Summary
Open sharing of clinical trial data has been proposed as a way to address the gap between the production of clinical evidence and the decision-making of physicians. Since a similar gap has already been addressed in the software industry by the open source software movement, we examine how the social and technical principles of the movement can be used to guide the growth of an open source clinical trial community.
Introduction
Despite the rapid increase in the volume of evidence being published (1), physicians make decisions without access to the evidence they need (2, 3), and with good reasons to be sceptical about the evidence they find (4). While there have been improvements made through mandatory registration, and ideas proposed for improving evidence translation throughout all phases of drug development and biomedical innovation (4–9), the gap between evidence and practice remains a grand challenge in healthcare (10). However, there is a group of people who have already successfully addressed a similar gap in another industry of comparable size. Hackers fundamentally altered the software industry by devising the principles that drive the open source software movement, countering the economic and cultural motivations that drove the production of closed source software, disengagement with user needs, and poor interoperability. Since the problems faced in the clinical evidence domain are essentially the same, can we learn from the open source software movement when guiding the growth of an emerging open source clinical trial community?
All information should be free
The open source software movement grew out of the intellectual curiosity of hackers and a fundamental belief that all information should be free (11). In this case, free meant more than just zero cost, it meant libre rather than just gratis. The ‘four freedoms’ of open source software included the freedom to run the program for any purpose, to study how the program works and change it, to redistribute copies, and to distribute modified copies (12).
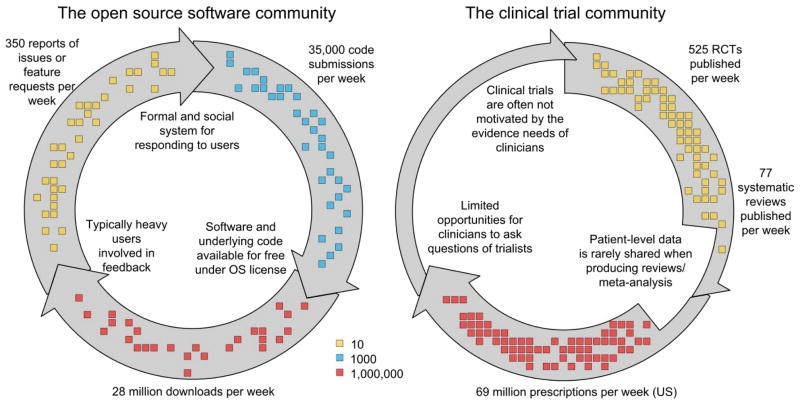
The reasons for the success of the open source software movement are the developers’ active engagement with users in software creation and testing phases, and the rapid filling of gaps in functionality by a decentralised community of developers (Fig. 1). The process is supported by online communities centred around repositories of code. GitHub, the largest code host in the world, started in 2008 and has 1.2 million users and hosts 3.5 million repositories. Source Forge is the second largest and most established, with over 326,000 diverse software projects, supported by a globally-distributed community of 2.7 million developers. Examples of well-known open source software include the widely used Firefox web browser; Apache (the most common web server); and the Android and Symbian operating systems, used on the majority of smart phones globally.
Figure 1.
The significant bottlenecks present in the translation of clinical trial evidence into practice have been resolved in the software industry via open source communities like the one facilitated by SourceForge.
Open source communities often out-perform their closed source counterparts when addressing the needs of users (13), arguably as a direct consequence of the dialogue with users and the interoperability that comes from transparency and developer interaction (14). Users of open source software are encouraged to become directly involved by reporting issues and gaps in functionality they wish to see addressed, even if they are unable to contribute directly to the development. In open source software this is described as “given enough eyes, all bugs are shallow” (14).
Individuals and organisations of all sizes are involved in open source software communities – their motivations range from career development, reputation and other signalling incentives (15, 16). Large companies see value in engaging with open source software development as a way to improve the “social contagion” of their products by engaging directly with heavy users (17), directly profiting from offering complementary services (16), and as a tool for recruiting better developers.
Open access to clinical data
The process of translating clinical evidence into practice is slowed by limited sharing of patient-level clinical data, bottlenecks in the dissemination of evidence and the inefficiencies associated with answering the wrong questions in the first place. The problems are attributed to biases affecting which trials are conducted and published (18), the slow dissemination of trustworthy evidence obscured by marketing delivered through the same channels, and the difficulty of constructing timely and reliable guidelines (19).
Despite these systemic problems, individual successes have come from expanding access to de-identified patient level data. The Framingham Heart Study resulted in 1872 publications between 1948 and 2007, and when access to genotypic and phenotypic data was opened to a much wider group of researchers this number increased by 19% to 2223 in just 4 years (20). In a separate example, the public release of genotypic information associated with an outbreak of Escherichia coli in May 2011 (40 deaths and 3000 cases of infection) led to genomic analyses on four continents (21). Within a week of public release, the genome had been assembled, assigned to an existing sequence type, and two dozen reports had been filed, providing information on virulence, resistance and phylogenetic lineage. In another example, a patient-level analysis of cholesterol treatment was produced collaboratively by researchers involved in 26 clinical trials, including 170,000 patients (22). The protocol was published in 1995 before any of the individual trials were completed, making this an early and successful example of broad collaboration and sharing.
There are strong similarities between the processes for collaborative engagement with software source code and patient-level data. There are differences, too, such as the expertise and infrastructure required when producing source code and clinical trial data, and the sources of funding that underpin the two systems.
Principles for an open source clinical trial community
The analogy between open source software development and the synthesis of clinical evidence suggests the following principles for an open source clinical trial community:
Clinical trialists, other researchers and clinicians are able to access and contribute to a repository of interoperable patient-level data that sits alongside the mandatory registration and provision of summary-level clinical trial results.
Infrastructure for repositories, data standards, and interaction amongst clinical trialists are provided to foster the growth of a decentralised community whose main aim is to rapidly identify and address gaps in clinical evidence.
Improved dialogue between trialists and physicians allows physicians to routinely ask new questions of the clinical trial community, and follow and discuss the clinical trial data aggregated to answer existing questions.
An open source clinical trial community requires data standards that allow trials to be recorded in a uniform way while retaining the flexibility to represent the variety of protocols and interventions that are currently seen in current clinical trial registries (23). The development of data standards can be slow and laborious but it is an area where much work has been done (24). Sim and colleagues (25) have led the design of data standards to capture machine-interpretable knowledge bases, as well as building a framework for storing summary information with enough detail for searching and aggregating across trials. Other examples of standards and developing communities that already exist to support the exchange of clinical trial data include the Study Data Tabulation Model (26), and Open mHealth (27). By leveraging the most flexible of the data standards, it should be possible to create, store and aggregate data sets without losing the rigour of the protocols established within each of the clinical trials.
We envision a set of tools similar to those provided by GitHub or SourceForge, which will allow clinical trial researchers to submit de-identified patient-level outcome data alongside the usual metadata expected when registering clinical trials (23). Each submitted module represents patient-level outcomes separated by study arms, alongside information about the inclusion criteria and the interventions applied. Modules with equivalent interventions may be combined and compared against alternatives to produce the equivalent of patient-level meta-analyses. Differences between groups of patients receiving the same intervention will be used to capture local effects not otherwise measurable outside of observational studies. The result would be an increase in the utility of the clinical trials through improved connectivity between, and access to, the data required for patient-level meta-analysis of controlled trials.
Patient-level meta-analyses are rarely performed (approximately 50 a year) despite being considered of high quality and holding a long list of advantages over meta-analyses based on summary information (28). The disadvantage specific to patient-level meta-analysis is the time consuming nature of coordination and data management – issues that would be largely resolved by standardising the data storage, standards, and avenues for collaboration and communication. As a consequence of sharing, it will also be possible to genuinely de-couple the collection of data and the analysis for study types ranging from controlled studies of single interventions up to more complex studies such as network meta-analyses. Under most open source licenses, attribution of published syntheses would automatically flow back to the individuals and groups contributing data, and it would be possible to extend open source licenses to include co-authorship privileges.
As is the case for software users in the open source software movement, the open source clinical trial community would facilitate the direct participation of physicians, when looking for decision support. As an open access repository, physicians would have access to the conclusions drawn from large patient-level meta-analyses, and find trust in the transparency of the analyses and underlying data. Physicians could ask new questions in the same way that software features are requested in open source communities – via an online submission system. As a consequence, a direct dialogue is created between those producing the evidence and the physicians using the evidence in their decision-making.
By providing useful ways to share patient-level data and directly engaging with the users of clinical evidence, the evidence developers will be able to better provide the evidence that physicians need, effectively re-use outcome data through patient-level meta-analysis, reduce bottle-necks associated with data access (29), and therefore reduce the burden of waste currently associated with the neglect of data in research (30).
Challenges ahead
Some of the technical challenges around building an open source community for clinical trials include applying methods to avoid or eliminate the potential for identifying individual patients (31), and implementing new ways to address quality standards, which can be low for decentralised contribution (5, 18). Less onerous challenges include providing the software infrastructure and tools that foster discussion, growth and dialogue with physicians, and providing tools that aid the self-governance of quality.
Privacy is clearly a concern when providing open access to patient-level data, when de-identification cannot be ensured (31). One solution is to require signed agreements as is currently done for access to existing longitudinal datasets. An alternative would be to generate statistically identical samples on request, allowing all necessary inferences to be made without compromising privacy.
Perhaps the most important challenge faced by the clinical trial community comes from the currently unbalanced value system in which publish-or-perish mentalities and marketing concerns often outweigh the value of making effective contributions to the support of clinical decision-making. Pharmaceutical companies may have an aversion to providing open access to patient-level data because it may reduce their ability to control the conclusions that are drawn, or to avoid dissemination of unfavourable results. In open source software the incentives for participation are understood, even by large companies (15, 16), and these values may also be discovered by pharmaceutical companies. Addressing this challenge in the clinical domain will depend on choosing or creating licenses that usefully capture contributions, and to confer recognition to those contributions in publications and career progression.
A recent international shift towards requiring open access to all publications resulting from publicly-funded clinical trials signals a move towards ‘not paying for research twice’. A further push, at least for trials with non-industry funding sources, may come from extending the mandate of open access publication to include open access to patient-level data for publicly-funded clinical trials.
Despite the technical and social challenges, significant movement towards crowdsourcing and open access to data has already been seen in early-phase drug development (7, 32, 33), apparently in response to the slowing of approvals that signalled a grand challenge for the field (34). The domain of clinical evidence faces its own grand challenge, and requires a similar push to close the gap between what physicians need and the ways in which trials are funded, undertaken and reported. By recognising the open source software community as a role model for improvement, it is possible to guide the principles, standards and tools that will catalyse the growth of a more efficient and socially responsible clinical trial community.
Acknowledgments
The authors acknowledge the valuable comments of four anonymous reviewers, and the funding support of the National Health and Medical Research Council (Program Grant 568612).
References
- 1.Bastian H, Glasziou P, Chalmers I. Seventy-Five Trials and Eleven Systematic Reviews a Day: How Will We Ever Keep Up? PLoS Medicine. 2010;7:e1000326. doi: 10.1371/journal.pmed.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotchen TA. Why the Slow Diffusion of Treatment Guidelines Into Clinical Practice? Archives of Internal Medicine. 2007;167:2394. doi: 10.1001/archinte.167.22.2394. [DOI] [PubMed] [Google Scholar]
- 3.Davidoff F, Miglus J. Delivering Clinical Evidence Where It’s Needed. JAMA: The Journal of the American Medical Association. 2011;305:1906. doi: 10.1001/jama.2011.619. [DOI] [PubMed] [Google Scholar]
- 4.Rasmussen N, Lee K, Bero L. Association of trial registration with the results and conclusions of published trials of new oncology drugs. Trials. 2009;10:116. doi: 10.1186/1745-6215-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickersin K, Rennie D. Registering Clinical Trials. JAMA: The Journal of the American Medical Association. 2003;290:516. doi: 10.1001/jama.290.4.516. [DOI] [PubMed] [Google Scholar]
- 6.Lutchen K, Ayers J, Gallagher S, Abu-Taleb L. Engineering Efficient Technology Transfer. Science Translational Medicine. 2011;3:110cm32. doi: 10.1126/scitranslmed.3003004. [DOI] [PubMed] [Google Scholar]
- 7.Munos BH, Chin WW. How to Revive Breakthrough Innovation in the Pharmaceutical Industry. Science Translational Medicine. 2011;3:89cm16. doi: 10.1126/scitranslmed.3002273. [DOI] [PubMed] [Google Scholar]
- 8.Norman TC, Bountra C, Edwards AM, Yamamoto KR, Friend SH. Leveraging Crowdsourcing to Facilitate the Discovery of New Medicines. Science Translational Medicine. 2011;3:88mr1. doi: 10.1126/scitranslmed.3002678. [DOI] [PubMed] [Google Scholar]
- 9.Terry SF, Terry PF. Power to the People: Participant Ownership of Clinical Trial Data. Science Translational Medicine. 2011;3:69cm3. doi: 10.1126/scitranslmed.3001857. [DOI] [PubMed] [Google Scholar]
- 10.Chalmers I, Glasziou P. Avoidable waste in the production and reporting of research evidence. The Lancet. 2009;374:86. doi: 10.1016/S0140-6736(09)60329-9. [DOI] [PubMed] [Google Scholar]
- 11.Levy S. Hackers: Heroes of the Computer Revolution. Anchor Press/Doubleday; Garden City, New York: 1984. p. 458. [Google Scholar]
- 12.The Free Software Foundation. [Accessed July 14, 2011];The Free Software Definition. 1996 http://www.gnu.org/philosophy/free.
- 13.Paulson JW, Succi G, Eberlein A. An Empirical Study of Open-Source and Closed-Source Software Products. IEEE Trans Softw Eng. 2004;30:246. [Google Scholar]
- 14.Raymond E. The cathedral and the bazaar. Knowledge, Technology & Policy. 1999;12:23. [Google Scholar]
- 15.Lerner J, Tirole J. Some Simple Economics of Open Source. The Journal of Industrial Economics. 2002;50:197. [Google Scholar]
- 16.Bonaccorsi A, Rossi C. Comparing motivations of individual programmers and firms to take part in the open source movement: From community to business. Knowledge, Technology & Policy. 2006;18:40. [Google Scholar]
- 17.Iyengar R, Van den Bulte C, Valente TW. Opinion Leadership and Social Contagion in New Product Diffusion. Marketing Science. 2011;30:195. [Google Scholar]
- 18.Bourgeois FT, Murthy S, Mandl KD. Outcome Reporting Among Drug Trials Registered in ClinicalTrials.gov. Annals of Internal Medicine. 2010;153:158. doi: 10.1059/0003-4819-153-3-201008030-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabana MD, et al. Why Don’t Physicians Follow Clinical Practice Guidelines? A Framework for Improvement. JAMA: The Journal of the American Medical Association. 1999;282:1458. doi: 10.1001/jama.282.15.1458. [DOI] [PubMed] [Google Scholar]
- 20.Framingham Heart Study. [Accessed July 14, 2011];Framingham Heart Study Bibliography. 2011 http://www.framinghamheartstudy.org/biblio/index.html.
- 21.Rohde H, et al. Open-Source Genomic Analysis of Shiga-Toxin–Producing E. coli O104:H4. New England Journal of Medicine. 2011;365:718. doi: 10.1056/NEJMoa1107643. [DOI] [PubMed] [Google Scholar]
- 22.Cholesterol Treatment Trialists’ (CTT) Collaboration. Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. The Lancet. 2010;376:1670. doi: 10.1016/S0140-6736(10)61350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zarin DA, Tse T, Williams RJ, Califf RM, Ide NC. The ClinicalTrials.gov Results Database - Update and Key Issues. New England Journal of Medicine. 2011;364:852. doi: 10.1056/NEJMsa1012065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coiera E. Building a National Health IT System from the Middle Out. Journal of the American Medical Informatics Association. 2009 May 1;16:271. doi: 10.1197/jamia.M3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sim I, Detmer DE. Beyond Trial Registration: A Global Trial Bank for Clinical Trial Reporting. PLoS Med. 2005;2:e365. doi: 10.1371/journal.pmed.0020365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood F, Guinter T. Evolution and Implementation of the CDISC Study Data Tabulation Model (SDTM) Pharmaceutical Programming. 2008;1:20. [Google Scholar]
- 27.Estrin D, Sim I. Open mHealth Architecture: An Engine for Health Care Innovation. Science. 2010;330:759. doi: 10.1126/science.1196187. [DOI] [PubMed] [Google Scholar]
- 28.Riley RD, Lambert PC, Abo-Zaid G. Meta-analysis of individual participant data: rationale, conduct, and reporting. BMJ. 2010;340 doi: 10.1136/bmj.c221. [DOI] [PubMed] [Google Scholar]
- 29.Gøtzsche PC, Jørgensen AW. Opening up data at the European Medicines Agency. BMJ. 2011;342 doi: 10.1136/bmj.d2686. [DOI] [PubMed] [Google Scholar]
- 30.Hanson B, Sugden A, Alberts B. Making Data Maximally Available. Science. 2011;331:649. doi: 10.1126/science.1203354. [DOI] [PubMed] [Google Scholar]
- 31.Loukides G, Denny JC, Malin B. The disclosure of diagnosis codes can breach research participants’ privacy. Journal of the American Medical Informatics Association. 2010;17:322. doi: 10.1136/jamia.2009.002725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boguski MS, Mandl KD, Sukhatme VP. Repurposing with a Difference. Science. 2009;324:1394. doi: 10.1126/science.1169920. [DOI] [PubMed] [Google Scholar]
- 33.Munos BH, Chin WW. A Call for Sharing: Adapting Pharmaceutical Research to New Realities. Science Translational Medicine. 2009;1:9cm8. doi: 10.1126/scitranslmed.3000155. [DOI] [PubMed] [Google Scholar]
- 34.Paul SM, et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nature Reviews Drug Discovery. 2010;9:203. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]