Abstract
Myc is an oncogenic transcription factor frequently dysregulated in human cancer. To identify pathways supporting the Myc oncogenic program, we employed a genome-wide RNAi screen for Myc-synthetic-lethal genes and uncovered a role for the SUMO-activating-enzyme (SAE1/2). Loss of SAE1/2 enzymatic activity drives synthetic lethality with Myc. Inactivation of SAE2 leads to mitotic catastrophe and cell death selectively upon Myc hyper-activation. Mechanistically, SAE2 inhibition switches a transcriptional subprogram of Myc from activated to repressed. A subset of these SUMOylation-dependent-Myc-switchers (SMS genes) is required for mitotic spindle function and to support the Myc oncogenic program. SAE2 is required for Myc-dependent tumor growth, and patient survival significantly correlates with SAE1/SAE2 levels in Myc-high tumors. These studies reveal a mitotic vulnerability of Myc-driven cancers, demonstrate that inhibiting sumoylation impairs Myc-dependent tumorigenesis, and suggest inhibiting SUMOylation may have therapeutic benefits for patients with Myc-driven cancer.
Cancers are driven by genomic alterations resulting in the activation of proto-oncogenes and inactivation of tumor suppressor genes. Removing oncogene function can often reverse the tumorigenic phenotype, a phenomena referred to as “oncogene addiction” (1, 2), and the cancer community has largely focused on exploiting oncogene-addiction by discovering and targeting cancer-causing oncogenes (3). However, many oncogenes (such as Ras and Myc) have proven difficult to inhibit pharmacologically, highlighting the need for complementary approaches. Recently, the concept of non-oncogene addiction (NOA) has postulated that oncogenic mutations confer dependencies on cellular processes that can be exploited therapeutically, and recent anti-cancer therapies have been shown to exploit NOA in humans (4–6). However, the genes and pathways underlying such oncogenic-support processes are largely unexplored, and since these genes are not themselves oncogenes or otherwise mutated in cancer, they cannot be discovered directly through physical analysis of cancer genomes/epigenomes. For instance, the underlying NOA pathways supporting the classical c-Myc oncogene (referred to herein as Myc) are poorly understood. The bHLHZip transcription factor Myc is frequently dysregulated by amplification, mutation, overexpression, or protein stabilization (7). Amplification/overexpression of Myc occurs in ~25% of breast cancer patients (8–11), and these cancers are particularly aggressive, conferring poor patient prognosis (12). Genetic experiments have shown that Myc is required for tumor maintenance and progression in several types of malignancy (13, 14). However, despite three decades of intensive studies, there remains no effective method to inhibit Myc in patients.
Oncogenic activation of Myc promotes a delicate balance in cells, conferring both pro- and anti-tumorigenic properties (2, 15, 16). This raises the interesting possibility that the balance between these opposing properties could be influenced by altering cellular pathways to reduce Myc oncogenic support pathways. To search for such pathways required for cells to tolerate the Myc oncogene, we performed a genome-wide genetic screen for Myc-synthetic lethal (MySL) shRNAs in human mammary epithelial cells (HMECs) engineered with an inducible c-Myc-estrogen receptor fusion transgene (Myc-ER HMECs) (fig S1A). Induction resulted in increased expression of known Myc targets (fig S1B) and a modest decrease in HMEC proliferation rate (fig S1C). HMECs are ER-negative, and in the absence of Myc-ER, do not respond to tamoxifen (fig S2). Using this system, we screened for shRNAs that alter cell fitness only in the presence of aberrant Myc signaling (screen design in Fig. 1A).
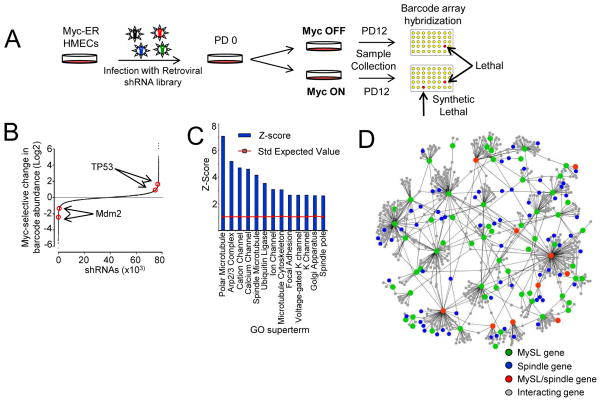
Figure 1. Genome-wide screen for Myc-Synthetic Lethal (MySL) candidates.
a) Identification of Myc-Synthetic Lethal (MySL) genes. Myc-ER HMECs were transduced with a genome-wide library of retroviral shRNAs in triplicate. At population doubling 0(PD0), cells were cultured +/− Myc -ER induction for 12 population doublings(PD12). To identify MySL candidates, relative barcode abundance from both conditions was compared to initial PD0 samples via barcode microarrays.
b) Identification of MySL shRNAs. The Myc-selective effect of all shRNAs from the genome-wide library are graphed (y-axis represents median difference between Myc-ER-on and Myc-ER-off groups (log2)), with a ratio < −1.0 indicating a decrease of at least twofold. shRNAs are shown on the x -axis (rank ordered by MySL effect).
c) Multiple cellular processes are required to tolerate the Myc oncogenic state. MySL candidates were analyzed for gene ontology (GO; Z-scores for enriched cellular components and processes).
d) MySL proteins engage in a highly connected interaction network that regulates the mitotic spindle. Protein-protein interactions between the top 100 MySL proteins were analyzed via HPRD. Green indicates a MySL protein, blue indicates a protein with a known role in mitotic spindle function, red indicates a MySL protein with a known role in spindle function, and gray indicates a protein that interacts with a MySL protein.
To identify MySL shRNAs, we transduced Myc-ER HMECs with a retroviral library of 74,905 shRNAs targeting 32,293 unique transcripts in 3 independent replicates. Transduced Myc-ER HMECs were propagated −/+ Myc-ER induction, and the relative change in shRNA-barcode abundance was measured in both cell states (Myc-OFF and Myc-ON). We identified 403 MySL shRNAs exhibiting >2-fold decrease in abundance in the Myc-ON state (relative to the Myc-OFF state) (p<0.02; Fig. 1B, fig. S3, table S1). Content analysis (Gene Ontology (GO)) indicated that these candidates were highly enriched for ion channels and enzymes functioning in ubiquitin-like protein conjugation (including SUMOylation) (p=0.002), thus implicating many new enzymatic targets for drug development against Myc-driven cancers (Fig. 1C). Notably, we observed that components of the mitotic spindle were highly enriched among MySL candidates (Fig. 1C). Furthermore, analysis of the MySL candidates using Human Protein Reference Database (HPRD) revealed a highly connected protein-protein interaction network, with many components of this network playing a role in the mitotic spindle (Fig. 1D; protein labels shown in fig. S4), suggesting that Myc hyper-activation may impose a stress on proper mitotic progression(17).
Among MySL candidates were several genes previously implicated in the survival of Myc hyper-activated cells (GSK3β, FBXW7, and PTK2; fig. S5A) (18–20). In addition, shRNAs targeting MDM2 exhibited Myc-synthetic lethality, while p53-targeting shRNAs enhanced proliferation in a Myc-selective manner (Fig. 1B and fig. S5B). Myc has been shown to promote p53 activation (2, 21–23) and these dependencies are consistent with the role of p53 defects in promoting Myc-induced tumorigenesis (2, 21, 22).
We also identified many candidates with previously unknown roles in Myc biology. To prioritize these, we rank ordered MySL genes using a modified 2-way ANOVA that we developed to summarize the effects of all shRNAs for a given gene. Using this method, we identified 8 genes exhibiting Myc-synthetic lethality with a p≤0.001 (table S2). The most significant candidate from both of these analyses was the SUMO-activating enzyme (SAE) subunit 2 (SAE2/UBA2) (p<0.00001), a critical component of the sole SUMO-activating enzyme necessary for SUMO-conjugation to proteins (24). Multiple SAE2-shRNAs exhibited Myc synthetic lethality in the primary screen (fig. S5C). The primary screen also identified SAE1, the heterodimeric partner of SAE2 (table S2). Thus, we focused on SAE and the potential synthetic lethal interaction between SUMOylation and Myc.
To explore the physiological significance of the Myc-SAE2 synthetic lethal interaction, Myc-ER HMECs were transduced with two independent shRNAs targeting SAE2 or a control shRNA and the effect of Myc activation on HMEC proliferation was measured. SAE2-targeting shRNAs depleted SAE2 protein (fig. S6A), and led to profound increase in doubling time upon Myc-induction (Fig. 2A, left graph; representative images in Fig. 2A right panels). Similar results were observed when a constitutive Myc transgene was expressed together with shSAE2, suggesting that these observations are not an artifact of the Myc-ER fusion system (fig. S7). Notably, two independent SAE2 shRNAs elicited a Myc-synthetic lethal phenotype (Fig. 2A), and restoration of SAE2 protein levels with a SAE2 wild-type cDNA suppressed the MySL phenotype of SAE2 shRNA (Fig. 2C–2D, described below), indicating that the Myc-SAE2 synthetic phenotype is not due to an RNAi off-target effect. Furthermore, multiple shRNAs targeting SAE1 and the downstream SUMO E2 conjugating enzyme UBE2I (UBC9) (fig. S6 B, C) were also synthetically lethal with Myc hyperactivation (Fig. 2A, middle and right graphs), demonstrating SUMOylation interference is synthetically lethal with hyper-activated Myc.
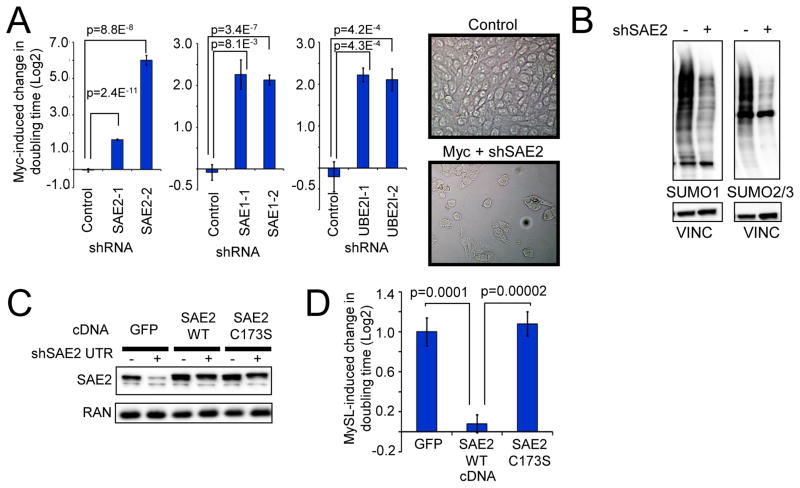
Figure 2. The E1 SUMO-activating enzyme is required to tolerate oncogenic MYC in HMECs.
a) Inactivation of SAE2, SAE1, or UBE2I is synthetically lethal with Myc hyper-activation. Myc-ER HMECs were infected with control, SAE2, SAE1, or UBE2I-targeting shRNAs and cultured −/+ Myc-ER induction. Cell number was quantified after 6 d. Data are represented as relative change in doubling time in Myc-On vs Myc-Off states and normalized to control shRNA (Average of at least 8 replicates for each shRNA). Representative images demonstrating altered morphology and decreased cell number in Myc/shSAE2 cells.
b) Total protein SUMOylation is decreased upon SAE2 knockdown. Myc-ER HMECs infected with control or SAE2-shRNA encoding virus were analyzed for SUMO1, SUMO2/3, and Vinculin (loading control) protein levels.
c) and d) SAE2 catalytic activity is required to tolerate Myc hyper-activation. Myc-ER HMECs transduced with a doxycycline (dox)-inducible shRNA targeting the SAE2 UTR (pInducer11-mir-SAE2 UTR-eGFP) were subsequently infected with virus expressing GFP, SAE2 WT or SAE2 C173S cDNAs. Western blots were performed to confirm depletion of SAE2 (c). Cells were cultured in −/+ Dox and −/+ Myc-ER-induction, and cell number was quantified after 8 d. The Y-axis indicates the relative change in growth of shSAE2 expressing cells due to Myc induction in the presence of the indicated transgenes (average of 8 replicates) (d).
Our data suggests SUMOylation is required to tolerate aberrant Myc activation. Depletion of SAE2 decreased levels of SUMO1- or SUMO2/3-modified proteins (Fig. 2B), indicating global impairment of SUMOylation in these cells. To determine whether SAE2 enzymatic activity is required to support Myc, Myc-ER HMECs were engineered with an inducible SAE2-shRNA (pINDUCER11-shSAE2) (25) together with constitutive shRNA-resistant cDNAs encoding WT SAE2, catalytically-inactive SAE2-C173S, or control eGFP. SAE2 WT and mutant cDNAs restored SAE2 to endogenous SAE2 levels (Fig. 2C). Restoration of WT SAE2 suppressed the MySL phenotype of SAE2 shRNA (Fig. 2D). However, SAE2-C173S failed to suppress the synthetic lethality of SAE2-shRNA (Fig. 2D), indicating that SAE2 enzymatic activity is required to prevent the Myc-SAE2 synthetic lethal interaction. Together, these data suggest SUMOylation is required for HMECs to tolerate aberrant Myc signaling.
A key question is how SAE2 depletion in the presence of Myc hyper-activation impairs proliferation. This could be due to changes in the cell cycle and/or cell death. Therefore, we examined the effects of Myc hyper-activation and SAE2-depletion on these processes. In SAE2-depleted cells, Myc-induction increased the number of cells with a G2/M DNA content (fig. S8A) accompanied by a concomitant accumulation of aberrant (>2N) DNA content (Fig. 3A, fig. S8A). These cell cycle defects were followed by a significant apoptotic response (Fig. 3B, fig. S8B–C). The increase in G2/M and >2N DNA content is characteristic of mitotic defects known to cause mitotic catastrophe and apoptosis. A potential mitotic defect is supported by our observation that MySL genes are significantly enriched for genes involved in the mitotic spindle (Fig. 1D), suggesting that Myc hyper-active cells might experience mitotic stress. To explore this, we examined mitotic spindles upon Myc hyper-activation in the presence or absence of SAE2 depletion. As hypothesized, Myc-active/SAE2-inactive HMECs exhibited significantly more spindle defects (defects in >25% of all mitoses) than Myc-alone or shSAE2-alone (Fig. 3C–D p=7E-7), including abnormal spindle number and lagging chromosomes. Consistent with previous literature, these spindle defects could explain the extensive aneuploidy and apoptosis observed. Collectively, these data suggest that the Myc-SAE2 genetic interaction results in dysregulation of the mitotic spindle that may contribute to synthetic lethality.
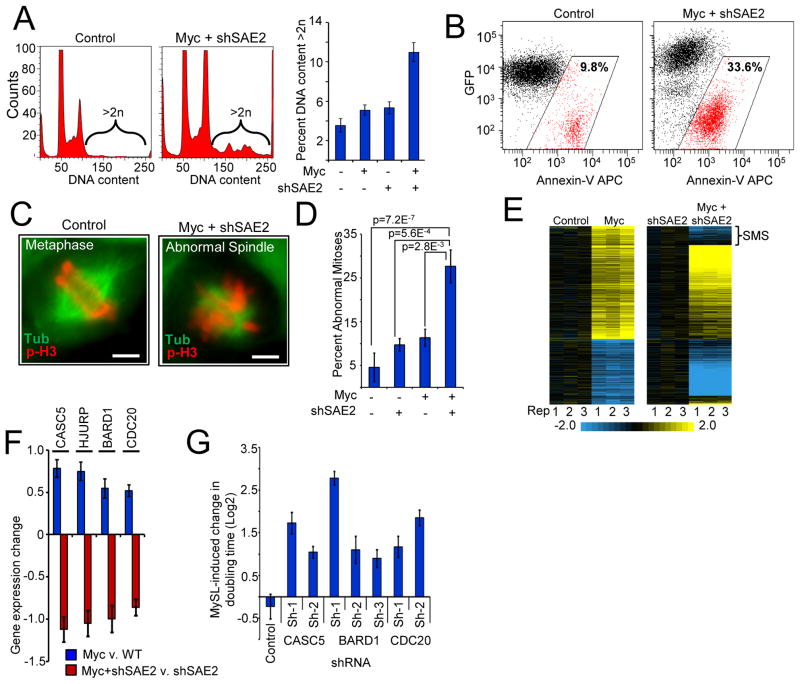
Figure 3. Inactivation of SAE2 switches the Myc transcriptional program and dysregulates mitotic fidelity and cell viability.
a) Ectopic Myc activation and SAE2 inactivation leads to increase in G2/M cells and aberrant chromosomal content. Myc-ER HMECs transduced with inducible shSAE2 were cultured −/+ Myc-ER-induction (24 h) and −/+ shSAE2 induction. Cells were analyzed for DNA content by flow cytometry (quantification of cells with >2N DNA, right panel).
b) Depletion of SAE2 induces apoptosis in cooperation with Myc hyper-activation. pINDUCER-mir-SAE2-eGFP Myc-ER HMECs were cultured −/+ Myc-ER-induction and −/+ shSAE2 induction (48 h). The cells were analyzed for apoptosis (Annexin-V) by flow cytometry.
c) and d) Myc-SAE2 genetic interaction leads to defects in the mitotic spindle. Myc-ER HMECs transduced with inducible shRNA-SAE2 were cultured −/+ Myc-ER-induction (16 h) and −/+ shSAE2 induction. Cells were stained for Tubulin (green) and phospho-H3 (red) to visualize mitotic defects. Images from c) were quantified for both total and abnormal mitotic events d). Data are represented as percent abnormal mitoses (at least 100 mitotic events counted per condition; p -values from Fisher’s exact test). Scale bar=5uM
e) Loss of SAE2 alters the transcriptional response to Myc. HMECs expressing Myc-ER and dox-inducible SAE2-shRNA were analyzed by gene expression profiling −/+ Myc-ER-induction and −/+ SAE2-shRNA induction. All mRNAs altered by Myc-ER-induction (p < 0.05, 2-fold) are shown. The effect of Myc-ER-induction on mRNA levels in the absence or presence of shRNA-SAE2-induction are shown (left and right panels, respectively). mRNAs that change their response to Myc in the presence or absence of shSAE2 are termed “Sumoylation-dependent Myc switchers,” or SMS genes.
f) Loss of SAE2 alters Myc control of spindle-regulatory genes. The effect of Myc in the absence or presence of shSAE2 (blue and red bars, respectively) is shown for top 4 of 17 SMS genes with known roles in spindle integrity and function (see Fig. S9B for list of 17).
g) SMS genes are required to tolerate Myc hyper-activation. Myc-ER HMECs transduced with shRNAs targeting the indicated SMS genes were cultured −/+ Myc-ER-induction for 6 d. Cell numbers were counted and analyzed as in Fig 2A.
We next sought to understand how Myc hyper-activation and SAE2 depletion results in the observed mitotic aberrations. Myc hyper-activation induces different cellular consequences (e.g. proliferation, apoptosis, senescence) depending on the genetic and epigenetic context. If this is due to the ability of Myc to regulate distinct transcriptional programs, loss of SAE2 may lead to mitotic dysfunction by altering Myc’s transcriptional program. Therefore, we employed gene expression profiling to define the transcriptional effects of Myc with or without SAE2 inactivation. Myc activation alone in HMECs led to significant changes in the level of 605 mRNAs (p<0.05; Fig. 3E, left panel). Surprisingly, 22.5% (86/383) of Myc-induced transcripts are not induced or become repressed in response to Myc when SAE2 is depleted (Fig. 3E, right panel), suggesting that a portion of the Myc transcriptional response is “switched” depending upon the status of SAE2 function. Because the expression of these genes switches from Myc-induced to Myc-repressed in a SAE2-dependent manner, we termed these genes SUMOylation-dependent Myc-switchers (or SMS genes).
Remarkably, MESH analysis revealed that SMS genes were significantly enriched for regulators of the mitotic spindle (p<4.9E-12) (fig. S9A), and mining of published literature revealed that 17 of 86 SMS genes have been shown genetically to participate in the assembly or integrity of mitotic spindles (26–31). Each of these spindle-related genes is induced by Myc hyper-activation (Fig. 3F, fig. S9B, blue bars) but exhibits a strong SAE2-dependent switch in their Myc-response to repression (Fig. 3F and fig. S9B, red bars). These observations highlight regulation of spindle assembly as a key vulnerability in cells harboring the Myc-active/SAE2-inactive state, and suggest that SMS genes may be key linchpins in the Myc-SAE2 synthetic lethal relationship. To test this hypothesis, we examined whether SMS genes known to play a role in the mitotic spindle are synthetically lethal with Myc. Remarkably, 3 of the top 4 SMS genes (CASC5, BARD1, CDC20) were synthetically lethal with Myc-hyper-activation, with depletion of SMS candidates leading to up to an 8-fold Myc-selective increase in cell doubling time (Fig. 3G, fig. S10A–C). These results suggest the SMS transcriptional subprogram may be required to tolerate the Myc oncogenic state.
The Myc-SAE2 synthetic lethal interaction suggests that Myc-driven cancers may be dependent on SAE2 and SUMOylation to support their tumorigenic phenotypes. To test this hypothesis in the context of human breast cancer, we first assessed the dependency of breast cancer-derived cell lines on Myc function. Human breast cancer cells were transduced with control or Myc-targeting shRNA viruses and tested for clonogenicity. Clonogenicity of SUM159 and MDA-MB-231 breast cancer cells was significantly impaired by Myc depletion, while MCF7 and SKBR3 breast cancer cells were unaffected (Fig. 4A). Therefore, we classified the SUM159 and MDA-MB-231 cells as Myc-dependent and the MCF7 and SKBR3 cells as Myc-independent.
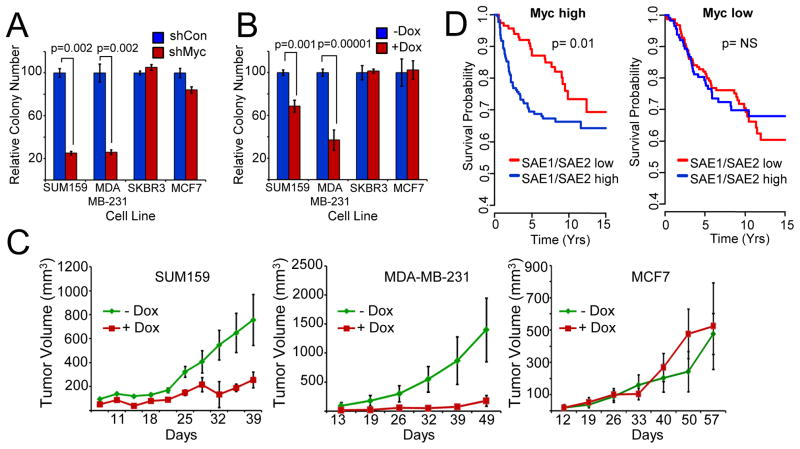
Figure 4. The E1 SUMO-activating enzyme is required to support MYC-dependent human breast cancers.
a) Myc-dependency in breast cancer cells. Breast cancer-derived cell lines infected with control-or Myc-shRNA lentivirus were analyzed for clonogenic growth. Macroscopic colonies were quantified and normalized to control-shRNA infected cells for each cell line.
b) Inactivation of SAE2 inhibits clonogenicity in Myc-dependent breast cancer cells. Breast cancer-derived cell lines infected with dox-inducible control-or SAE2 -shRNA lentivirus were analyzed for clonogenic growth +/− dox.
c) Inactivation of SAE2 inhibits tumorigenicity of Myc-dependent tumors. Myc-dependent (SUM159 and MDA-MB-231, left and middle panels, respectively) or Myc-independent (MCF7, right panel) breast cancer cells infected with dox-inducible SAE2-targeting shRNA lentivirus were transplanted into nude mice. Recipient animals were treated +/− dox and xenograft volume was measured over time.
d) Low SUMO-Activating-Enzyme expression correlates with patient metastasis-free survival selectively in Myc-high breast cancers. The expression of SAE1/SAE2 is inversely correlated with increased metastasis-free survival in patients with Myc-high tumors (p=0.001, log-rank test). Tumors with the highest and lowest tertile of Myc mRNA expression were considered “Myc high” and “Myc low,” respectively. Patients with the highest-and lowest -tertile of SAE1/SAE2 mRNA expression are shown as blue and red lines, respectively.
To determine whether Myc-dependent breast cancer cells are similarly dependent on SAE2 function, we transduced each breast cancer cell line with an inducible SAE2-shRNA lentivirus. SAE2 protein was significantly depleted in a doxycycline-dependent manner in each cell line (fig. S11A). Notably, SAE2 depletion led to a significant decrease in clonogenicity of the Myc-dependent breast cancer cells, but had no effect on Myc-independent cells (Fig. 4B). Similarly, depletion of SAE2 also reduced the growth rate of Myc-dependent breast cancer cells, as determined by a multi-color competition assay (fig. S11B). In contrast, SAE2-shRNA had no or modest effects on the proliferation of several normal cell types (fig. S12). Collectively, these results suggest that SAE2 is required for the growth and fitness of Myc-dependent breast cancer cells.
Because our data indicate that SAE2 supports the transformed state of Myc-dependent breast cancer cells, we hypothesized that SAE2 may be essential for in vivo tumorigenicity of these cells. To test this hypothesis, Myc-dependent (SUM159 and MDA-MB-231) and Myc-independent (MCF7) breast cancer cells engineered with a dox-inducible SAE2-shRNA were transplanted into immunocompromised mice and tumor volume was measured over time. To circumvent the effects of SAE2 depletion on in vitro proliferation from confounding the tumorigenicity analyses, mice were treated +/− dox only after tumor transplantation. As shown in Figure 4C, SAE2 depletion significantly inhibits tumor growth of Myc-dependent SUM159 and MDA-MB-231 tumors in vivo (left and middle panels), but had no significant effect on Myc-independent MCF7 tumors (right panel). SAE2-depletion also increased survival time as compared to the –dox treated animals (fig. S13). Furthermore, the tumors emerging in doxycycline contain significantly fewer GFP/shSAE2-expressing cells, consistent with a selection against tumor cells depleted of SAE2 during tumor growth (fig. S14). Together, our data suggest that SAE2 function is required for tumorigenicity of Myc-dependent breast cancers.
Our functional data suggest that breast cancers with aberrant Myc activation are dependent on the core SUMO-activating enzyme SAE1/SAE2. In patients, this hypothesis predicts that breast cancers with low expression of the SUMO-activating enzyme may exhibit a less aggressive clinical behavior selectively in Myc-high breast cancers, since these cancer cells would be less able to tolerate Myc hyper-activation. To test this prediction, we compiled breast cancer datasets (n=1297 patients) for which there was gene expression data (Affymetrix U133 platform only) and a common endpoint criterion (metastatic recurrence) (32–39). Tumors were stratified based on Myc expression levels, with 432 and 429 tumors defined as Myc-high and Myc-low, respectively. We then determined if levels of SAE1/SAE2 were associated with patient outcome (metastasis-free survival) in the Myc-high or Myc-low groups. Strikingly, in patients with Myc-high tumors, those with lower-level expression of SAE1/SAE2 had significantly better metastasis-free survival than those with higher SAE1/SAE2 (Fig. 4D, left panel, p=0.01 log-rank test). In contrast, lower-level expression of SAE1/SAE2 did not correlate with outcome in patients with Myc-low tumors (Fig. 4D, right panel). This suggests that Myc hyper-activation leads to an increased dependency on SAE1/SAE2 in human breast cancers. Together with our functional data, these observations strongly support the hypothesis that SAE1/SAE2 and the downstream Myc-SMS effectors enable breast cancers to tolerate oncogenic Myc activation, and suggest that inactivation of the SUMO-activating enzyme represents a new therapeutic strategy for patients with Myc-driven breast cancer.
The inability to identify small molecule inhibitors of important oncogenes such as Ras and Myc has hampered cancer therapeutics. Recently, the emerging concept of non-oncogene addiction (NOA) has postulated that oncogenic mutations provide dependencies on cellular pathways that can be exploited for therapeutic advantage (4). We sought to exploit NOA genetically by identifying genes whose inactivation is synthetically lethal with Myc hyper-activation. With this strategy, we discovered a critical role for the E1 SUMO–activating enzyme (SAE) in enabling cells to tolerate Myc hyper-activation, and further provide multiple lines of evidence suggesting the SAE is a potential therapeutic target in Myc-dependent breast cancer and other Myc-driven malignancies. Similar to other core cellular processes that are targets for recently developed anti-cancer agents (e.g. the proteasome) (5, 40), SAE1 and SAE2 represent new enzymatic examples of NOA, and thus illustrate the power of unbiased genetic screens in the identification of new directions for cancer therapeutics.
Mechanistically, we show that loss of SUMOylation leads to substantial mitotic catastrophe and cell death by switching a subprogram of Myc transcriptional targets that support mitotic spindle function. Thus, inactivation of SAE2 mimics the mitotic disruption caused by spindle poisons, but in a genotype-specific way (i.e. selectively in cells that harbor oncogenic Myc activation). Notably, mitotic interference is a mainstay of cancer therapeutics, and agents such as taxanes that disrupt proper spindle function are used to treat a wide variety of cancers. However, a major limitation of this class of therapeutics is their significant toxicity to non-tumor organ systems, thus limiting their therapeutic window. Our finding that inhibiting SUMOylation can mimic spindle poisons selectively when Myc is aberrantly activated makes the important prediction that drugs targeting the SUMO pathway may have the anti-tumor effects of spindle poisons with substantially less side effects on normal tissues, a prediction that warrants further investigation.
Myc promotes a balance of pro- and anti-tumorigenic properties, and mutations in Myc can shift this balance in pro- and anti-oncogenic Myc functions, demonstrating that distinct transcriptional (or other biochemical) functions of Myc may be segregated (15, 16). Based on our observations, we propose that the Myc transcriptional program can be shifted to favor the anti-oncogenic state. Specifically, our data suggest that the inactivation of SAE2 drives synthetic lethality with the Myc oncogene by altering a subprogram of Myc transcriptional targets that supports proper mitosis and thus cell viability, a subprogram we term SUMOylation-dependent Myc switchers, or SMS genes. This SMS program is highly enriched in proteins that control spindle integrity, and the Myc-SAE2 synthetic lethal interaction elicits significant aberrations in the mitotic spindle and eventual cell death. These findings suggest that SUMOylation plays a critical role in Myc’s ability to promote oncogenesis, in part by cooperating with Myc to maintain expression of Myc target genes involved in mitotic fidelity. These observations highlight the idea that altering distinct subprograms of Myc transcription (by SAE2 inactivation or other mechanisms) may be exploited as a therapeutic strategy in Myc-driven cancers, and more broadly, suggest that subverting transcriptional programs may be a general strategy in treating cancers driven by oncogenic transcription factors that are notoriously difficult to target therapeutically.
Supplementary Material
Acknowledgments
We thank C. Bland for critical manuscript input. We thank T. Mitchell, W. Choi, S. Songyang, and the BCM C-BASS and CCSC Cores for reagents and technical assistance. J.D.K and K.L.M. are supported by NIH training grants T32HD05520/T32CA090221-09 and DOD pre-doctoral fellowship W81XWH-10-1-0354, respectively. The Golding lab is supported by NIH grant R01GM082837, HFSP grant RGY70/2008, Welch Foundation GrantQ-1759 and NSF Grant082265(PFC: Center for the Physics of Living Cells). This work was supported by a Susan G. Komen for the Cure grant (KG090355) to T.F.W., S.P.O.R.E developmental grant (P50 CA058183) to T.F.W., SU2C-AACR Breast Cancer program grant to R.S. and C.K.O., and U.S. Army Innovator Award (W81XWH0410197) to S.J.E. S.J.E. is an Investigator with the Howard Hughes Medical Institute. T.F.W. is a scholar of The V Foundation and The Mary Kay Ash Foundation for Cancer Research.
REFERENCES AND NOTES
- 1.Weinstein IB. Science. 2002 Jul 5;297:63. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 2.Lowe SW, Cepero E, Evan G. Nature. 2004 Nov 18;432:307. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 3.Jones S, et al. Science. 2008 Sep 26;321:1801. [Google Scholar]
- 4.Luo J, Solimini NL, Elledge SJ. Cell. 2009 Mar 6;136:823. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Annu Rev Med. 2006;57:33. doi: 10.1146/annurev.med.57.042905.122625. [DOI] [PubMed] [Google Scholar]
- 6.Farmer H, et al. Nature. 2005 Apr 14;434:917. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 7.Soucek L, Evan G. Curr Opin Genet Dev. 20(Feb):91. doi: 10.1016/j.gde.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deming SL, Nass SJ, Dickson RB, Trock BJ. Br J Cancer. 2000 Dec;83:1688. doi: 10.1054/bjoc.2000.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van de Vijver MJ, et al. N Engl J Med. 2002 Dec 19;347:1999. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 10.Chin K, et al. Cancer Cell. 2006 Dec;10:529. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Adler AS, et al. Nat Genet. 2006 Apr;38:421. doi: 10.1038/ng1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Olopade OI. Expert Rev Anticancer Ther. 2008 Oct;8:1689. doi: 10.1586/14737140.8.10.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soucek L, et al. Nature. 2008 Oct 2;455:679. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boxer RB, Jang JW, Sintasath L, Chodosh LA. Cancer Cell. 2004 Dec;6:577. doi: 10.1016/j.ccr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 15.Hemann MT, et al. Nature. 2005 Aug 11;436:807. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang DW, Claassen GF, Hann SR, Cole MD. Mol Cell Biol. 2000 Jun;20:4309. doi: 10.1128/mcb.20.12.4309-4319.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menssen A, et al. Cell Cycle. 2007 Feb 3;6:339. doi: 10.4161/cc.6.3.3808. [DOI] [PubMed] [Google Scholar]
- 18.Rottmann S, Wang Y, Nasoff M, Deveraux QL, Quon KC. Proc Natl Acad Sci U S A. 2005 Oct 6; doi: 10.1073/pnas.0505114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, et al. Cancer Cell. 2004 May;5:501. doi: 10.1016/s1535-6108(04)00113-8. [DOI] [PubMed] [Google Scholar]
- 20.Beierle EA, et al. J Biol Chem. 2007 Apr 27;282:12503. doi: 10.1074/jbc.M701450200. [DOI] [PubMed] [Google Scholar]
- 21.Zindy F, et al. Genes Dev. 1998 Aug 1;12:2424. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vafa O, et al. Mol Cell. 2002 May;9:1031. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 23.Hermeking H, Eick D. Science. 1994 Sep 30;265:2091. doi: 10.1126/science.8091232. [DOI] [PubMed] [Google Scholar]
- 24.Hay RT. Mol Cell. 2005 Apr 1;18:1. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Meerbrey KL, et al. Proc Natl Acad Sci U S A. 2011 Mar 1;108:3665. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joukov V, et al. Cell. 2006 Nov 3;127:539. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 27.Song L, Rape M. Mol Cell. 2010 May 14;38:369. doi: 10.1016/j.molcel.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiyomitsu T, Obuse C, Yanagida M. Dev Cell. 2007 Nov;13:663. doi: 10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. Proc Natl Acad Sci U S A. Jan 26;107:1349. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higgins J, et al. BMC Cell Biol. 2010;11:85. doi: 10.1186/1471-2121-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sillje HH, Nagel S, Korner R, Nigg EA. Curr Biol. 2006 Apr 18;16:731. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 32.Loi S, et al. Proc Natl Acad Sci U S A. 107(Jun 1):10208. [Google Scholar]
- 33.Wang Y, et al. Lancet. 2005 Feb 19–25;365:671. [Google Scholar]
- 34.Desmedt C, et al. Clin Cancer Res. 2007 Jun 1;13:3207. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 35.Miller LD, et al. Proc Natl Acad Sci U S A. 2005 Sep 20;102:13550. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt M, et al. Cancer Res. 2008 Jul 1;68:5405. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, et al. Breast Cancer Res Treat. 2009 Jul;116:303. doi: 10.1007/s10549-008-0183-2. [DOI] [PubMed] [Google Scholar]
- 38.Minn AJ, et al. Proc Natl Acad Sci U S A. 2007 Apr 17;104:6740. doi: 10.1073/pnas.0701138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minn AJ, et al. Nature. 2005 Jul 28;436:518. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lord CJ, Ashworth A. Curr Opin Pharmacol. 2008 Aug;8:363. doi: 10.1016/j.coph.2008.06.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.