Background: Albumin has a long serum half-life, which is regulated by FcRn.
Results: A cluster of conserved tryptophan residues of FcRn is required for binding to albumin and anti-FcRn albumin blocking antibodies.
Conclusion: The FcRn-albumin interaction is pH-dependent but hydrophobic in nature.
Significance: This study provides mechanistic insight into how FcRn binds albumin and regulates its long half-life.
Keywords: Albumin, Antibody, Biodegradation, Bioengineering, Fc Receptor, pH Regulation, FcRn, Half-life, Hydrophobic
Abstract
Albumin is the most abundant protein in blood and plays a pivotal role as a multitransporter of a wide range of molecules such as fatty acids, metabolites, hormones, and toxins. In addition, it binds a variety of drugs. Its role as distributor is supported by its extraordinary serum half-life of 3 weeks. This is related to its size and binding to the cellular receptor FcRn, which rescues albumin from intracellular degradation. Furthermore, the long half-life has fostered a great and increasing interest in utilization of albumin as a carrier of protein therapeutics and chemical drugs. However, to fully understand how FcRn acts as a regulator of albumin homeostasis and to take advantage of the FcRn-albumin interaction in drug design, the interaction interface needs to be dissected. Here, we used a panel of monoclonal antibodies directed towards human FcRn in combination with site-directed mutagenesis and structural modeling to unmask the binding sites for albumin blocking antibodies and albumin on the receptor, which revealed that the interaction is not only strictly pH-dependent, but predominantly hydrophobic in nature. Specifically, we provide mechanistic evidence for a crucial role of a cluster of conserved tryptophan residues that expose a pH-sensitive loop of FcRn, and identify structural differences in proximity to these hot spot residues that explain divergent cross-species binding properties of FcRn. Our findings expand our knowledge of how FcRn is controlling albumin homeostasis at a molecular level, which will guide design and engineering of novel albumin variants with altered transport properties.
Introduction
Albumin is a product of hepatocytes and is the most abundant protein in blood (34–54 g/liter). It serves as versatile transporter of a wide range of endogenous and exogenous compounds such as metal ions, hormones, fatty acids, metabolites, toxins, and drugs (1). Similar to all serum proteins, its serum concentration is determined by its rate of synthesis and its size above the renal clearance threshold. However, a third feature of albumin that is only shared with IgG antibodies (Abs)5 is a greatly extended persistence in the circulatory system, which in both cases is caused by their interaction with the neonatal Fc receptor (FcRn) (1–5).
As implied by its name, FcRn was first recognized as the neonatal transporter of maternal IgG from mother's milk across the intestinal barrier to the blood of rats (6). It also proved to be the transporter of IgG across the maternofetal barrier in both humans and rodents (7–9). A large body of subsequent evidence has revealed that FcRn is expressed and functionally operative in a broad range of cells and tissues throughout life (10, 11), transporting IgG across epithelial and endothelial barriers, enhancing IgG-mediated antigen presentation by dendritic cells and phagocytosis by neutrophils (12–16), and substantially extending the serum persistence of IgG (17–20).
As FcRn has also been demonstrated to prolong the serum half-life of albumin (5, 20), it acts as a regulator of the circulatory half-life of two totally unrelated proteins. This is evident from the fact that mice lacking FcRn have serum levels of IgG and albumin 4–5 and 2–3-fold lower than normal mice, respectively, as do mice where FcRn is conditionally deleted in endothelial and hematopoietic cells (5, 20–22). Genetic linkage in humans is also found by the rare human disease, familial hypercatabolic hypoproteinemia, which is characterized by abnormally low levels of both ligands that correlates with FcRn expression deficiency (23).
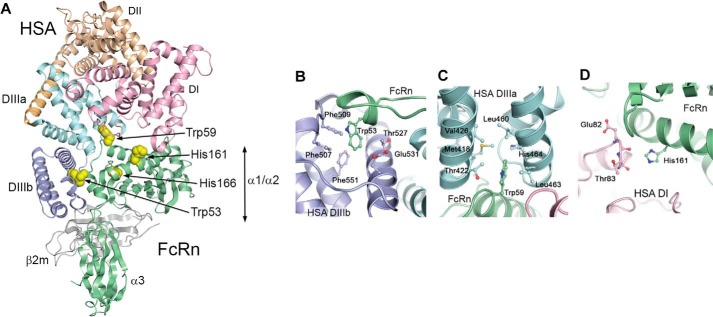
FcRn is a major histocompatibility class I-related molecule consisting of a unique transmembrane heavy chain (HC) with three extracellular domains (α1, α2, and α3) that are non-covalently bound to the common soluble β2-microglobulin. Crystal structures of the extracellular part of FcRn show that the amino-terminal α1-α2 platform is made up of eight antiparallel β-pleated strands topped by two long α-helices followed by the α3-domain (24–26). The soluble β2-microglobulin is tightly bound to the α1-α2 platform and the α3-domain is proximal to the membrane (Fig. 1A). Although major histocompatibility class I molecules bind peptides in their peptide-binding groove that is located between the two α-helices on the α1-α2 platform, the corresponding groove on FcRn is closed (24–26). Instead, FcRn has evolved to bind IgG and albumin.
FIGURE 1.
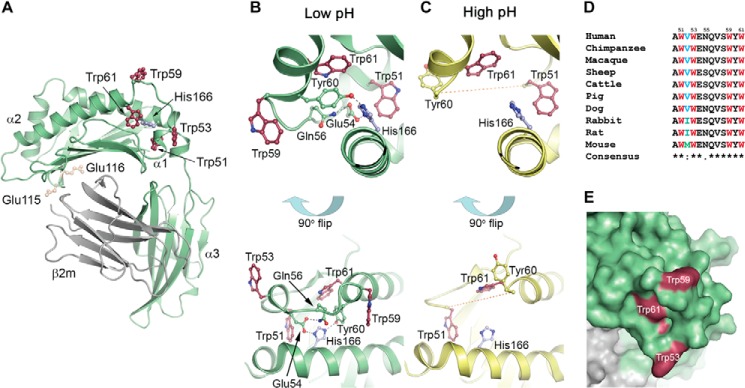
The structure of hFcRn and location of the pH-dependent flexible loop containing a cluster of tryptophans. A, overall structure of the extracellular part of hFcRn. The His-166 residue (blue ball-and-stick) within the α2-domain regulates a pH-dependent flexible loop (residues 51–61) within the α1-domain, which contains four tryptophan residues Trp-51, Trp-53, Trp-59, and Trp-61 (red ball-and-stick). The binding site for albumin is indicated relative to the IgG binding site that involves Glu-115 and Glu-116 (yellow ball-and-stick). The hFcRn HC is shown in green, and the β2-microglobulin (β2m) subunit is shown in gray. B, close up view of the FcRn HC loop at low pH (4.2) (25) where the positively charged His-166 makes charge-stabilized hydrogen bonds with Glu-54 and Tyr-60, structuring the loop of surrounding tryptophan residues. C, close up view of the FcRn HC loop at high pH (8.2) (26) where the uncharged His-166 loses the interactions with Glu-54 and Tyr-60, and the loop becomes flexible and structurally disordered (shown as a dashed line). D, alignment of a stretch of amino acids (residues 50–61) of the α1-domain of FcRn from 10 species. Asterisks indicate fully conserved amino acid residues. The four tryptophan residues are fully conserved, whereas a non-conserved amino acid is found in position 52. E, Trp-53 and Trp-59 are fully exposed at the surface of hFcRn, whereas Trp-61 is partially exposed, and Trp-51 is buried in the hydrophobic core of the molecule.
A hallmark of the interaction is that both IgG and albumin bind FcRn in a strictly pH-dependent manner, with binding at acidic pH (6.5–5.5) and no binding or release at neutral pH (2–5, 19, 24). This is fundamental for FcRn rescue of both ligands from lysosomal degradation. The underlying cellular mechanism has been deduced from live-cell imaging studies of the FcRn-IgG complex (11, 27–30), which demonstrate that FcRn resides predominantly within acidic endosomes where it encounters IgG taken up by fluid-phase endocytosis. The acidic milieu triggers binding of IgG via its Fc fragment to FcRn followed by exocytosis at the cell surface, where exposure to the near neutral pH of the blood results in release of IgG. Albumin is likely to follow the same rescue pathway, while proteins that do not bind the receptor are eliminated by lysosomal degradation.
Site-directed mutagenesis and inspection of the atomic resolution structure of rat FcRn in complex with rat IgG2a Fc have revealed that conserved histidine residues located at the IgG Fc elbow region (His-310 and His-435) regulate the strict pH dependence of the interaction (18, 19, 24), as histidines, positively charged at acidic pH, interact with negatively charged residues on FcRn. The more recently identified albumin interaction has not been mapped in the same detail. However, we and others have shown that the principal site for FcRn is located to the C-terminal domain III (DIII) of albumin, whereas domain I (DI) seems to modulate the interaction (2–4, 31). The pH dependence of the interaction is regulated by three conserved histidine residues, two were found within subdomains DIIIa (His-464) and DIIIb (His-535), and the last was in a loop connecting the two (His-510). These residues are involved in pH-sensitive interactions at the interfaces and internally in each protein (2, 32). Furthermore, His-166 in the α2-domain plays a regulatory role by stabilizing a flexible loop within the α1-domain. The loop is disordered at basic pH but forms an ordered structure at acidic pH (2, 32).
Here, we used a panel of monoclonal antibodies directed towards human FcRn in combination with site-directed mutagenesis and structural modeling to characterize the binding sites for human serum albumin (HSA) and antibodies blocking HSA binding. Our studies reveal the hFcRn epitopes that are crucial for binding of the antibodies, which overlap with amino acid residues that are required for binding to HSA. Furthermore, we show that the FcRn-albumin interaction is not only pH-dependent but also hydrophobic in nature. Binding is explained by a cluster of tryptophans within a pH-sensitive loop of the α1-domain of FcRn that is structurally ordered at acidic pH, and not at neutral pH, whereas divergent cross-species binding properties of albumin and the blocking antibodies are determined by structural differences in proximity to these hot spot residues. We structurally provide explanations for why the anti-hFcRn antibodies block HSA binding and show selective cross-species FcRn binding. The studies increase our understanding of the FcRn-albumin interaction at the atomic level that will facilitate the design of albumin variants with extended serum half-life for therapeutic applications.
EXPERIMENTAL PROCEDURES
Construction and Production of FcRn Variants
pcDNA3-GST-hβ2-microglobulin vectors encoding GST-tagged hFcRn variants (WT, E115A/E116A, E54Q, Q56A, H161A, and H166A), mouse FcRn (mFcRn) and rat FcRn have been described previously (2, 3, 33–35). In addition, cDNA sequences encoding the three extracellular domains of macaque (NCBI no. AAL92101), pig (NP_999362) and dog FcRn (XP_533618) HCs in addition to hFcRn mutants (H161E, H161Q, V52I, V52M, W51A, W53A, W59A, and W61A) were ordered from GenScript and subcloned into the same vector system. Also, cDNA encoding dog β2-microglobulin (NCBI no. ARF74978) was ordered from GenScript and subcloned into pcDNA3. All vectors were transiently transfected into HEK293E cells, and receptors were purified using a GSTrap FF column following the procedure described previously (36).
For production of truncated monomeric His-tagged mFcRn and hFcRn, a Baculovirus expression vector system was used, as described previously (8, 37). Viral stocks encoding His-tagged receptors were kind gifts from Dr. Sally Ward (University of Texas, Southwestern Medical Center, Dallas, TX). Receptors were purified using a HisTrap HP column supplied with Ni2+ ions (GE Healthcare). Prior to use, the column was pre-equilibrated with 1× PBS with 0.05% sodium azide. The pH of the supernatant containing mFcRn or hFcRn was adjusted with 1× PBS/0.05% sodium azide (pH 10.9) to pH 7.2, before being applied to the column with a flow rate of 5 ml/min. The column was washed using 200 ml of 1× PBS followed by 50 ml of 25 mm imidazole/1× PBS, pH 7.3. Bound receptors were eluted with 50 ml of 250 mm imidazole/1× PBS (pH 7.2–7.4). The collected protein was up-concentrated and buffer-changed to 1× PBS using Amicron Ultra-10 columns (Millipore). A HiLoad 26/600 Superdex 200 prep grade column (GE Healthcare) was used to isolate the monomeric fraction following the manufacturer's instructions. Fractions eluted were concentrated using Amicon Ultra columns (Millipore) and stored at 4 °C.
Production of MSA and HSA Variants
Recombinant mouse serum albumin (MSA), HSA, and HSA-K500A were produced in Saccharomyces cerevisiae, as described previously (2, 38).
Preparation of hFcRn-specific Antibodies
Mouse monoclonal Abs with hFcRn specificity (39) were produced in CELLine bioreactors (Sartorius), purified with HiTrap Protein G HP columns (GE Healthcare), buffer-changed to PBS using Spectra/Por dialysis membrane (Spectrum Labs), and concentrated using VIVASPIN ultrafiltration spin columns (Sartorius). Antibody purities exceeding 99% were determined by SDS-PAGE, and their concentrations were determined by their absorption at 280 nm using the molar extinction coefficient 210,000 m−1 cm−1 for IgG.
ELISA
Titrated amounts of mouse monoclonal Abs (0.5–4 μg/ml) with specificity for hFcRn were coated in microtiter wells (Nunc) and incubated overnight at 4 °C. Then wells were blocked with PBS/4% skim milk for 1 h at room temperature and washed four times in PBS/0.005% Tween 20 (PBS/T) pH 7.4. GST-tagged FcRn variants (1 μg/ml) were diluted in PBS/T/4% skimmed milk, pH 7.4, added to the wells, and incubated for 1.5 h at room temperature, prior to washing as described above. A horseradish peroxidase-conjugated anti-GST antibody (GE Healthcare), diluted (1:4000) in PBS/T/4% skimmed milk, pH 7.4, was added and incubated for 1 h. After washing as above, bound receptors were detected using tetramethylbenzidine substrate (Calbiochem). The absorbance was measured at 620 nm (or 450 nm when 100 μl of 1 m HCl was added) using the Sunrise spectophotometer (TECAN). The same ELISA was used to assay titrated amounts of HSA variants (1.56–200 μg/ml) coated in wells using PBS, pH 6.0, in all steps.
For binding to human IgG1, 3-iodo-4-hydroxy-5-nitrophenacetyl-conjugated bovine serum albumin (2 μg/ml) was coated in ELISA wells, and incubated with titrated amount of WT anti-3-iodo-4-hydroxy-5-nitrophenacetyl human IgG1 (0.0009–2 μg/ml). Subsequently, GST-tagged hFcRn variants were added and incubated for 1 h. The wells were blocked and washed as described above, using PBS, pH 6.0, in all steps.
SPR
Surface plasmon resonance (SPR) was conducted using a Biacore 3000 instrument (GE Healthcare) with CM5 sensor chips coupled with HSA (∼1,500 resonance units), hFcRn-GST variants (∼1,000 resonance units), or anti-hFcRn Abs (∼500 resonance units) using amine-coupling chemistry as described by the manufacturer. The coupling was performed by injecting 10 μg/ml of each protein into 10 mm sodium acetate, pH 4.5 (GE Healthcare), using the amine coupling kit (GE Healthcare). Phosphate buffer (67 mm phosphate buffer, 0.15 m NaCl, 0.005% Tween 20) at pH 6.0 or 7.4, or HBS-P buffer (0.01 m HEPES, 0.15 m NaCl, 0.005% surfactant P20) at pH 7.4 were used as running buffer and dilution buffer. For competitive binding to immobilized HSA or ADM31, 200 nm of soluble hFcRn was injected alone or in the presence of the Abs (200 nm), at either pH 6.0 or 7.4. Binding kinetics for His-tagged monomeric mFcRn and hFcRn toward immobilized Abs were determined by injecting titrated amounts (10–0.15 μm) of the receptors over immobilized Abs at pH 7.4, whereas kinetics for binding of HSA and MSA to immobilized hFcRn variants were determined by injecting titrated amounts of albumin over immobilized receptors at pH 6.0. All SPR experiments were conducted at 25 °C with a flow rate of 50 μl/min. Binding data were zero-adjusted, and the reference cell value subtracted. The Langmuir 1:1 ligand binding model and the steady-state affinity model provided by the BIAevaluation software (version 4.1) were used to determine the binding kinetics.
Structural Analysis
Coordinates were retrieved from the Protein Data Bank database: hFcRn Protein Data Bank high pH form, code 1EXU (26); hFcRn low pH form, Protein Data Bank code 3M17 (25); rat FcRn, Protein Data Bank code 1FRT (26); and hFcRn in complex with an HSA variant (HSA13) containing four mutations within DIII (V418M/T420A/E505G/V547A), Protein Data Bank code 4K71 (32). The structures of hFcRn and HSA were inspected using PyMOL (Schrödinger, Inc.).
RESULTS
His-166 Stabilizes a Loop with a Cluster of Tryptophans
We have previously demonstrated that His-166, a fully conserved residue localized to the α2-domain of the FcRn HC (Fig. 1A and supplemental Fig. S1), is crucial for binding to albumin, as mutation of this residue in both the mouse and human receptor eliminates binding (2, 3). Inspection of two crystal structures of hFcRn (25, 26), one solved at acidic and another at basic pH, shows that His-166 at acidic pH (pH 4.2) is engaged in a network of intramolecular interactions involving charge-stabilized hydrogen bonds with residues (Glu-54 and Tyr-60) found in a surface-exposed loop within the α1-domain encompassing residues 51–60 (Fig. 1B). In contrast, this loop is structurally disordered in the crystal structure solved at basic pH (pH 8.5) (Fig. 1C), which is explained by loss of the positive charge on His-166 as pH approaches neutrality (2, 25, 26). Furthermore, the loop contains a cluster of four tryptophan residues; Trp-51, Trp-53, Trp-59, and Trp-61 (Fig. 1B), which are conserved across species (Fig. 1D). Trp-53 and Trp-59 are fully exposed on the surface of the loop, Trp-61 is partly exposed and Trp-51 is buried inside the hFcRn structure (Fig. 1E). Notably, the loop is located on the opposite side of the binding site for IgG on the α1-α2 platform (Fig. 1A).
The Tryptophans Are Crucial for Albumin Binding
To investigate how each of the tryptophans contribute to binding of albumin, we performed an alanine scan, and the designed hFcRn mutants, each with one tryptophan changed to alanine, were produced as recombinant soluble forms in HEK293E cells (Fig. 2A). All mutants were secreted from the cells equal to the wild-type (WT). SPR was used to screen for binding to HSA by immobilizing the receptor and injecting equal amounts of HSA at pH 6.0. The resulting sensorgrams show a striking impact of mutating the tryptophans, as the hFcRn variants showed no (Trp-53 and Trp-61) or only minor (Trp-51 and Trp-59) binding compared with the WT (Fig. 2, B–F). HSA bound only weakly to the hFcRn-H166A mutant, which was included as a control (Fig. 2G).
FIGURE 2.
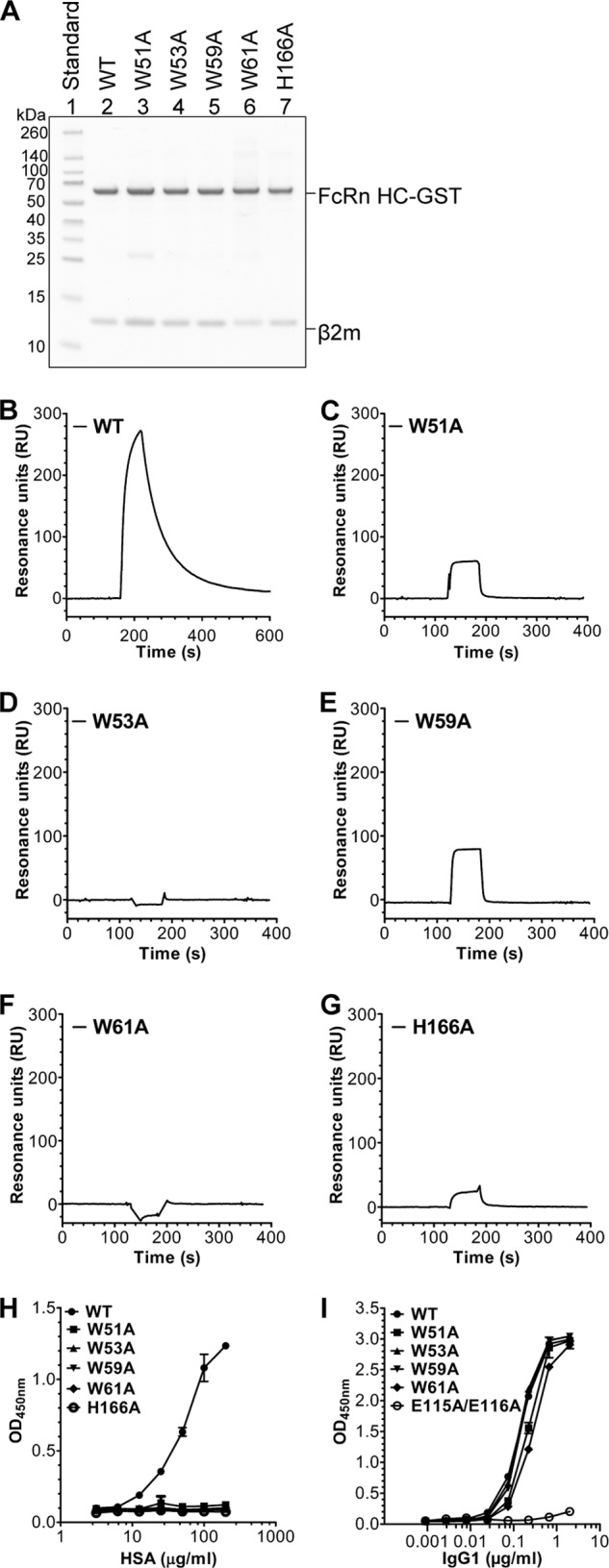
Mutation of the hFcRn tryptophan residues eliminates binding to albumin. A, SDS-PAGE gel migration of recombinant GST-tagged hFcRn WT and variants showing the expected molecular sizes of the GST-fused HC and β2-microglobulin (β2m). Representative SPR sensorgrams showing binding of immobilized WT hFcRn (B), hFcRn-W51A (C), hFcRn-W53A (D), hFcRn-W59A (E), hFcRn-W61A (F), and hFcRn-H166A (G) to WT HSA at pH 6.0. ELISA binding of WT hFcRn and the tryptophan mutants to titrated amounts of HSA (H) and human IgG1 (I) at pH 6.0 (n = 3). All data are presented as mean ± S.D.
Similarly, using ELISA, none of the Trp mutants were shown to bind HSA (Fig. 2H), whereas binding to human IgG1 was retained for all variants (Fig. 2I). Importantly, an hFcRn mutant, harboring two substitutions (E115A/E116A) at the core of the IgG-binding site (Fig. 1A), did not bind IgG1 (Fig. 2I). Thus, the conserved tryptophans are crucial for albumin binding.
Antibodies Blocking Albumin Binding to hFcRn
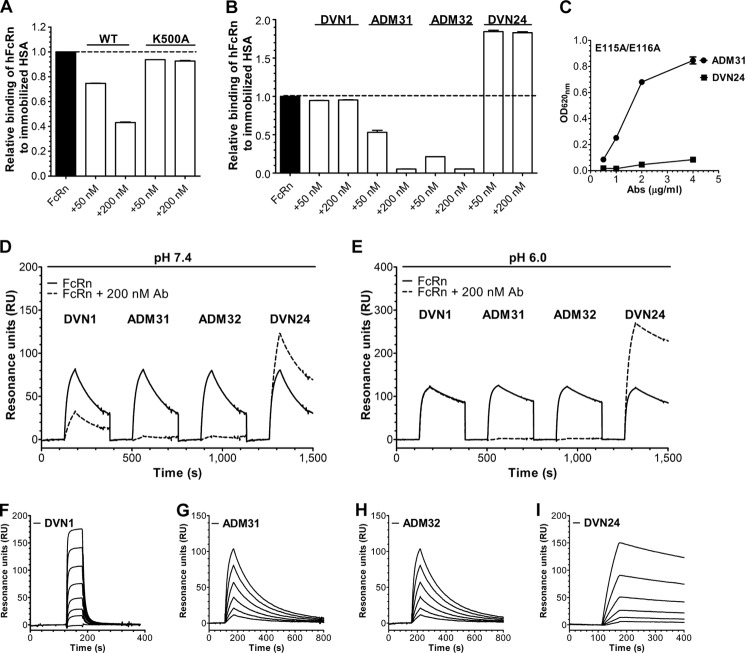
To further dissect the FcRn-albumin interaction, we took advantage of a panel of nine monoclonal Abs directed toward hFcRn (39), and SPR was used to test whether the Abs interfered with albumin binding to recombinant hFcRn at pH 6.0. HSA was immobilized on the chip followed by injection of soluble hFcRn in the presence of the Abs. Initially, binding of hFcRn to immobilized HSA was shown to be inhibited by soluble HSA, but not by HSA with Lys-500 mutated to alanine (K500A) (Fig. 3A and supplemental Fig. S2, A and B), which is in agreement with previous data showing 30-fold reduced binding of the mutant to hFcRn (2). Screening of the Abs then demonstrated that ADM31 and ADM32 blocked HSA binding and thus bound to the HSA binding site on FcRn. All other Abs bound the receptor at other sites, as additive binding of HSA and Ab was observed, except for DVN1 and DVN22, which did not bind hFcRn at acidic pH at all (Fig. 3B and supplemental Fig. S2, C–K).
FIGURE 3.
Identification of monoclonal Abs that block albumin binding to hFcRn. Competitive SPR analysis where monomeric hFcRn was injected alone or in the presence of WT HSA and HSA-K500A (A), or DVN1, ADM31, ADM32, and DVN24 (B) over immobilized WT HSA at pH 6.0. Relative binding was calculated by setting the maximum binding response of WT hFcRn toward WT HSA to 1.0. C, ELISA binding of hFcRn-E115A/E116A to titrated amounts of DVN24 and ADM31 coated in ELISA wells at pH 7.4 (n = 3). All data are presented as mean ± S.D. Competitive SPR analysis where monomeric hFcRn was injected alone or in the presence of DVN1, ADM31, ADM32, and DVN24 over immobilized ADM31 at pH 7.4 (D) and pH 6.0 (E). Representative sensorgrams showing binding of titrated amounts of monomeric hFcRn injected over immobilized DVN1 (F), ADM31 (G), ADM32 (H), and DVN24 (I) at pH 7.4. The binding kinetic constants are summarized in Table 1.
Furthermore, binding of DVN24 was fully dependent on Glu-115 and Glu-116 within the α2-domain (Fig. 1A), as mutating these positions (E115A/E116A) completely abolished binding (Fig. 3C). Thus, DVN24 binds to the IgG binding site on FcRn.
Next, we investigated whether binding of the Abs was affected by pH and whether they shared binding epitopes on hFcRn. ADM31 was immobilized on the chip, and soluble hFcRn was injected together with either DVN1, ADM31, ADM32, or DVN24. Although ADM32 competed with ADM31 for binding at both pH 6.0 and pH 7.4 (Fig. 3, D and E), DVN1 only did so at pH 7.4 (Fig. 3D). As expected, DVN24 did not interfere with binding (Fig. 3, D and E). A total of three Abs that bind to overlapping sites as albumin were thus identified. Two (ADM31 and ADM32) bound both at acidic and neutral pH, whereas one (DVN1) bound at neutral pH only.
Binding kinetics of the nine Abs were determined by injecting titrated amounts of monomeric hFcRn over immobilized Abs at pH 7.4, which generated distinct binding kinetics of each of the Abs (Fig. 3, F–I, data not shown). Specifically, ADM31 and ADM32 as well as DVN24 bound strongly to hFcRn, whereas DVN1 showed fast binding kinetics. The kinetic constants of the Abs are summarized in Table 1.
TABLE 1.
SPR derived binding kinetics
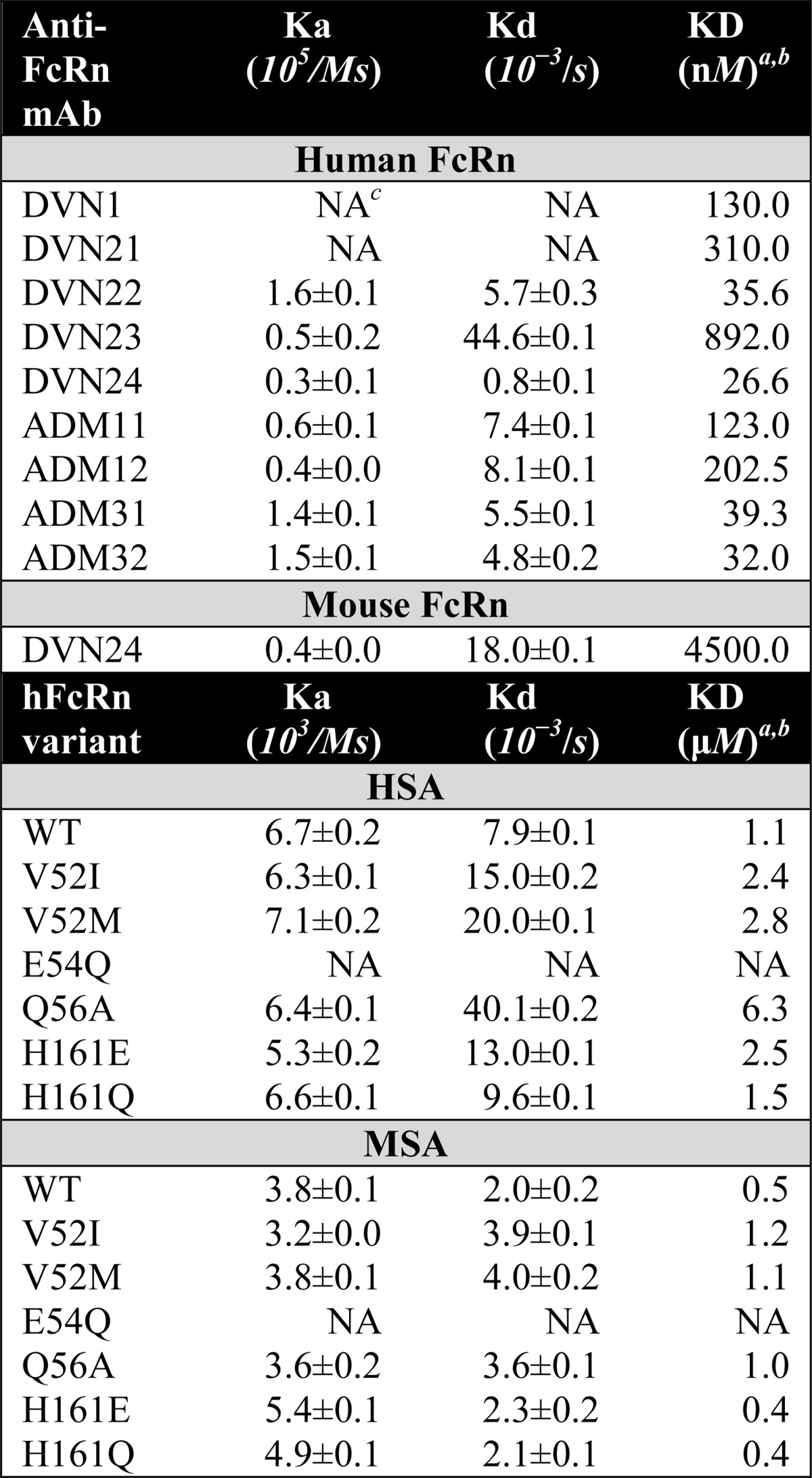
a The kinetic rate constants were obtained using a simple first-order. (1:1) Langmuir bimolecular interaction model. The kinetic values represent the average of triplicates.
b The steady-state affinity constants were obtained using an equilibrium (Req) binding model supplied by the BIAevaluation software (version 4.1). The affinities derived from equilibrium binding data represent the average of triplicates.
c NA, not acquired because of fast binding kinetics.
Trp-59 Is Required for Antibody Binding
To further characterize the binding site of the Abs, we investigated the importance of each of the four tryptophans for Ab binding by ELISA at pH 7.4. Titrated amounts of the Abs were coated in wells followed by addition of equal amounts of WT hFcRn and hFcRn mutants, W51A, W53A, W59A, W61A, and H166A. The results show that Trp-59 is fundamental for binding to ADM31, ADM32, and DVN1, as mutation at this position eliminated binding to all three Abs (Fig. 4, A–C). Mutation of Trp-61 completely abolished binding to DVN1, whereas binding to ADM31 and ADM32 was reduced. DVN1 binding was also strongly affected by mutating Trp-51, which had no or little effect on ADM31 and ADM32 binding. Finally, removal of Trp-53 had little or no effect on binding of either ADM31 or ADM32. Mutation of His-166 reduced binding to all three Abs (Fig. 4, A–C), whereas binding of DVN24 was not affected by removal of any of the tryptophans or His-166 (Fig. 4D).
FIGURE 4.
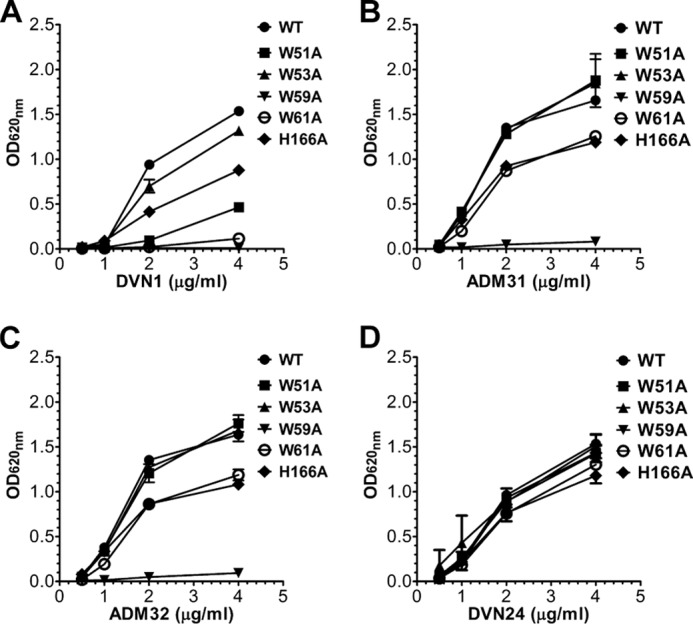
Trp-59 is required for binding to albumin-blocking anti-hFcRn Abs. Binding of WT hFcRn and the tryptophan mutants to titrated amounts of DVN1 (A), ADM31 (B), ADM32 (C), and DVN24 (D) coated in ELISA wells at pH 7.4 (n = 3). All data are presented as mean ± S.D.
Species-dependent Antibody Binding
Despite the fact that His-166 and the four tryptophans are fully conserved among species (Fig. 1E and supplemental Fig. S1), large cross-species differences in albumin binding exist. For instance, we have previously shown that hFcRn binds more strongly to MSA than to HSA and that mFcRn binds weakly to HSA (33). Thus, we tested Ab binding to various FcRn species. Recombinant forms of macaque, pig, dog, mouse, and rat FcRn were produced in HEK293E cells (Fig. 5A) and subsequently tested for binding to the Abs by ELISA at pH 7.4. Macaque and pig FcRn bound ADM31 and ADM32 with similar strength as hFcRn, whereas DVN1 did not bind pig (Fig. 5, B–D). Furthermore, dog FcRn bound with intermediate affinity to ADM31 and ADM32 but not to DVN1. Strikingly, no detectable binding of any of the Abs was detected toward the rat and mouse receptor (Fig. 5, B–J). Lack of binding to mFcRn was also confirmed by SPR, except for weak detectable binding of DVN24 (Table 1, data not shown). All other Abs also showed strictly species dependent and selective binding (Fig. 5, B–J). The relative binding of the Abs to the FcRn species is summarized in Table 2.
FIGURE 5.
The monoclonal Abs bind in a species-dependent manner to FcRn. A, SDS-PAGE gel migration of recombinant GST-tagged human, macaque, pig, dog, mouse, and rat FcRn showing the expected molecular sizes of the GST-fused HCs and β2-microglobulin (β2m). ELISA binding of human, macaque, pig, dog, mouse, and rat FcRn toward DVN1 (B), ADM31 (C), ADM32 (D), DVN24 (E), ADM11 (F), ADM12 (G), DVN21 (H), DVN22 (I), and DVN23 (J) at pH 7.4 (n = 3). All data are presented as mean ± S.D. An overview of relative binding of the Abs to the FcRn species is given in Table 2.
TABLE 2.
Cross-species binding properties of anti-FcRn monoclonal antibodies
Relative binding was determined based on ELISA screening (Fig. 5, B–J), and data are presented as equal binding as hFcRn (+++), partly reduced binding (++), poor binding (+), or no binding (−).
| FcRn/Ab | DVN1 | DVN21 | DVN22 | DVN23 | DVN24 | ADM11 | ADM12 | ADM31 | ADM32 |
|---|---|---|---|---|---|---|---|---|---|
| Human | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Macaque | +++ | +++ | + | ++ | +++ | +++ | +++ | +++ | +++ |
| Pig | − | − | ++ | + | +++ | +++ | +++ | +++ | +++ |
| Dog | − | − | − | + | +++ | +++ | +++ | ++ | ++ |
| Rat | − | − | − | − | − | − | − | − | − |
| Mouse | − | − | − | − | − | − | − | − | − |
His-161 Is Fundamental for Binding to Albumin-blocking Antibodies
To obtain a molecular explanation for the large cross-species binding differences of the antibodies, we inspected the HC sequences and crystal structures of rat and hFcRn and found that 10 of 12 residues in the loop encompassing the tryptophans are conserved (Fig. 1D). In contrast, position 52 is not conserved such that human has a valine, whereas rat and mouse have an isoleucine or a methionine, respectively. In addition, in structural proximity to the loop, a non-conserved residue is found at position 161 within the α2-domain (Fig. 6A). Although hFcRn has a histidine, rat and mouse have a negatively charged glutamic acid, and dog and rabbit have a glutamine (Fig. 6B). Interestingly, we have previously shown that a H161A mutation in hFcRn reduces binding to HSA (3). Based on these findings, we constructed four hFcRn mutants, V52I, V52M, H161E, and H161Q (Fig. 6C), and screened purified receptors for binding to the Abs by ELISA. Although neither the V52I nor the V52M mutation affected binding to the Abs (Fig. 6, D–F), binding of hFcRn-H161E (rat/mouse-like) to all three Abs was completely eliminated (Fig. 6, D–F), reflecting the binding pattern of the rat and mouse receptors. Likewise, the H161Q mutant (dog/rabbit-like) showed slightly reduced binding to ADM31 and ADM32, whereas DVN1 did not bind, reflecting the binding pattern of the dog receptor (Fig. 6, D–F, Fig. 5, B–D). In addition, the H161A mutation completely abolished binding to DVN1 and reduced binding to ADM31 and ADM32 (Fig. 6, D–F). All mutants bound equally well as WT hFcRn to DVN24 (Fig. 6G). Thus, non-conserved variations at position 161 within the α2-domain are responsible for species-dependent binding of DVN1, ADM31, and ADM32.
FIGURE 6.
Selective FcRn cross-species binding to the monoclonal Abs depends on His-161. A, close-up of the structural areas of hFcRn and rat FcRn encompassing the α1-domain loop with the tryptophan residues (red), hFcRn Val-52 and rat FcRn Ile-52 (cyan), and the α2-domain α-helix with hFcRn His-161 (blue), and rat FcRn Glu-163 (orange). The figures were made using the available crystal structures of hFcRn (25) and rat FcRn (24). B, an alignment of the stretch of amino acids of the α-helix containing His-166 in the α-2 domain of FcRn from 10 different species. A non-conserved amino acid variation is found at position 161 (human numbering). C, SDS-PAGE gel migration of recombinant GST-tagged hFcRn WT and mutants (V52I, V52M, H161E, and H161Q) showing the expected molecular sizes of the GST-fused HC and β2-microglobulin (β2m). Binding of WT hFcRn and mutants (V52I, V52M, W59A, H161A, H161E, and H161Q) to DVN1 (D), ADM31 (E), ADM32 (F), and DVN24 (G) at pH 7.4. Relative binding of the Abs to the hFcRn variants was calculated from an ELISA screen (data not shown), where binding of WT hFcRn to the Abs was set to 1.0 (n = 3). All data are presented as mean ± S.D.
Non-conserved Variations Modulate Albumin Binding
Next, we used SPR to investigate how these non-conserved amino acids affected binding to HSA. Titrated amounts of HSA were injected over immobilized hFcRn variants at pH 6.0. We found that the variation at position 52 modulates binding, as both hFcRn-V52I and V52M showed 2-fold reduced binding to HSA compared with WT hFcRn (Table 1 and Fig. 7, B and C). In addition, mutation of Glu-54 (E54Q) and Gln-56 (Q56A), which stabilize the loop (Fig. 1, B and C), either eliminated or reduced binding by 6-fold, respectively (Table 1 and Fig. 7D). Furthermore, the H161E mutation decreased binding by 2-fold, whereas the H161Q mutation gave a slight reduction in binding (Table 1 and Fig. 7, E and F). Thus, non-conserved residues in both the α1 and α2-domains contribute to the differences in cross-species binding of albumin.
FIGURE 7.
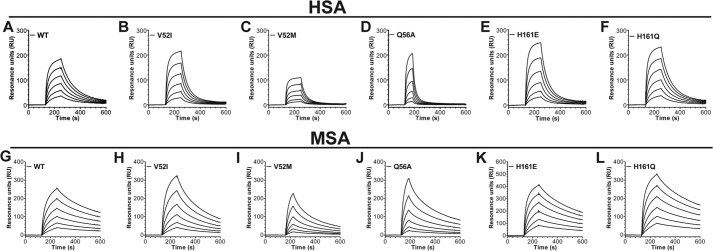
Non-conserved FcRn residues modulate albumin binding. Representative SPR sensorgrams show binding of titrated amounts of WT HSA to immobilized WT hFcRn (A), hFcRn-V52I (B), hFcRn-V52M (C), hFcRn-Q56A (D), hFcRn-H161E (E), and hFcRn-H161Q (F) at pH 6.0. Representative SPR sensorgrams show binding of titrated amounts of WT MSA to immobilized WT hFcRn (G), hFcRn-V52I (H), hFcRn-V52M (I), hFcRn-Q56A (J), hFcRn-H161E (K), and hFcRn-H161Q (L) at pH 6.0. The binding kinetic constants are summarized in Table 1.
The Impact of Non-conserved FcRn Residues on Binding to MSA
The fact that hFcRn binds more strongly to MSA than to HSA (33) prompted us to investigate how binding to MSA was affected by the hFcRn mutations. Using SPR, titrated amounts of MSA were injected over immobilized receptor variants at pH 6.0, and calculation of binding kinetics revealed that mutation of amino acid residues within the Trp loop, V52I, V52M, and Q56A, reduced binding affinity by 2-fold, whereas the hFcRn-E54Q variant gave rise to almost complete loss of binding (Fig. 7, G–J; Table 1).
As the HSA blocking antibodies require a histidine at position 161, this raised the question whether cross-species variations at this position modulate binding to MSA. Again, binding was assessed by SPR, which showed that replacement of His-166 with H161E or H161Q did not have any major impact on the binding kinetics (Fig. 7, K–L; Table 1). Thus, although binding of the HSA-blocking antibodies is fully dependent on His-161, the impact of this residue on binding to MSA and HSA is only minor.
FcRn Tryptophans Contact a Hydrophobic Platform on Albumin
Based on the obtained data, we inspected a recently published crystal structure of hFcRn in complex with a HSA variant containing four amino acid substitutions (HSA13) that binds less pH-dependent to the receptor (32). The exposed and conserved residues Trp-59 and Trp-53 are found to be in close proximity to hydrophobic regions in DIIIa and DIIIb of HSA, respectively (Fig. 8, A–C). Trp-53 contacts a hydrophobic platform on HSA formed by the conserved phenylalanine residues Phe-502, Phe-507, and Phe-551 as well as Thr-527, whereas Trp-59 is inserted in a narrow cleft formed by two α-helices in DIIIa as well as a loop in DI. The cleft is formed by the hydrophobic residues Met-418, Leu-460, and Leu-463 (Fig. 8C). In addition, Val-52, Glu-54, and Gln-56 of hFcRn make direct contacts with amino acid residues within DIII.
FIGURE 8.
Hydrophobic interactions between hFcRn tryptophan residues and HSA DIII stabilize the complex. A, an overview of the crystal structure of the hFcRn-HSA13 complex showing the positions of residues Trp-53, Trp-59, His-161, and His-166 (yellow spheres) in the α1-α2 domain platform of hFcRn, and their positions relative to the three α-helical domains in HSA; DI (pink), DII (orange), and DIII (cyan/blue). The DIII is split into subdomains DIIIa (cyan) and DIIIb (blue). B, a close-up of the region around Trp-53 showing interactions with hydrophobic HSA residues Phe-507, Phe-509, and Phe-551. The hydrophobic part of Thr-527 and Glu-531 also defines the hydrophobic pocket into which Trp-53 is inserted. C, a close-up of the region around Trp-59 showing the hydrophobic surroundings, with HSA residues Met-418, Thr-422, Leu-460, and Leu-463 being closest to Trp-59. Also, His-464 and Val-426 contribute to the hydrophobic pocket. D, a close-up of His-161 in hFcRn that points toward a loop in the HSA DII domain, containing residues Glu-82 and Thr-83. β2m, β2-microglobulin.
Furthermore, His-166 of hFcRn makes no direct contact with HSA but stabilizes the Trp loop via a network of hydrogen bonds, in accordance with our previous model (2). In this model, we also postulated that hFcRn makes contacts with the N-terminal DI of HSA. In line with this is the fact that His-161 of the hFcRn-HSA13 complex points toward a loop in HSA13 DI, which contains residues Glu-82 and Thr-83 (Fig. 8D). However, there are no direct hydrogen bond contacts between the side chains. Depending on the side chain conformation of His-161, possible hydrogen bond donors in HSA are the backbone carbonyls of Glu-82 and Thr-83.
DISCUSSION
FcRn is a versatile cellular receptor for the two most abundant proteins in blood, IgG and albumin. Their biodistribution, high concentration, and long serum half-life depend on FcRn-mediated transcytosis or rescue from intracellular degradation. Therefore, it is essential to unravel how FcRn binds both ligands at the atomic level. In addition, as IgG and albumin variants and fusions are increasingly utilized as diagnostics and therapeutics (40, 41), such knowledge will guide design of novel bio-pharmaceuticals. Although the FcRn-IgG interaction has been studied for more than two decades, the more recently discovered interaction with albumin is less well understood. Thus, we aimed for a mechanistic understanding of how hFcRn binds HSA and anti-hFcRn antibodies that block HSA binding. In this report, we identify key amino acid residues on hFcRn that are required for binding to HSA and the blocking antibodies.
Previously, we demonstrated that mutation of His-166 within the α2-domain of hFcRn eliminated binding to HSA without affecting IgG binding (3). His-166 is only partly exposed on the surface, as it is engaged in an intramolecular network of interactions that stabilize an exposed loop within the α1-domain (2, 32). This happens in a pH-dependent manner, where an ordered structure of the loop is obtained at acidic pH only. In line with this, targeting of charged residues within this loop (E54 and Q56) by mutagenesis eliminates or reduces binding to albumin (2).
Moreover, inspection of available crystal structures revealed that His-166 of hFcRn (His-168 in rat FcRn) is surrounded by a cluster of four tryptophan residues that are located within the flexible loop. Two of these tryptophans are fully exposed (Trp-53 and Trp-59), one is partly exposed (Trp-61) and one is buried (Trp-51). As all four tryptophans are conserved among species, we hypothesized that they are important for binding, and performed alanine scanning in hFcRn. Binding studies then revealed that each of them attenuated binding to HSA.
Furthermore, we took advantages of a panel of nine monoclonal Abs directed toward hFcRn, which were made by immunizing mice lacking expression of FcRn with spleen cells from mice that transgenically express hFcRn (39). Two of the generated Abs (ADM31 and ADM32) were shown to block HSA binding to recombinant hFcRn at both pH 6.0 and 7.4, whereas one (DVN1) bound to an overlapping binding site at pH 7.4 only. Furthermore, at pH 7.4, all three Abs competed for binding. The binding behavior of the Abs is in agreement with previous cellular studies (39).
Next, we aimed to identify their binding epitopes, and found them to have slightly different binding profiles. None bound hFcRn-W59A, all were affected by the mutation of Trp-61, whereas only DVN1 was dependent on the presence of Trp-51. None of these required the presence of Trp-53, which was crucial for HSA binding. Thus, the binding sites for the antibodies overlap with that of HSA, without being identical.
His-161, which is exposed on the surface of an α-helix on the α2-domain, is in close structural proximity to Trp-59 and was shown to be important for binding to all three Abs, and crucial for binding of DVN1. Substitution of His-161 with alanine, glutamine, or glutamic acid eliminated binding to DVN1. The charge of His-161 changes with pH and thus may explain why DVN1 binds pH-dependent. In addition, Trp-61 and Trp-51 may also be less exposed when the α1-domain loop is stabilized by His-166 at acidic pH and contribute to this pH dependence. As His-161 is in contact with DI of HSA (32), the albumin-blocking antibodies mask this interaction.
To gain structural insights, we inspected the crystal structure of the hFcRn-HSA13 complex, which revealed that the surface-exposed Trp-53 and Trp-59 make direct contact with HSA13 at acidic pH, whereas Trp-51 and Trp-61 stabilize the ends of the hFcRn loop comprising these residues. Thus, the co-crystal structure confirms our findings and together show that the FcRn-albumin interaction is not only pH-dependent but also hydrophobic in nature. This is supported by isothermal titration calorimetry analysis showing that the interaction has hydrophobic features as evidenced by a large positive change in entropy (4). Furthermore, the co-crystal structure shows that the exposed Trp-53 in hFcRn is contacting the HSA partner on a hydrophobic platform in DIIIb of HSA. The platform is comprised of phenylalanine residues in the loop that correspond to residues 500–510, which connects the subdomains DIIIa and DIIIb of HSA as well as the next to last C-terminal α-helix. We recently showed that both these structural areas are of importance for binding of HSA to hFcRn (2). The platform is formed by the loop residues Phe-502, Phe-507, and Phe-551. Trp-53 makes direct contact with the platform by hydrophobic stacking with the three fully conserved phenylalanines.
Furthermore, Trp-59 is interacting with the hydrophobic residues Met-418, Leu-460, and Leu-463 in DIIIa. The hydrophobic interface involving Trp-53 and Trp-59 is surrounded by the conserved histidines that mediate the strict pH dependence of the interaction. Notably, a binding pocket is located near the hydrophobic platform, and binding of compounds to this pocket may well affect FcRn binding. Indeed, a recent study shows that bound long fatty acids negatively modulate binding to hFcRn (32).
In addition, a crystal structure of the hFcRn in complex with WT HSA was very recently made available (42). The HSA model was divided into separate parts to find a solution during the molecular replacement procedure. Overall, this model resembles that of the hFcRn-HSA13 complex, with the exception of the local conformation changes induced by the four amino acids of the HSA13 variant.
Large differences exist across species in the FcRn-albumin interaction, which compromise preclinical in vivo evaluations (33). Specifically, mFcRn binds only weakly to HSA, whereas hFcRn binds more strongly to MSA than to HSA. Here, we demonstrate that binding of albumin blocking Abs require the presence of His-161. Neither mouse nor rat FcRn bind the Abs, and both have a glutamic acid at the corresponding position (Glu-163), whereas the surrounding residues are otherwise very similar in human, mouse and rat FcRn. Also, dog FcRn, with a glutamine at position 161, showed reduced Ab binding, and in line with this, hFcRn-H161Q bound the Abs with reduced affinity. Moreover, we previously showed that amino acid differences in the vicinity of His-166 of hFcRn (His-168 of mFcRn) modulate binding to albumin (33). Notably, although Oganesyan and colleagues (42) suggest from the hFcRn-HSA WT co-crystal structure that His-161 is the only histidine that mediates strict pH-dependent binding to HSA, we find this unlikely as this position is not conserved across species. We have also previously shown that mutation of His-161 (H161A) reduced affinity toward HSA, but it still bound pH dependently (3).
Although the tryptophans within the α1-domain loop are fully conserved, position 52 varies among species; human has a valine, rat has an isoleucine, and mouse has a methionine in this position. Despite the fact that these amino acids are similar in size and chemical properties, replacing Val-52 of hFcRn with an isoleucine or a methionine-reduced binding to both HSA and MSA. This shows that only minor changes in the composition of the loop affect albumin binding, which further emphasizes the importance of this pH-stabilized loop in albumin binding.
In this study, all recombinant soluble hFcRn variants were secreted in equal amounts from HEK293E cells, and SDS-PAGE analysis demonstrated that the mutants migrated as distinct bands similar to that of the WT. Furthermore, measurements of their stability by differential scanning fluorimetry gave rise to melting temperatures ranging from 53.9–55.7 °C (data not shown). In addition, the amino acid substitutions that were shown to decrease binding to HSA did not disrupt binding to IgG. Thus, the results strongly indicate that the single point substitutions of the hFcRn heavy chain do not have a large impact on the global stability of the proteins. However, we cannot exclude the possibility that the mutations affect the local structural stability.
The long serum half-life of albumin makes albumin ideal as a fusion partner or carrier for drugs (40, 41). This is an attractive approach as the therapeutic efficacy of many small drugs is hampered by their very short in vivo half-life. This study provides an increased understanding of the FcRn-HSA interaction at the atomic level that may pave the way for design of albumin variants with extended serum half-life.
At last, there is an urgent need for anti-hFcRn antibodies for cellular and in vivo studies. However, there are no such well characterized and high affinity monoclonal antibodies commercially available. Thus, the panel of anti-hFcRn antibodies described in this work should be of great interest and, in particular, the antibodies that bind hFcRn pH independently and block ligand binding at acidic pH.
Supplementary Material
Acknowledgments
We are grateful to Sathiaruby Sivaganesh and Kristin Støen Gunnarsen for excellent technical assistance.
This work was supported in part by the Research Council of Norway through its Centres of Excellence funding scheme (Project 179573). I. S., J. T. A., B. D., J. C., and D. S. are co-inventors of pending patent applications related to the data described in this work.

This article contains supplemental Figs. S1 and S2.
- Ab
- antibody
- FcRn
- neonatal Fc receptor
- HC
- heavy chain
- DIII
- domain III
- HSA
- human serum albumin
- mFcRn
- mouse FcRn
- hFcRn
- human FcRn
- SPR
- surface plasmon resonance
- MSA
- mouse serum albumin.
REFERENCES
- 1. T. P. (1996) All About Albumin: Biochemistry, Genetics and Medical Applications, Academic Press, San Diego, CA [Google Scholar]
- 2. Andersen J. T., Dalhus B., Cameron J., Daba M. B., Plumridge A., Evans L., Brennan S. O., Gunnarsen K. S., Bjørås M., Sleep D., Sandlie I. (2012) Structure-based mutagenesis reveals the albumin-binding site of the neonatal Fc receptor. Nat. Commun. 3, 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andersen J. T., Dee Qian J., Sandlie I. (2006) The conserved histidine 166 residue of the human neonatal Fc receptor heavy chain is critical for the pH-dependent binding to albumin. Eur. J. Immunol. 36, 3044–3051 [DOI] [PubMed] [Google Scholar]
- 4. Chaudhury C., Brooks C. L., Carter D. C., Robinson J. M., Anderson C. L. (2006) Albumin binding to FcRn: distinct from the FcRn-IgG interaction. Biochemistry 45, 4983–4990 [DOI] [PubMed] [Google Scholar]
- 5. Chaudhury C., Mehnaz S., Robinson J. M., Hayton W. L., Pearl D. K., Roopenian D. C., Anderson C. L. (2003) The major histocompatibility complex-related Fc receptor for IgG (FcRn) binds albumin and prolongs its lifespan. J. Exp. Med. 197, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simister N. E., Mostov K. E. (1989) Cloning and expression of the neonatal rat intestinal Fc receptor, a major histocompatibility complex class I antigen homolog. Cold Spring Harb. Symp. Quant. Biol. 54, 571–580 [DOI] [PubMed] [Google Scholar]
- 7. Story C. M., Mikulska J. E., Simister N. E. (1994) A major histocompatibility complex class I-like Fc receptor cloned from human placenta: possible role in transfer of immunoglobulin G from mother to fetus. J. Exp. Med. 180, 2377–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Firan M., Bawdon R., Radu C., Ober R. J., Eaken D., Antohe F., Ghetie V., Ward E. S. (2001) The MHC class I-related receptor, FcRn, plays an essential role in the maternofetal transfer of γ-globulin in humans. Int. Immunol. 13, 993–1002 [DOI] [PubMed] [Google Scholar]
- 9. Medesan C., Radu C., Kim J. K., Ghetie V., Ward E. S. (1996) Localization of the site of the IgG molecule that regulates maternofetal transmission in mice. Eur. J. Immunol. 26, 2533–2536 [DOI] [PubMed] [Google Scholar]
- 10. Roopenian D. C., Akilesh S. (2007) FcRn: the neonatal Fc receptor comes of age. Nat. Rev. Immunol. 7, 715–725 [DOI] [PubMed] [Google Scholar]
- 11. Ward E. S., Ober R. J. (2009) Chapter 4: Multitasking by exploitation of intracellular transport functions the many faces of FcRn. Adv. Immunol. 103, 77–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baker K., Qiao S. W., Kuo T. T., Aveson V. G., Platzer B., Andersen J. T., Sandlie I., Chen Z., de Haar C., Lencer W. I., Fiebiger E., Blumberg R. S. (2011) Neonatal Fc receptor for IgG (FcRn) regulates cross-presentation of IgG immune complexes by CD8-CD11b+ dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 108, 9927–9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dickinson B. L., Badizadegan K., Wu Z., Ahouse J. C., Zhu X., Simister N. E., Blumberg R. S., Lencer W. I. (1999) Bidirectional FcRn-dependent IgG transport in a polarized human intestinal epithelial cell line. J. Clin. Invest. 104, 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qiao S. W., Kobayashi K., Johansen F. E., Sollid L. M., Andersen J. T., Milford E., Roopenian D. C., Lencer W. I., Blumberg R. S. (2008) Dependence of antibody-mediated presentation of antigen on FcRn. Proc. Natl. Acad. Sci. U.S.A. 105, 9337–9342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spiekermann G. M., Finn P. W., Ward E. S., Dumont J., Dickinson B. L., Blumberg R. S., Lencer W. I. (2002) Receptor-mediated immunoglobulin G transport across mucosal barriers in adult life: functional expression of FcRn in the mammalian lung. J. Exp. Med. 196, 303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vidarsson G., Stemerding A. M., Stapleton N. M., Spliethoff S. E., Janssen H., Rebers F. E., de Haas M., van de Winkel J. G. (2006) FcRn: an IgG receptor on phagocytes with a novel role in phagocytosis. Blood 108, 3573–3579 [DOI] [PubMed] [Google Scholar]
- 17. Ghetie V., Hubbard J. G., Kim J. K., Tsen M. F., Lee Y., Ward E. S. (1996) Abnormally short serum half-lives of IgG in β 2-microglobulin-deficient mice. Eur. J. Immunol. 26, 690–696 [DOI] [PubMed] [Google Scholar]
- 18. Ghetie V., Popov S., Borvak J., Radu C., Matesoi D., Medesan C., Ober R. J., Ward E. S. (1997) Increasing the serum persistence of an IgG fragment by random mutagenesis. Nat. Biotechnol. 15, 637–640 [DOI] [PubMed] [Google Scholar]
- 19. Kim J. K., Firan M., Radu C. G., Kim C. H., Ghetie V., Ward E. S. (1999) Mapping the site on human IgG for binding of the MHC class I-related receptor, FcRn. Eur. J. Immunol. 29, 2819–2825 [DOI] [PubMed] [Google Scholar]
- 20. Montoyo H. P., Vaccaro C., Hafner M., Ober R. J., Mueller W., Ward E. S. (2009) Conditional deletion of the MHC class I-related receptor FcRn reveals the sites of IgG homeostasis in mice. Proc. Natl. Acad. Sci. U.S.A. 106, 2788–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kobayashi K., Qiao S. W., Yoshida M., Baker K., Lencer W. I., Blumberg R. S. (2009) An FcRn-dependent role for anti-flagellin immunoglobulin G in pathogenesis of colitis in mice. Gastroenterology 137, 1746–1756.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roopenian D. C., Christianson G. J., Sproule T. J., Brown A. C., Akilesh S., Jung N., Petkova S., Avanessian L., Choi E. Y., Shaffer D. J., Eden P. A., Anderson C. L. (2003) The MHC class I-like IgG receptor controls perinatal IgG transport, IgG homeostasis, and fate of IgG-Fc-coupled drugs. J. Immunol. 170, 3528–3533 [DOI] [PubMed] [Google Scholar]
- 23. Wani M. A., Haynes L. D., Kim J., Bronson C. L., Chaudhury C., Mohanty S., Waldmann T. A., Robinson J. M., Anderson C. L. (2006) Familial hypercatabolic hypoproteinemia caused by deficiency of the neonatal Fc receptor, FcRn, due to a mutant β2-microglobulin gene. Proc. Natl. Acad. Sci. U.S.A. 103, 5084–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burmeister W. P., Huber A. H., Bjorkman P. J. (1994) Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature 372, 379–383 [DOI] [PubMed] [Google Scholar]
- 25. Mezo A. R., Sridhar V., Badger J., Sakorafas P., Nienaber V. (2010) X-ray crystal structures of monomeric and dimeric peptide inhibitors in complex with the human neonatal Fc receptor, FcRn. J. Biol. Chem. 285, 27694–27701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. West A. P., Jr., Bjorkman P. J. (2000) Crystal structure and immunoglobulin G binding properties of the human major histocompatibility complex-related Fc receptor(,). Biochemistry 39, 9698–9708 [DOI] [PubMed] [Google Scholar]
- 27. Ober R. J., Martinez C., Lai X., Zhou J., Ward E. S. (2004) Exocytosis of IgG as mediated by the receptor, FcRn: an analysis at the single-molecule level. Proc. Natl. Acad. Sci. U.S.A. 101, 11076–11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ober R. J., Martinez C., Vaccaro C., Zhou J., Ward E. S. (2004) Visualizing the site and dynamics of IgG salvage by the MHC class I-related receptor, FcRn. J. Immunol. 172, 2021–2029 [DOI] [PubMed] [Google Scholar]
- 29. Prabhat P., Gan Z., Chao J., Ram S., Vaccaro C., Gibbons S., Ober R. J., Ward E. S. (2007) Elucidation of intracellular recycling pathways leading to exocytosis of the Fc receptor, FcRn, by using multifocal plane microscopy. Proc. Natl. Acad. Sci. U.S.A. 104, 5889–5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ward E. S., Martinez C., Vaccaro C., Zhou J., Tang Q., Ober R. J. (2005) From sorting endosomes to exocytosis: association of Rab4 and Rab11 GTPases with the Fc receptor, FcRn, during recycling. Mol. Biol. Cell 16, 2028–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Andersen J. T., Daba M. B., Sandlie I. (2010) FcRn binding properties of an abnormal truncated analbuminemic albumin variant. Clin. Biochem. 43, 367–372 [DOI] [PubMed] [Google Scholar]
- 32. Schmidt M. M., Townson S. A., Andreucci A. J., King B. M., Schirmer E. B., Murillo A. J., Dombrowski C., Tisdale A. W., Lowden P. A., Masci A. L., Kovalchin J. T., Erbe D. V., Wittrup K. D., Furfine E. S., Barnes T. M. (2013) Crystal structure of an HSA/FcRn complex reveals recycling by competitive mimicry of HSA ligands at a pH-dependent hydrophobic interface. Structure 21, 1966–1978 [DOI] [PubMed] [Google Scholar]
- 33. Andersen J. T., Daba M. B., Berntzen G., Michaelsen T. E., Sandlie I. (2010) Cross-species binding analyses of mouse and human neonatal Fc receptor show dramatic differences in immunoglobulin G and albumin binding. J. Biol. Chem. 285, 4826–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andersen J. T., Justesen S., Fleckenstein B., Michaelsen T. E., Berntzen G., Kenanova V. E., Daba M. B., Lauvrak V., Buus S., Sandlie I. (2008) Ligand binding and antigenic properties of a human neonatal Fc receptor with mutation of two unpaired cysteine residues. FEBS J. 275, 4097–4110 [DOI] [PubMed] [Google Scholar]
- 35. Andersen J. T., Pehrson R., Tolmachev V., Daba M. B., Abrahmsén L., Ekblad C. (2011) Extending half-life by indirect targeting of the neonatal Fc receptor (FcRn) using a minimal albumin binding domain. J. Biol. Chem. 286, 5234–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berntzen G., Lunde E., Flobakk M., Andersen J. T., Lauvrak V., Sandlie I. (2005) Prolonged and increased expression of soluble Fc receptors, IgG and a TCR-Ig fusion protein by transiently transfected adherent 293E cells. J. Immunol. Methods 298, 93–104 [DOI] [PubMed] [Google Scholar]
- 37. Popov S., Hubbard J. G., Kim J., Ober B., Ghetie V., Ward E. S. (1996) The stoichiometry and affinity of the interaction of murine Fc fragments with the MHC class I-related receptor, FcRn. Mol. Immunol. 33, 521–530 [DOI] [PubMed] [Google Scholar]
- 38. Andersen J. T., Cameron J., Plumridge A., Evans L., Sleep D., Sandlie I. (2013) Single-chain variable fragment albumin fusions bind the neonatal Fc receptor (FcRn) in a species-dependent manner: implications for in vivo half-life evaluation of albumin fusion therapeutics. J. Biol. Chem. 288, 24277–24285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Christianson G. J., Sun V. Z., Akilesh S., Pesavento E., Proetzel G., Roopenian D. C. (2012) Monoclonal antibodies directed against human FcRn and their applications. mAbs 4, 208–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Andersen J. T., Sandlie I. (2009) The versatile MHC class I-related FcRn protects IgG and albumin from degradation: implications for development of new diagnostics and therapeutics. Drug Metab. Pharmacokinet. 24, 318–332 [DOI] [PubMed] [Google Scholar]
- 41. Sleep D., Cameron J., Evans L. R. (2013) Albumin as a versatile platform for drug half-life extension. Biochim. Biophys. Acta 1830, 5526–5534 [DOI] [PubMed] [Google Scholar]
- 42. Oganesyan V., Damschroder M. M., Cook K. E., Li Q., Gao C., Wu H., Dall'Acqua W. F. (2014) Structural insights into neonatal Fc receptor-based recycling mechanisms. J. Biol. Chem. 289, 7812–7824 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.