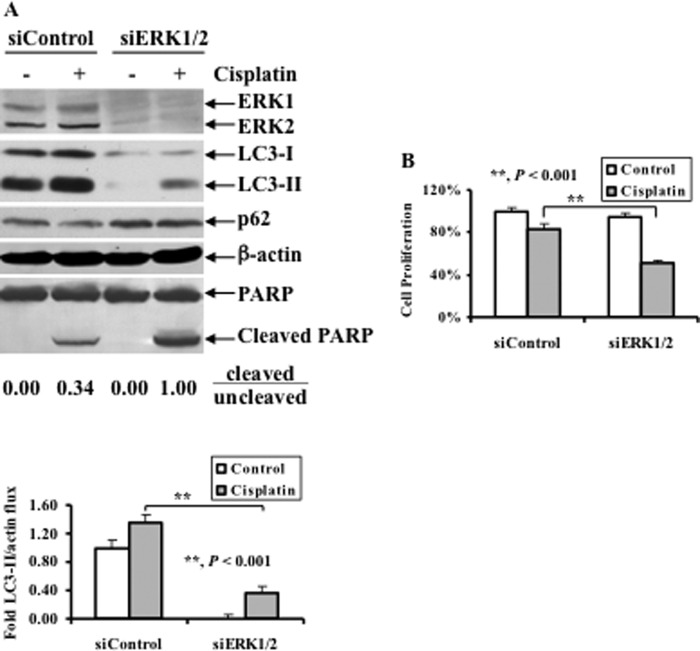
Knockdown of ERK by siRNA Decreases Cisplatin-induced LC3-II while Increasing Cisplatin-induced Cell Death
To address the direct role of ERK in autophagy induction, OV433 cells were transfected with either nontarget (siControl) or ERK siRNAs (siERK1/2), and the effects of ERK knockdown on LC3-I/II levels and PARP cleavage were examined. Fig. 4A shows that total ERK in cells transfected with ERK siRNA was decreased significantly compared with cells transfected with control siRNA. As expected, cisplatin treatment increased LC3-II levels in cells transfected with control siRNA. In contrast, knockdown of ERK significantly decreased LC3-II while increasing p62, and such changes were correlated with increased PARP cleavage (the ratio of cleaved to uncleaved PARP was 1.00 in cells transfected with ERK siRNA whereas such a ratio was 0.34 in cells transfected with control siRNA) (Fig. 4A). These data suggest that ERK knockdown decreases cisplatin-induced autophagy. To determine the effect of autophagy inhibition by knockdown of ERK on cisplatin sensitivity, OV433 cells transfected with ERK or control siRNAs were treated with 20 μm cisplatin, and growth inhibition was determined by MTT assays. Fig. 4B shows that knockdown of ERK significantly increased cisplatin-induced growth inhibition, as exemplified by 50% inhibition of cells transfected with ERK siRNA versus 20% inhibition of cells transfected with control siRNA. Thus, these data suggest that ERK-mediated autophagy contributes to cisplatin resistance in ovarian cancer cells.
FIGURE 4.
Effect of ERK1/2 knockdown on cisplatin-induced LC3 levels, PARP cleavage, and growth inhibition. A, Western blot analyses of ERK1/2, LC3, p62, and PARP cleavage (upper panel) and quantification of LC3-II (lower panel). OV433 cells transfected with either control nontarget siRNAs (siControl) or ERK1/2 siRNAs (siERK1/2) were left untreated or treated with 20 μm cisplatin for 24 h. Quantification of PARP cleavage detected by Western blots was performed using densitometry and ratios of cleaved to uncleaved PARP are indicated. Data represent mean ± S.D. (error bars) of three independent experiments. B, MTT analyses of growth inhibition. OV433 cells were treated as described in A. Data represent mean ± S.D. of three independent experiments. **, p < 0.001, statistically significant.
Pharmacological Inhibition of Autophagy or Knockdown of Atg5 Enhances Cisplatin-induced Growth Inhibition
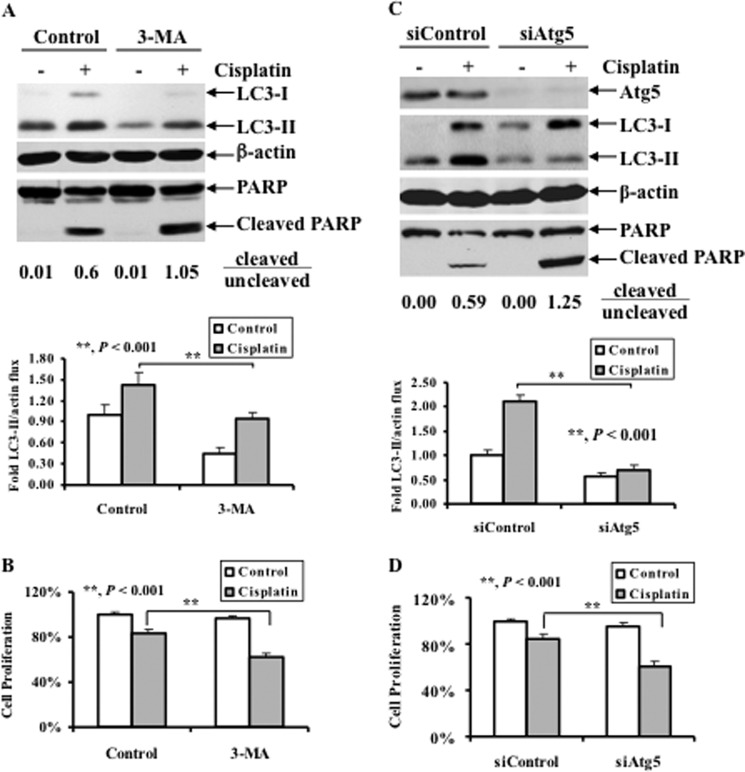
To investigate a role for autophagy in cisplatin sensitivity, we evaluated the effect of pharmacological inhibition of autophagy on cisplatin sensitivity. Specifically, OV433 cells were left untreated or treated with cisplatin in the presence or absence of 3-MA, a pharmacological inhibitor of the class III-PI3K that acts at the levels of the formation of pre-autophagosomes (36), and the levels of LC3-II and PARP cleavage were determined. Fig. 5A shows that 3-MA significantly inhibited cisplatin-induced LC3-II compared with that of untreated cells. Interestingly, 3-MA also enhanced cisplatin-induced PARP cleavage (Fig. 5A) and growth inhibition (Fig. 5B), suggesting that pharmacological inhibition of autophagy promotes cisplatin-induced apoptosis.
FIGURE 5.
Effect of pharmacological inhibition of autophagy or knockdown of Atg5 on cisplatin-induced apoptosis and cisplatin sensitivity. A, Western blot analyses of LC3 and PARP cleavage (upper panel) and quantification of LC3-II (lower panel). OV433 cells were left untreated or treated with 20 μm cisplatin in the presence or absence of 1 mm 3-MA for 24 h. Quantification of PARP cleavage was described in Fig. 4A. B, MTT analyses of growth inhibition. OV433 cells were treated as in A. C, Western blot analyses of the levels of Atg5, LC3, and PARP cleavage (upper panel) and quantification of LC3-II (lower panel). OV433 cells transfected with either control nontarget siRNAs (siControl) or Atg5 siRNAs (siAtg5) were left untreated or treated with 20 μm cisplatin for 24 h. Quantification of PARP cleavage was described above. D, MTT analyses of growth inhibition. OV433 cells were treated as in C. Data represent mean ± S.D. (error bars) of three independent experiments. **, p < 0.001, statistically significant.
Although 3-MA is a pharmacological inhibitor for class III-PI3K that is required for the formation of autophagosomes, it is possible that 3-MA may affect other kinases that regulate cancer cell growth independent of autophagy. To address the direct role of autophagy in the regulation of cisplatin resistance, we assessed the effect of knockdown of Atg5, a key component of the autophagy pathway, on cisplatin sensitivity. Accordingly, OV433 cells transfected with either nontarget control siRNA (siControl), or siRNA against Atg5 (siAtg5) were assessed for cisplatin sensitivity. As shown in Fig. 5C, knockdown of Atg5 abolished cisplatin-induced LC3-II whereas such a change was not observed in cells transfected with control siRNA. Furthermore, upon cisplatin treatment, Atg5 knockdown led to enhance PARP cleavage compared with cells transfected with control siRNA. Importantly, cisplatin was able to inhibit growth of Atg5 knockdown cells by 40% whereas such inhibition dropped to 20% of cells transfected with control siRNA (Fig. 5D). Collectively, these data suggest that cisplatin-induced autophagy involves the activation of the Atg5-mediated autophagy pathway and that this autophagy pathway plays a role in cell survival.
ERK-mediated Autophagy Is Involved in Acquired Cisplatin-resistant Cells, and Its Blockage Sensitizes Resistant Cells to Cisplatin-induced Apoptosis
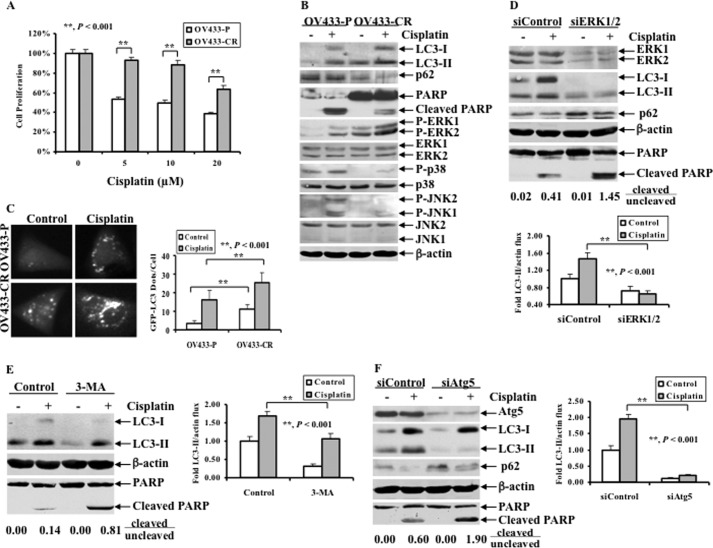
The drawback of using cisplatin as an anti-cancer agent is that cancer cells initially sensitive to cisplatin develop acquired resistance (37). Thus, acquired resistance is an urgent issue that needs to be addressed. To assess whether ERK-mediated autophagy plays a role in acquired cisplatin resistance in ovarian cancer cells, we tested this mechanism in a pair of ovarian cancer cells: parental OV433 (OV433-P) cells and their derivative cisplatin-resistant OV433 cells (OV433-CR). OV433-CR cells were generated by chronically exposing parental OV433 cells to cisplatin for more than 6 months (28). Fig. 6A shows that OV433-CR cells were much more resistant than OV433-P to cisplatin. In OV433-P cells, cisplatin treatment induced PARP cleavage whereas such cleavage was insignificant in OV433-CR (Fig. 6B). Interestingly, OV433-CR expressed a higher level of the basal and cisplatin-induced phosphorylated ERK but not JNK and p38 compared with OV433-P cells (Fig. 6B). Inversely, OV433-CR expressed a lower level of p62 compared with OV433-P cells (Fig. 6B). Importantly, OV433-CR cells expressed higher levels of basal and cisplatin-induced LC3-II compared with OV433-P cells under the same treatment conditions (Fig. 6B). Furthermore, when GFP-LC3 was transfected into OV433-P and OV433-CR cells in the absence of cisplatin treatment, we detected a significant increase in LC3 punctate dots in OV433-CR cells but not in OV433-P cells, which were proportionally increased upon cisplatin treatment (Fig. 6C). These results suggest that OV433 cells, once develop acquired cisplatin resistance, can gain a high level of autophagic activity. Next, we asked whether this mechanism could be targeted for the improvement of cisplatin sensitivity. We knocked down ERK by siRNA in OV433-CR cells and found that knockdown of ERK significantly decreased LC3-II levels while increasing p62 levels (Fig. 6D), suggesting that an autophagic response was decreased. Inversely, ERK knockdown-mediated down-regulation of LC3-II led to an increase in cisplatin-induced PARP cleavage. The ratio of cleaved to uncleaved PARP was 1.45 in ERK siRNA-transfected cells whereas such a ratio was 0.41 in cells transfected with control siRNA. Consistently, pharmacological inhibition of autophagy by 3-MA also increased cisplatin-induced PARP cleavage while significantly decreasing the levels of LC3-II in OV433-CR cells (Fig. 6E). Similarly, knockdown of Atg5 also significantly decreased LC3-II levels but increased p62 levels and PARP cleavage in OV433-CR cells. Because OV433-CR cells are very resistant to cisplatin, our data strongly suggest that autophagy plays a critical role in acquired cisplatin resistance and that inhibition of the autophagic activity may be a viable strategy to overcome cisplatin resistance in human ovarian cancer cells.
FIGURE 6.
ERK activation, autophagy induction, and effect of autophagy inhibition on apoptosis in acquired cisplatin-resistant ovarian cancer cell line. A, MTT analyses of growth inhibition. OV433-P and OV433-CR cells were left untreated or treated with cisplatin at the indicated concentrations for 72 h. B, Western blot analyses of LC3, p62, P-ERK1/2, ERK1/2, P-p38, p38, p-JNK, JNK, and PARP. OV433-P and OV433-CR cells were left untreated or treated with 20 μm cisplatin for 24 h. C, representative fluorescent images of GFP-LC3 (left panel) and statistical analysis of GFP-LC3 dots (right panel). OV433-P and OV433-CR cells transfected with GFP-LC3 were selected with G418 for 3 weeks to isolate clones that stably express GFP-LC3. The resulting cells were left untreated or treated with cisplatin (20 μm, 20 h). Bright dots denote autophagosomes. Bars represent mean ± S.D. (error bars) of triplicate samples with >20 cells analyzed per sample. D, Western blot analyses of ERK1/2, LC3, p62, and PARP cleavage (upper panel) and quantification of LC3-II (lower panel). OV433-CR cells transfected with either control nontarget siRNAs (siControl) or ERK1/2 siRNAs (siERK1/2) were left untreated or treated with 20 μm cisplatin for 24 h. Quantification of PARP cleavage was described above. E, Western blot analyses of LC3, and PARP cleavage (left panel) and quantification of LC3-II (right panel). OV433-CR cells were left untreated or treated with 20 μm cisplatin in the presence or absence of 1 mm 3-MA for 24 h. Cleaved PARP was quantified as described above. F, Western blot analyses of Atg5, LC3, p62 and PARP (left panel) and quantification of LC3-II (right panel). OV433-CR cells transfected with siRNA against Atg5 (siAtg5) or control siRNA (siControl) were left untreated or treated with 20 μm cisplatin for 24 h. Cleaved PARP was quantified as described above. Data represent mean ± S.D. of three independent experiments. **, p < 0.001, statistically significant.
DISCUSSION
Over the past decade, much effort to study the genetic alterations in epithelial ovarian cancer has led to identification of many genetic changes. The hope that this would lead to more effective, targeted therapies is yet to be realized, as platinum-based (e.g. cisplatin-based) chemotherapy remains the first line treatment for this cancer type. Despite the obvious benefits of cisplatin-based treatment of ovarian cancer, almost all patients receiving cisplatin-based drugs eventually relapse and die from their metastatic disease. Therefore, a better understanding of the mechanism of cisplatin resistance is of clinical significance. In this study, we showed that cisplatin treatment induces an autophagic response that may counteract cisplatin-induced cell killing. We showed that cisplatin-induced autophagy is mediated by the ERK pathway, which requires the involvement of the Atg5 autophagy pathway. Importantly, we showed that pharmacological inhibition of autophagic activity or knockdown of Atg5 enhances cisplatin-induced apoptosis in ovarian cancer cells. Furthermore, our data showed that inhibition of autophagy enhances cisplatin-induced apoptosis in cells that have developed acquired resistance, suggesting that apoptosis and autophagy are two opposing cell signals that simultaneously regulate cisplatin sensitivity.
Autophagy is induced by conditions causing nutrient/energy depletion, cellular damage, and stress, which plays an important role not only in cancer development, but also in cancer treatment (38, 39). An extensive literature documents the prosurvival functions of autophagy, but there are cases where autophagy appears to have a pro-death role (40, 41). Recent studies indicate that autophagy induction contributed to chemoresistance including cisplatin resistance in cancer cells including lung and liver cancers (22, 23). It has been shown that acute cisplatin treatment activates an autophagic response that serves as a survival factor (22, 23). In lung cancer cells, autophagy has been suggested to play a role in acquired cisplatin resistance (24, 25). However, the role of autophagy in cisplatin resistance in ovarian cancer cells has not been determined. We showed that increased LC3-II levels are correlated with cisplatin resistance. We also showed that the pharmacological inhibition of the autophagic activity at the level of autophagosome formation or knockdown of Atg5, one of the key components of the autophagy pathway, increased cisplatin-induced growth inhibition. These data suggest that autophagy induction is a survival signal to counteract cisplatin-induced cell death.
The mechanisms by which autophagy is regulated are not fully understood. Emerging evidence suggests that all three MAPK subfamilies can regulate autophagy (30–35). For example, several studies indicate that ERK promotes autophagy (34, 35). In the case of JNK, it phosphorylates Bcl-2 upon starvation, thus disrupting Bcl-2/Beclin-1 binding and facilitating Beclin-1 interaction with Vps34 (33). As for p38, its role in the regulation of autophagy is contradictory. On one hand, p38 promotes autophagy in response to glucose (42, 43). On the other hand, pharmacological inhibition of p38 activity with SB202190 or genetic knockdown with p38 siRNA induces autophagy in colorectal cancer cells (30). The basis for these differences is not clear, but may in part be cell context-dependent or influenced by the type of autophagic stimuli. Consistent with the role of MAPKs in the regulation of autophagy, we showed that cisplatin treatment activates all major MAPK pathways. We also showed that the ERK pathway, but not the JNK or p38 pathway, involves cisplatin-induced autophagy because pharmacological inhibition of ERK but not JNK and p38 blocked cisplatin-induced autophagy. Thus, we conclude that cisplatin-induced autophagy is through the ERK pathway.
Autophagy induction requires the expression of autophagy genes including Atg5 (44–46). Atg5 is a member of the autophagy-related gene family that plays an important role in regulating autophagy (47, 48). Several studies indicated that Atg5 also regulates apoptosis. It has been shown that Atg5 can interact with FADD (Fas-associated protein with death domain) to mediate interferon-γ-induced death (49). Another study showed that overexpression of Atg5 sensitizes cells to various drugs whereas its knockdown decreases cell death (50). The underlying mechanism is believed to be the following: apoptotic stimuli cause Atg5 cleavage by calpains 1 and 2 to generate a 24-kDa truncated form of Atg5. The latter translocates to the mitochondria where it binds to Bcl-XL, leading to inactivation of Bcl-XL anti-apoptotic activity, cytochrome c release, and subsequent apoptosis (50). Therefore, Atg5 serves as a molecular switch between autophagy and apoptosis. In this study, we believed that Atg5-mediated autophagy is a survival signal because knockdown of Atg5 enhanced cisplatin-induced growth inhibition through enhanced cisplatin-induced apoptosis. The discrepancy between this study and the study by Yousefi et al. (50) can be due to the cell types and chemotherapeutics used. Nevertheless, these studies indicate that Atg5 serves as a switch between apoptosis and autophagy.
Acquired cisplatin resistance is a clinical problem that leads to treatment failure. Although we showed that activation of autophagy is a survival signal that counteracts cisplatin-induced cell death, the question is if this mechanism is clinically relevant. We shows that OV433-CR cells express higher levels of phosphorylated ERK and have a higher autophagic activity compared with their counterpart OV433-P cells. Higher levels of autophagic activity are correlated with decreased cisplatin-induced apoptosis. Importantly, inhibition of autophagy induction sensitized acquired resistant cells to cisplatin-induced cell death. Therefore, we conclude that inhibition of autophagy can render resistant cells sensitive to cisplatin.
In conclusion, we demonstrate that autophagy plays a critical role in cisplatin resistance in ovarian cancer cells. We show that cisplatin treatment activates the ERK survival pathway and subsequently promotes autophagy, leading to cisplatin resistance (Fig. 7). We also show that inhibition of ERK activation or inhibition of autophagy increases cisplatin-induced growth inhibition. Importantly, pharmacological inhibition of an autophagic response or knockdown of Atg5 can sensitize cells that have developed acquired resistance to cisplatin-induced apoptosis. Thus, this study suggests that targeting the autophagy pathway may present an opportunity to overcome cisplatin resistance in ovarian cancer.
FIGURE 7.
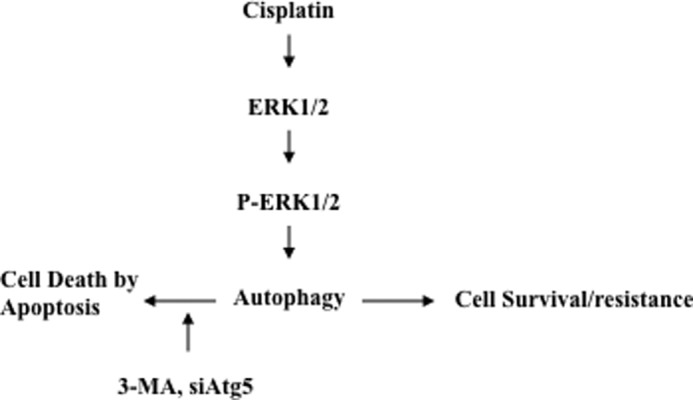
Proposed model for the mechanism by which cisplatin activates ERK1/2, which subsequently promotes an autophagic response, leading to cisplatin resistance in ovarian cancer cells.
Acknowledgments
We thank Dr. Nelly Auersperg for the IOSE358 cells and Dr. Hong-Gang Wang for GFP-LC3 expression vector.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R21CA178111-01 through the NCI.
- Atg
- autophagy-related gene
- CREB
- cAMP-responsive element-binding protein
- LC3
- microtubule-associated protein 1A/1B-light chain 3
- 3-MA
- 3-methyladenine
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PARP
- poly(ADP-ribose) polymerase.
REFERENCES
- 1. Cho K. R., Shih IeM. (2009) Ovarian cancer. Annu. Rev. Pathol. 4, 287–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bryant C. S., Kumar S., Spannuth W., Shah J. P., Munkarah A. R., Deppe G., Alvarez R. D., Morris R. T. (2011) Feasibility of extension of platinum-free interval with weekly bolus topotecan and subsequent platinum retreatment outcomes in recurrent ovarian cancer. Arch. Gyn. Obstet. 283, 361–367 [DOI] [PubMed] [Google Scholar]
- 3. Tummala M. K., McGuire W. P. (2005) Recurrent ovarian cancer. Clin. Adv. Hematol. Oncol. 3, 723–736 [PubMed] [Google Scholar]
- 4. Go R. S., Adjei A. A. (1999) Review of the comparative pharmacology and clinical activity of cisplatin and carboplatin. J. Clin. Oncol. 17, 409–422 [DOI] [PubMed] [Google Scholar]
- 5. Kelland L. (2007) The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 7, 573–584 [DOI] [PubMed] [Google Scholar]
- 6. Boeckman H. J., Trego K. S., Turchi J. J. (2005) Cisplatin sensitizes cancer cells to ionizing radiation via inhibition of nonhomologous end joining. Mol. Cancer Res. 3, 277–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Henkels K. M., Turchi J. J. (1997) Induction of apoptosis in cisplatin-sensitive and -resistant human ovarian cancer cell lines. Cancer Res. 57, 4488–4492 [PubMed] [Google Scholar]
- 8. Agarwal R., Kaye S. B. (2003) Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat. Rev. Cancer 3, 502–516 [DOI] [PubMed] [Google Scholar]
- 9. Johnson S. W., Shen D., Pastan I., Gottesman M. M., Hamilton T. C. (1996) Cross-resistance, cisplatin accumulation, and platinum-DNA adduct formation and removal in cisplatin-sensitive and -resistant human hepatoma cell lines. Exp. Cell Res. 226, 133–139 [DOI] [PubMed] [Google Scholar]
- 10. Cullen K. J., Newkirk K. A., Schumaker L. M., Aldosari N., Rone J. D., Haddad B. R. (2003) Glutathione S-transferase pi amplification is associated with cisplatin resistance in head and neck squamous cell carcinoma cell lines and primary tumors. Cancer Res. 63, 8097–8102 [PubMed] [Google Scholar]
- 11. Dabholkar M., Vionnet J., Bostick-Bruton F., Yu J. J., Reed E. (1994) Messenger RNA levels of XPAC and ERCC1 in ovarian cancer tissue correlate with response to platinum-based chemotherapy. J. Clin. Invest. 94, 703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perego P., Giarola M., Righetti S. C., Supino R., Caserini C., Delia D., Pierotti M. A., Miyashita T., Reed J. C., Zunino F. (1996) Association between cisplatin resistance and mutation of p53 gene and reduced bax expression in ovarian carcinoma cell systems. Cancer Res. 56, 556–562 [PubMed] [Google Scholar]
- 13. Beale P. J., Rogers P., Boxall F., Sharp S. Y., Kelland L. R. (2000) BCL-2 family protein expression and platinum drug resistance in ovarian carcinoma. Br. J. Cancer 82, 436–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Altomare D. A., Wang H. Q., Skele K. L., De Rienzo A., Klein-Szanto A. J., Godwin A. K., Testa J. R. (2004) AKT and mTOR phosphorylation is frequently detected in ovarian cancer and can be targeted to disrupt ovarian tumor cell growth. Oncogene 23, 5853–5857 [DOI] [PubMed] [Google Scholar]
- 15. Kuo M. T., Chen H. H., Song I. S., Savaraj N., Ishikawa T. (2007) The roles of copper transporters in cisplatin resistance. Cancer Metastasis Rev. 26, 71–83 [DOI] [PubMed] [Google Scholar]
- 16. Yang Z. J., Chee C. E., Huang S., Sinicrope F. (2011) Autophagy modulation for cancer therapy. Cancer Biol. Ther. 11, 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carew J. S., Nawrocki S. T., Cleveland J. L. (2007) Modulating autophagy for therapeutic benefit. Autophagy 3, 464–467 [DOI] [PubMed] [Google Scholar]
- 18. Maycotte P., Thorburn A. (2011) Autophagy and cancer therapy. Cancer Biol. Ther. 11, 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klionsky D. J. (2007) Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8, 931–937 [DOI] [PubMed] [Google Scholar]
- 20. Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clavé C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Droge W., Dron M., Dunn W. A., Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fesus L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., Gonzalez-Estevez C., Gorski S., Gottlieb R. A., Haussinger D., He Y. W., Heidenreich K., Hill J. A., Hoyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jaattela M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovacs A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., Lopez-Otin C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Melendez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Munz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nurnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D. C., Ryan K. M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P. O., Seleverstov O., Settleman J., Shacka J. J., Shapiro I. M., Sibirny A., Silva-Zacarin E. C., Simon H. U., Simone C., Simonsen A., Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Talloczy Z., Tanaka K., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcategui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mizushima N. (2007) Autophagy: process and function. Genes Dev. 21, 2861–2873 [DOI] [PubMed] [Google Scholar]
- 22. O'Donovan T. R., O'Sullivan G. C., McKenna S. L. (2011) Induction of autophagy by drug-resistant esophageal cancer cells promotes their survival and recovery following treatment with chemotherapeutics. Autophagy 7, 509–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zou Z., Wu L., Ding H., Wang Y., Zhang Y., Chen X., Chen X., Zhang C. Y., Zhang Q., Zen K. (2012) MicroRNA-30a sensitizes tumor cells to cis-platinum via suppressing beclin 1-mediated autophagy. J. Biol. Chem. 287, 4148–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ren J. H., He W. S., Nong L., Zhu Q. Y., Hu K., Zhang R. G., Huang L. L., Zhu F., Wu G. (2010) Acquired cisplatin resistance in human lung adenocarcinoma cells is associated with enhanced autophagy. Cancer Biother. Radiopharm. 25, 75–80 [DOI] [PubMed] [Google Scholar]
- 25. Sirichanchuen B., Pengsuparp T., Chanvorachote P. (2012) Long-term cisplatin exposure impairs autophagy and causes cisplatin resistance in human lung cancer cells. Mol. Cell. Biochem. 364, 11–18 [DOI] [PubMed] [Google Scholar]
- 26. Wang J., Zhou J. Y., Wu G. S. (2007) ERK-dependent MKP-1-mediated cisplatin resistance in human ovarian cancer cells. Cancer Res. 67, 11933–11941 [DOI] [PubMed] [Google Scholar]
- 27. Wang J., Zhou J. Y., Wu G. S. (2011) Bim protein degradation contributes to cisplatin resistance. J. Biol. Chem. 286, 22384–22392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang J., Zhou J. Y., Zhang L., Wu G. S. (2009) Involvement of MKP-1 and Bcl-2 in acquired cisplatin resistance in ovarian cancer cells. Cell Cycle 8, 3191–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Comes F., Matrone A., Lastella P., Nico B., Susca F. C., Bagnulo R., Ingravallo G., Modica S., Lo Sasso G., Moschetta A., Guanti G., Simone C. (2007) A novel cell type-specific role of p38α in the control of autophagy and cell death in colorectal cancer cells. Cell Death Differ. 14, 693–702 [DOI] [PubMed] [Google Scholar]
- 31. Corcelle E., Nebout M., Bekri S., Gauthier N., Hofman P., Poujeol P., Fénichel P., Mograbi B. (2006) Disruption of autophagy at the maturation step by the carcinogen lindane is associated with the sustained mitogen-activated protein kinase/extracellular signal-regulated kinase activity. Cancer Res. 66, 6861–6870 [DOI] [PubMed] [Google Scholar]
- 32. Pattingre S., Bauvy C., Codogno P. (2003) Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J. Biol. Chem. 278, 16667–16674 [DOI] [PubMed] [Google Scholar]
- 33. Wei Y., Pattingre S., Sinha S., Bassik M., Levine B. (2008) JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell 30, 678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang J., Whiteman M. W., Lian H., Wang G., Singh A., Huang D., Denmark T. (2009) A non-canonical MEK/ERK signaling pathway regulates autophagy via regulating Beclin 1. J. Biol. Chem. 284, 21412–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tang D., Kang R., Livesey K. M., Cheh C. W., Farkas A., Loughran P., Hoppe G., Bianchi M. E., Tracey K. J., Zeh H. J., 3rd, Lotze M. T. (2010) Endogenous HMGB1 regulates autophagy. J. Cell Biol. 190, 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu Y. T., Tan H. L., Shui G., Bauvy C., Huang Q., Wenk M. R., Ong C. N., Codogno P., Shen H. M. (2010) Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J. Biol. Chem. 285, 10850–10861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang D., Lippard S. J. (2005) Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 4, 307–320 [DOI] [PubMed] [Google Scholar]
- 38. Mathew R., Kongara S., Beaudoin B., Karp C. M., Bray K., Degenhardt K., Chen G., Jin S., White E. (2007) Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 21, 1367–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jin S., White E. (2007) Role of autophagy in cancer: management of metabolic stress. Autophagy 3, 28–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Scarlatti F., Maffei R., Beau I., Codogno P., Ghidoni R. (2008) Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 15, 1318–1329 [DOI] [PubMed] [Google Scholar]
- 41. Denton D., Nicolson S., Kumar S. (2012) Cell death by autophagy: facts and apparent artefacts. Cell Death Differ. 19, 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moruno-Manchón J. F., Pérez-Jiménez E., Knecht E. (2013) Glucose induces autophagy under starvation conditions by a p38 MAPK-dependent pathway. Biochem. J. 449, 497–506 [DOI] [PubMed] [Google Scholar]
- 43. Matsuzawa T., Kim B. H., Shenoy A. R., Kamitani S., Miyake M., Macmicking J. D. (2012) IFN-γ elicits macrophage autophagy via the p38 MAPK signaling pathway. J. Immunol. 189, 813–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 45. Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M. D., Klionsky D. J., Ohsumi M., Ohsumi Y. (1998) A protein conjugation system essential for autophagy. Nature 395, 395–398 [DOI] [PubMed] [Google Scholar]
- 46. Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Kominami E., Tanaka K., Chiba T. (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kametaka S., Matsuura A., Wada Y., Ohsumi Y. (1996) Structural and functional analyses of APG5, a gene involved in autophagy in yeast. Gene 178, 139–143 [DOI] [PubMed] [Google Scholar]
- 48. Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 49. Pyo J. O., Jang M. H., Kwon Y. K., Lee H. J., Jun J. I., Woo H. N., Cho D. H., Choi B., Lee H., Kim J. H., Mizushima N., Oshumi Y., Jung Y. K. (2005) Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J. Biol. Chem. 280, 20722–20729 [DOI] [PubMed] [Google Scholar]
- 50. Yousefi S., Perozzo R., Schmid I., Ziemiecki A., Schaffner T., Scapozza L., Brunner T., Simon H. U. (2006) Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 8, 1124–1132 [DOI] [PubMed] [Google Scholar]