Abstract
Amphiphilic self-assembling peptides are functional materials, which, depending on the amino acid sequence, the peptide length, and the physicochemical conditions, form a variety of nanostructures including nanovesicles, nanotubes, and nanovalves. We designed lipid-like peptides with an aspartic acid or lysine hydrophilic head and a hydrophobic tail composed of six alanines (i.e., ac-A6K-CONH2, KA6-CONH2, ac-A6D-COOH, and DA6-COOH). The resulting novel peptides have a length similar to biological lipids and form nanovesicles at physiological conditions. AFM microscopy and light scattering analyses of the positively charged lipid-like ac-A6K-CONH2, KA6-CONH2 peptide formulations showed individual nanovesicles. The negatively charged ac-A6D-COOH and DA6-COOH peptides self-assembled into nanovesicles that formed clusters that upon drying were organized into necklace-like formations of nanovesicles. Encapsulation of probe molecules and release studies through the peptide bilayer suggest that peptide nanovesicles may be good candidates for sustained release of pharmaceutically active hydrophilic and hydrophobic compounds. Lipid-like peptide nanovesicles represent a paradigm shifting system that may complement liposomes for the delivery of diagnostic and therapeutic agents.
Keywords: lipid-like peptides, designer peptide surfactants, liposome alternatives, tunable peptides, controlled release
Introduction
Molecular self-assembly has enabled the fabrication of nanostructures and the development of advanced functional materials.1 The design and synthesis of biologically inspired molecules with self-assembling properties has significantly advanced the field of biomaterials and includes peptide, oligonucleotide, and polysaccharide systems.2 Depending on the amino acid sequence, self-assembling peptides have varying properties and have been tested in biomedicine as permissive biological scaffolds for regenerative medicine and drug delivery.3−8 These short, self-assembling peptides are non-toxic, non-immunogenic, and degrade to natural amino acids which can be physiologically metabolized.
Self-assembly of amphiphilic lipid-like peptides leads to the formation of stable nanotubes, nanodoughnuts, nanovalves, nanovesicles, or micelles similar to lipids and chemical surfactants.9 The type and size of the self-assembled peptide nanostructures depend on the peptide concentration, the peptide specific critical micelle concentration (CMC), the amino acid sequence, geometrical constraints (defined by the size of the amino acid side groups), the ionic strength, and the pH of the medium.9,10 These factors affect peptide alignment, packing density, and strength of the intermolecular interactions between the monomers, which determine the formation of hierarchical supramolecular structures of different morphologies and properties.9,11 Since their discovery in 2002,9 self-assembling lipid-like peptides have been studied by many groups; these studies improved our understanding and highlighted the importance of the system.12−16 The development of lipid-like self-assembling peptides with surfactant properties has opened new avenues for applications in biotechnology for the stabilization of membrane proteins more effectively than commercial detergents11,17,18 and in nanotechnology for the construction of energy conversion devices.19
Lipid-like self-assembling peptides are amenable to molecular design allowing modifications in the number, type, and order of amino acids on the peptide chain as well as incorporation of active peptide sequences to facilitate cell penetration or reactive chemical groups such as fluorescent dyes or biotin. The ease of production and the wide scope of modification allow for the synthesis of designer sequences with “tailor-made” tunable properties. In this work, we set out to investigate the physicochemical of cationic and anionic lipid-like peptides, and determine the release kinetics of model drug compounds through peptide formulations as a first step towards the development of a peptide-based drug delivery system.
Materials and Methods
Lipid-like Self-Assembling Peptides
Ac-A6K-CONH2, KA6-CONH2, ac-A6D-COOH, and DA6-COOH were received in powder (SynBioSci, Livermore, CA). The purity of the peptides was 90–94% as determined by electrospray ionization-quadrupole-time-of-flight (ESI-Q-TOF) mass spectrometry. Peptides were dispersed in PBS pH 7.4, probe sonicated for 10 min to facilitate dispersion, and equilibrated for 15 min to allow for self-association of the monomers.
CMC of Lipid-like Peptides
DLS was used to determine the peptides’ CMC (PDDLS/Batch setup, Precision Detectors, Franklin, MA). Solutions of different peptide concentrations in PBS pH 7.4 were filtered through 0.45 μm pore size filters prior to measuring. Scattered light was detected at 90° and the number of photons was recorded and processed by Precision Deconvolve. The solvent viscosity and the refractive index of the buffer were taken as 0.894 cP and 1.33, respectively, at 25 °C.
Particle Size Determination
Normalized intensity (I)–time (t) autocorrelation functions g(2)(q,t) = ⟨I(q,t)I(q,0)⟩/⟨I(q,0)⟩2 of the peptide vesicle dispersions in PBS at 25 °C were measured (n ≥ 4) over a broad time scale (10–7–104 s) using a full multiple τ digital correlator with 280 channels spaced quasi-logarithmically (ALV–5000/FAST) and the 671 nm line of a diode pumped solid state laser (operated at a power <5 mW). The scattered light was collected by a single-mode optical fiber, transferred to an avalanche photo-detector and then to the digital correlator for analysis. In dilute suspensions, the normalized electric field (E)–time autocorrelation function g(1)(q,t) = ⟨E(q,t)E*(q,0)⟩/⟨E(q,0)⟩2 is related to the experimentally recorded g(2)(q,t)2 through the Siegert relation:20g(2)(q,t) = B[1 + f*|g(1)(g,t)|2] where B describes the long delay time behavior of g(2)(q,t) and f* is an instrumental factor (in our system f* ∼ 0.95). Hence, g(1)(t) (for simplicity we drop the q-dependence) was analyzed as a weighted sum of independent exponential contributions g(1)(t) = ∫L(τ) exp(−t/τ)dτ = ∫L(ln τ) exp(−t/τ)d ln τ. The distribution of relaxation times L(ln τ) = τL(τ) was obtained by inverse Laplace transformation of g(1)(q,t) using the CONTIN algorithm.21 The apparent hydrodynamic radii of the suspended particles were determined using the Stokes–Einstein relation Rh = kBT/6πηD, where kB is the Boltzmann constant, η is the viscosity of the solvent, and D is the diffusion coefficient of the particle which is equal to D = (1/τ)q2, where τ is the relaxation time of g(1)(q,t).
AFM of Peptide Nanovesicles
3 μL of the peptide vesicle dispersions (20 mg/mL of peptides in PBS filtered through 0.45 μm filters) were deposited on mica (G250-2, rms ∼ 0.4 nm; Agar Scientific Ltd, Essex, U.K.), rinsed with 200 μL of water (Millipore) after 1 min of equilibration and dried in nitrogen gas stream for 5 min or in air for 1 h. Images were obtained immediately in air using a Bruker MultiMode NanoScope 3D Controller Scanning Probe Microscope (Digital Instruments) operated in tapping mode and soft silicon probes (FESP; nominal length lnom = 225 μm, width wnom = 28 μm, tip radius Rnom = 8 nm, resonant frequency υnom = 75 kHz, spring constant knom = 2.8 N m–1; Veeco Instruments SAS, France). Images were collected from two samples of each peptide system at random spot surface sampling (at least eight areas).
Release through Peptide Nanovesicles
Peptides in powder were mixed to a final concentration of 5 mg/mL with 1 mM CF solution in PBS and the suspension was probe-sonicated for 10 min. After equilibration for 1 h, 1 mL of the nanovesicle suspension was placed in Microcon YM-10 membrane tubes (10 kDa cut-off). Non-encapsulated CF was removed by centrifugation at 11 000 rpm for 20 min. Then the vesicles were re-suspended in PBS pH 7.4 to a final volume of 1 mL and were incubated at 25 °C. CF released through peptide nanovesicles (n = 4) was collected by centrifugation of the membrane tubes at 11 000 rpm for 20 min, the fluorescence intensity of the filtrate was measured, fresh PBS pH 7.4 was added to a final volume of 1 mL, and the suspension was incubated. This process was repeated at specific time points, and a graph was created to show the time course of released CF through peptide nanovesicles. All measurements were carried out in a PerkinElmer LS-50B spectrophotometer using 1 cm quartz cuvettes. The excitation wavelength was at 470 nm, and the emission maximum was observed at 520 nm.
Nile red was used as model compound to study the uptake of hydrophobic molecules by lipid-like peptide nanovesicles. Peptide powder (5 mg) was added to 1 mL PBS pH 7.4 containing 3.14 μM Nile red (i.e., maximum concentration of Nile red in water),25 the suspension was probe sonicated for 10 min, placed in Microcon YM-10 membrane tubes (n = 4) and incubated at 25 °C. To determine the release kinetics we measured fluorescence intensity of the peptide nanovesicle suspension, removed the released Nile red by centrifugation of the membrane tubes at 11 000 rpm for 20 min and added fresh PBS pH 7.4 to 1 mL. This process was repeated at specific time points. Fluorescence emission was recorded from 550 to 700 nm using 1 cm quartz cuvettes (excitation was at 545 nm).
Effect of Lipid-like Peptides on Cell Viability
Caco-2 cells (passage 40) were cultured at 37°C in growth medium (i.e., DMEM, supplemented with 10% v/v FBS, 1% w/v non-essential amino acids and 100 μg/mL penicillin and streptomycin) in 5% v/v CO2 humidified atmosphere. The medium was changed every 2–3 days until cells reached 80% confluency in culture, and then, cells were trypsinized and subcultured. The effect of lipid-like peptides on cell viability and proliferation was studied using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium (MTT) assay (Trevigen Inc., MD). Caco-2 cells grown to a density of 4 × 103 cells/cm2 in 96-well plates were incubated for 3 and 24 h in growth medium containing 0.2 or 1.0 mg/mL of lipid-like peptides. Then, 10 μL of MTT reagent were added, and the plates were incubated for 3 h at 37 °C. Absorbance of the formazan product was determined at 600 nm with a Teknika ELISA plate reader after 1 h.
Results and Discussion
A class of self-assembling peptides (i.e., ac-A6K-CONH2, KA6-CONH2, ac-A6D-COOH, and DA6-COOH) was designed to mimic lipids of biological membranes. These peptides have a hydrophobic tail composed of six alanines, a hydrophilic head, which is an amino acid with charged side group and a length of ∼2.5 nm (Figure 1). As lipid-based systems, addition of lipid-like peptides to water or an electrolyte solution results in formation of a turbid suspension due to self-assembly of the peptide monomers to minimize the interaction between hydrophobic domains and polar environment.9 We determined the CMC of lipid-like peptides in phosphate buffer saline (PBS, 100 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, 2.7 mM KCl at pH 7.4) to be 0.12 mg/mL for A6K-CONH2, 0.09 mg/mL for KA6-CONH2, 0.08 mg/mL for ac-A6D-COOH, and 0.06 mg/mL for DA6-COOH.
Figure 1.
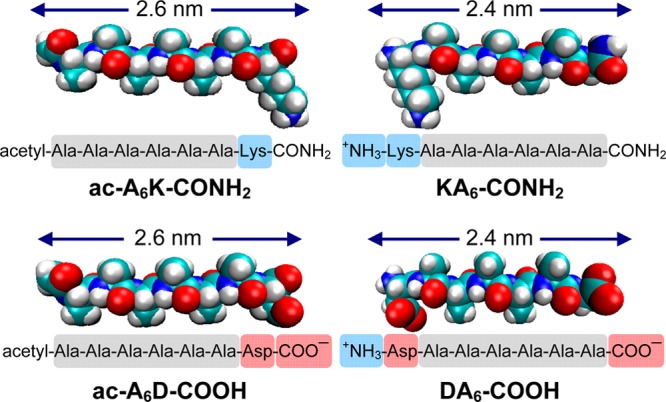
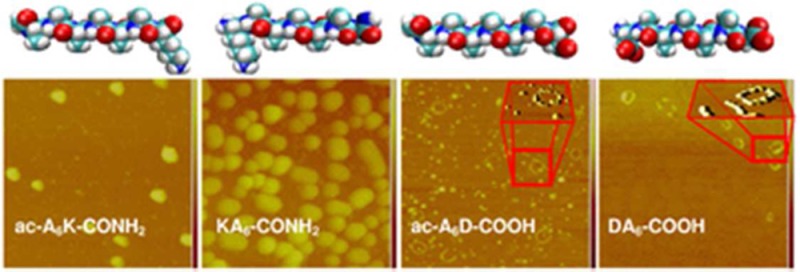
Molecular modeling, amino acid sequence, and charge distribution of lipid-like self-assembling peptides. The peptide length is similar to biological phospholipids. The hydrophobic domain of the peptides consists of six alanines. Color code: cyan, carbon; red, oxygen; blue, nitrogen; white, hydrogen. Illustrations were generated by VMD. Highlighted domains represent amino acids with positive charge (blue), negative charge (red), and hydrophobic side chains (grey). Using pKa values from the literature the net charge of the peptides at pH 7.4 was calculated to be ac-A6K-CONH2 (+1), KA6-CONH2 (+2), ac-A6D-COOH (−2), DA6-COOH (−1).
Atomic Force Microscopy (AFM) Imaging
We used AFM to study the morphology of lipid-like peptide assemblies. Previously, we reported that self-assembly of lipid-like peptides results in uncontrolled formation of various supramolecular structures including vesicles, micelles, and nanotubes.9,10 Herein, the experimental conditions typically resulted in nanovesicle formation (Figure 2). Size distribution analyses of the peptide nanovesicles showed that self-assembly of the lysine-containing, positively charged ac-A6K-CONH2 and KA6-CONH2 peptides results in larger particles (126 and 169 nm, respectively) compared to those observed from association of the negatively charged ac-A6D-COOH and DA6-COOH (28 and 44 nm, respectively) (Table 1, Figure 2 insets). This is due to the smaller side chain of aspartic acid compared to lysine, which allows for better packing of ac-A6D-COOH and DA6-COOH peptides in the nanovesicle bilayer as suggested previously by molecular modeling studies.9
Figure 2.
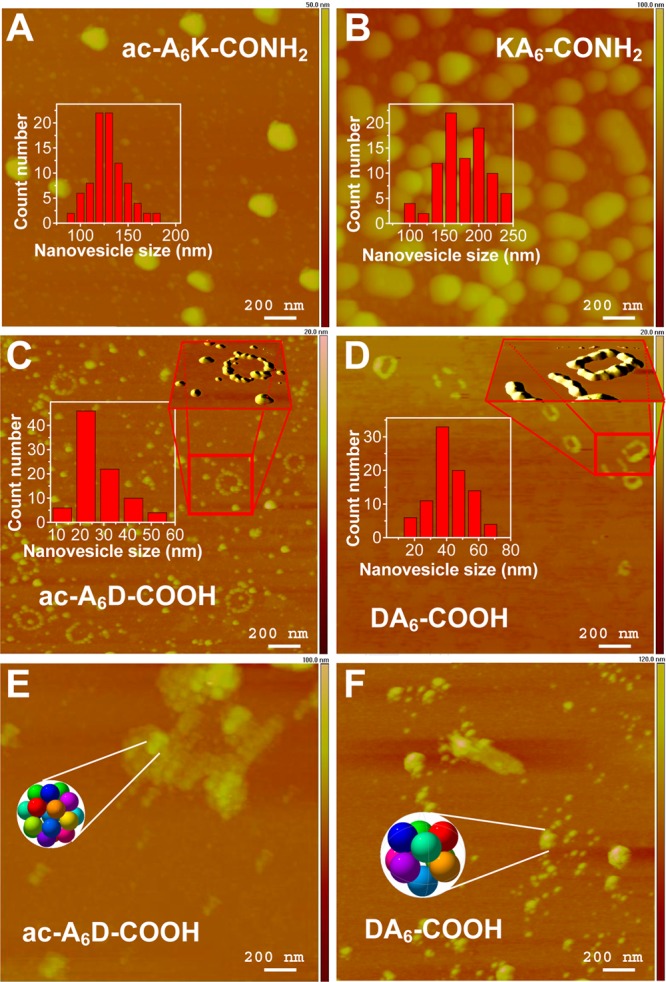
AFM of lipid-like peptide nanovesicles on mica. (A–D Insets) size distribution histograms of peptide nanovesicles were generated from AFM image analysis. (C and D) ac-A6D-COOH and DA6-COOH peptide nanovesicle necklace-like formation upon quick drying of the samples under nitrogen gas. (C and D Insets) ImageJ image processing showing the surface topology of the necklace-like formations. (E and F) ac-A6D-COOH and DA6-COOH peptide nanovesicle clusters formed upon slow air drying of the samples. (E and F Insets) Graphical representation of 20 and 9 tightly packed ac-A6D-COOH and DA6-COOH nanovesicles was generated by http://www.randomwalk.de/sphere/insphr/spheresinsphr.html.
Table 1. Size of Individual Lipid-Like Peptide Nanovesicles, Peptide Nanovesicle Necklace-Like Formations and Peptide Nanovesicle Clusters As Determined by AFM (Dry State) and DLS Curve Fitting (Solution).
| AFM analysis |
DLS analysis | |||
|---|---|---|---|---|
| peptide | vesicle diam. (nm) | necklace diam. (nm) | cluster diam. (nm) | particle diam. (nm) |
| ac-A6K-CONH2 | 126 ± 23 | 122 (30–173) | ||
| KA6-CONH2 | 169 ± 29 | 164 (65–350) | ||
| ac-A6D-COOH | 28 ± 9 | 200 ± 11 | 113 ± 26 | 99 (40–320) |
| DA6-COOH | 44 ± 11 | 159 ± 26 | 135 ± 24 | 137 (70–690) |
AFM imaging of ac-A6D-COOH and DA6-COOH peptide assemblies revealed dispersed nanovesicles as well as necklace-like ultra-structures (Figure 2 C–D). Such formations are observed at the micrometer scale in colloidal systems, when a drop of the suspension evaporates on a solid surface and have been described by two dimensional crystallization laws,22 or the coffee-ring effect.23 It may be that such phenomena are reproduced at the nanometer scale due to evaporation of the solvent and repulsion of like-charged peptide nanovesicles. The diameter of necklaces composed of ac-A6D-COOH and DA6-COOH nanovesicles is 200 and 159 nm, respectively (Table 1). Image analysis showed that ac-A6D-COOH necklaces have 20 ± 5 individual (average diameter 28 nm) ac-A6D-COOH nanovesicles per necklace whereas necklaces composed of larger (average diameter 44 nm) DA6-COOH nanovesicles have 9 ± 3 nanovesicles per necklace.
These observations prompted inquiry about the mechanism of necklace formation. Changing the sample drying conditions (from quick nitrogen gas drying to slow air drying for 1 h) revealed metastable nanovesicle clusters (Figure 2 E–F) suggesting that prior to necklace formation ac-A6D-COOH and DA6-COOH peptide nanovesicles formed loosely bound clusters with diameter 113 and 135 nm, respectively. It is likely that during quick sample drying under nitrogen gas the negatively charged peptide nanovesicle clusters disassemble and spread on the like-charged mica surface leading to nanovesicle necklace-like formations. Theoretical calculations assuming tight packing of hard spheres, revealed that 20 ac-A6D-COOH nanovesicles with diameter 28 nm and 9 DA6-COOH nanovesicles with diameter 44 nm form clusters with diameter 97 and 120 nm, respectively.24 These values correlate well with the AFM determined sizes of the ac-A6D-COOH (i.e., 113 nm) and DA6-COOH (i.e., 135 nm) nanovesicle clusters.
Dynamic Light Scattering (DLS)
The hydrodynamic radii, Rh, of the peptide nanovesicle suspensions were determined by DLS from which the intensity-time correlation functions and the corresponding inverse Laplace transform analyses of lipid-like peptide nanovesicles in PBS were calculated (Figure 3). Analysis of the light scattering data yielded monomodal peptide vesicle size distributions except in the case of KA6-CONH2 suspensions in which peak analysis showed the presence of particles with average diameter of 164 and 906 nm; the latter probably represent aggregates of individual nanovesicles.
Figure 3.
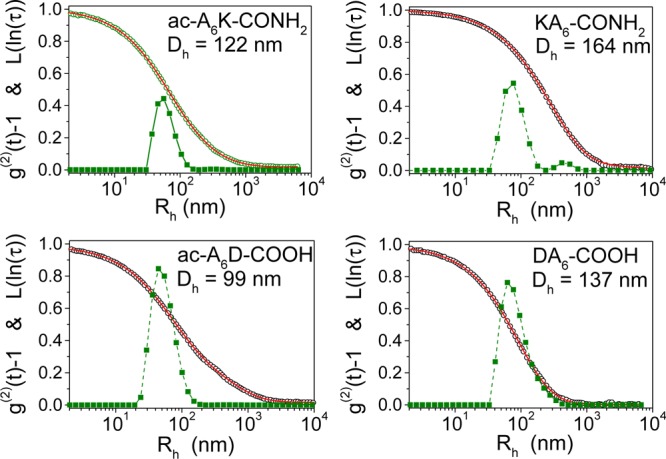
Inverse Laplace transform analysis of the time correlation functions of lipid-like peptide nanovesicles in PBS. Open circles, experimental data; solid lines, best fit curve obtained by inverse Laplace transform; solid squares, volume distributions of the hydrodynamic radii. The hydrodynamic diameters for each distribution are also shown.
DLS measurements of the ac-A6K-CONH2 and KA6-CONH2 nanovesicle diameter are in agreement with the size of mica deposited nanovesicles as determined by AFM. However, DLS analysis of ac-A6D-COOH and DA6-COOH peptide nanovesicle suspensions revealed the presence of particles with diameter 99 and 137 nm, respectively (Figure 3). These values deviate from the nanovesicle diameter determined by AFM for dispersed ac-A6D-COOH and DA6-COOH nanovesicles but resemble the size of the nanovesicle clusters suggesting that these clusters are stable in PBS in which DLS measurements were performed (Table 1, Figure 3). Nanovesicle cluster spreading on the mica surface accounts for the higher cluster diameter determined by AFM compared with that of the DLS analysis.
The hydrodynamic diameter of the nanovesicles (Table 1) appears to be slightly smaller than that measured by AFM which may be due to slight deformation of the nanovesicles caused by adsorption on the mica surface and/or AFM tip broadening effects. Therefore, the size of the nanovesicles measured by AFM (dry state) correlates well with the hydrodynamic diameter determined by DLS. This suggests that nanovesicle shrinking is negligible during drying that precedes AFM scanning.
Release of Hydrophilic Molecules through Peptide Nanovesicles
To determine whether peptide nanovesicles can be used in drug delivery applications, we measured the release kinetics of the hydrophilic fluorescent probe CF through the vesicle bilayer. Encapsulated CF is strongly quenched, and therefore, only the released CF contributes to the fluorescence emitted. Figure 4A shows that positively charged ac-A6K-CONH2 peptide nanovesicles retained CF entrapped for more than 6 h whereas negatively charged ac-A6D-COOH nanovesicles released CF in ∼3 h. Nanovesicles composed of the positively charged KA6-CONH2 and the negatively charged DA6-COOH peptides released CF within ∼1 h.
Figure 4.
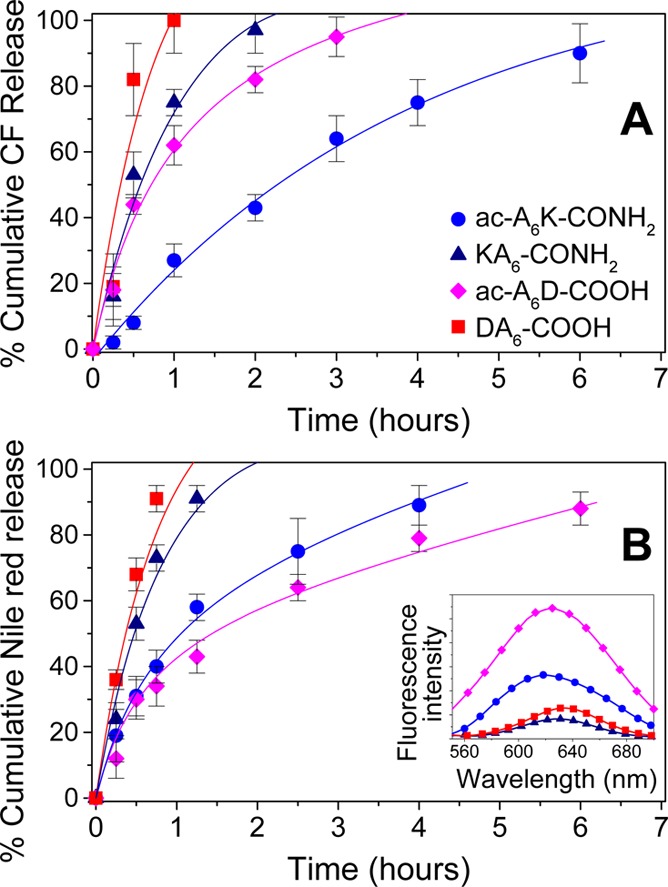
Release kinetics of (A) carboxyfluorescein, CF and (B) Nile red through (●) ac-A6K-CONH2, (▲) KA6-CONH2, (⧫) ac-A6D-COOH and (■) DA6-COOH peptide nanovesicles in PBS. (B Inset) Nile Red emission spectra upon interaction with the lipid-like peptide bilayer in PBS. All data points represent the average of four samples.
This suggests that electrostatic interactions do not affect CF encapsulation and release through the nanovesicles because (i) the less positively charged ac-A6K-CONH2 nanovesicles retained the negatively charged CF better than the more positively charged KA6-CONH2 nanovesicles (KA6-CONH2 carries two positive charges while ac-A6K-CONH2 peptide has one positive charge, Figure 1) and (ii) the negatively charged DA6-COOH nanovesicles did not retain CF better compared to the more negatively charged ac-A6D-COOH nanovesicles (DA6-COOH overall carries one negative charge at the C-terminal whereas ac-A6D-COOH carries two negative charges at the C-terminal). These results suggest that peptide nanovesicles can be used for encapsulation and controlled release of hydrophilic compounds.
Hydrophobic Molecule Uptake and Release through Peptide Nanovesicles
We next studied the hydrophobic character of the peptide nanovesicle bilayer using Nile red as probe molecule, which is hydrophobic and often used to assess liposome bilayer stability.25 The fluorescence emitted by Nile red in water is weak and shows maximum at ∼660 nm. The intensity increases and the emission maximum is blue shifted when Nile red is buried in a hydrophobic environment shielded from the polar solvent.25 Figure 4B, inset, shows the emission spectra of Nile red incorporated into lipid-like peptide nanovesicles. Depending on the peptide formulation emission maxima were observed between 621 and 633 nm, which suggests that assembly of the peptides’ hydrophobic tails results in the formation of a bilayer with hydrophobic properties. The fluorescence intensity was higher in the case of Nile red interacting with ac-A6D-COOH nanovesicles compared to that emitted by Nile red incorporated in ac-A6K-CONH2 nanovesicles. This result suggests that the ac-A6D-COOH bilayer accommodated more Nile red molecules with better shielding from water compared to Nile red inside the ac-A6K-CONH2 bilayer. Ac-A6D-COOH and ac-A6K-CONH2 have similar CMCs and therefore, the observed differences in the emitted fluorescence of nanovesicle incorporated Nile red is likely due to better packing of the ac-A6D-COOH peptide monomers in the bilayer. This results in the presentation of a more hydrophobic environment for incorporating Nile red in the ac-A6D-COOH bilayer compared to the ac-A6K-CONH2 bilayer. The fluorescence intensity of Nile red interacting with KA6-CONH2 and DA6-COOH nanovesicles was significantly lower suggesting less Nile red incorporation in these peptides’ bilayers. Furthermore, we studied the release kinetics through peptide nanovesicles, and we found slow Nile red release through ac-A6D-COOH and ac-A6K-CONH2 nanovesicles reaching significant levels after 6 and 4 h, respectively (Figure 4B). KA6-CONH2 and DA6-COOH nanovesicles appeared to be less stable and released Nile red within 2 and 1 h, respectively. Notably, the release kinetics of Nile red through the peptide nanovesicles follow the order of Nile red fluorescence emission incorporated inside the hydrophobic environment of the peptide bilayer (Figure 4B and inset). As in the case of CF release, we observed that ac-A6K-CONH2 and ac-A6D-COOH nanovesicles retain Nile red for prolonged periods of time suggesting that these peptide nanovesicles can be used for sustained delivery of hydrophilic and hydrophobic compounds.
Cell Viability
To determine the effect of lipid-like peptides on Caco-2 cell viability, we used the MTT assay. Figure 5 shows a constant increase in cell numbers after 3 and 24 h incubation with and without 0.2 or 1.0 mg/mL lipid-like peptides (P < 0.05). These results suggest that ac-A6K-CONH2, KA6-CONH2, ac-A6D-COOH, and DA6-COOH lipid-like peptides do not affect Caco-2 cell proliferation compared with the control.
Figure 5.
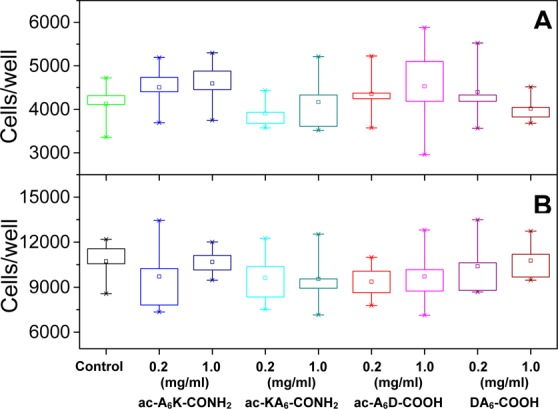
Effect of 0.2 and 1.0 mg/mL lipid-like peptides on Caco-2 cell viability after (A) 3 h and (B) 24 h in culture. In each box chart, the bottom (×) shows the minimum value and marks the 0th percentile. The bottom of the box marks the 25th percentile and the top of the box marks the 75th percentile. The square symbol (□) in the box marks the mean. The top (×) shows the maximum value and 100th percentile. n = 4 sample points. The differences in the number of viable cells at each condition compared to the control are significant (P < 0.05).
Conclusions
Lipid-like peptides were designed to mimic natural lipids having a hydrophilic head and a hydrophobic domain. Although all peptides form nanovesicles some of them do not retain model drug probe molecules for prolonged times. Peptide design by altering amino acid sequence and charge distribution provides a means to control the loading capacity and the drug release kinetics through the nanovesicles. Of the two leading lipid-like peptide nanovesicle systems (i.e., ac-A6K-CONH2 and ac-A6D-COOH), the latter may be more suitable for drug delivery. Negatively charged drug carriers, such as the ac-A6D-COOH nanovesicles, are preferable for intravenous administration because they result in electrostatic repulsions with the like-charged surface of blood cells and vessel walls, which allows for prolonged circulation in the bloodstream.
Liposomes were proposed as drug delivery carriers in the 1970s. A significant amount of work has been done to increase liposomal stability in serum, prolong drug release and reduce side effects associated with immunogenicity and toxicity of liposomes. Lipid-like peptides are nontoxic, nonimmunogenic and may encapsulate or incorporate and slowly release both hydrophilic and hydrophobic drug molecules and may present a platform to append to existing liposomal drug release systems.
Peptide self-assembly is similar to that of lipids and fatty acids. However, peptides differ from these systems because the peptide bilayer is stabilized by a combination of hydrophobic interactions of the hydrophobic amino acids’ side groups and hydrogen bonding of the peptides’ polar backbones. Therefore, the bilayer’s internal chemistry differs between liposomes and peptide nanovesicles. However, lipid-like peptides readily mix with lipids to form hybrid peptide/lipid liposome systems.26 The incorporation of lipid-like peptides in liposomes conferred functionality and modulated the liposome bilayer curvature and stability of the formulation. Furthermore, lipid-like peptides can be easily modified and tailored to incorporate other molecules such as sugars and functional motifs, including cell signaling and cell penetrating peptides to allow the synthesis of cell targeting drug delivery systems.
Depending on their sequence peptide nanovesicles can retain and slowly release both hydrophilic and hydrophobic compounds. We believe that these simple, inexpensive, and nontoxic peptides will open new paths in the field of vesicle-mediated drug delivery systems.
Acknowledgments
The Center for Environmental Health Sciences (CEHS; Grant No. NIEHS ES002109) at MIT is acknowledged for mass spectrometric analysis and technical support. We also thank Dr. M. Spanakis for expert technical assistance with cell assay.
Glossary
Abbreviations:
- ac
acetyl group
- AFM
atomic force microscopy
- CMC
critical micelle concentration
- DLS
dynamic light scattering
- CF
5,6-carboxyfluorescein
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Whitesides G. M.; Mathias J. P.; Seto C. T. Molecular Self-Assembly and Nanochemistry—A Chemical Strategy for the Synthesis of Nanostructures. Science 1991, 254, 1312–1319. [DOI] [PubMed] [Google Scholar]
- Koutsopoulos S. Molecular Fabrications of Smart Nanobiomaterials and Applications in Personalized Medicine. Adv. Drug Delivery Rev. 2012, 64, 1459–1476. [DOI] [PubMed] [Google Scholar]
- Kisiday J.; Jin M.; Kurz B.; Hung H.; Semino C.; Zhang S.; Grodzinsky A.J. Self-Assembling Peptide Hydrogel Fosters Chondrocyte Extracellular Matrix Production and Cell Division: Implications for Cartilage Tissue Repair. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 9996–10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsopoulos S.; Zhang S. Long-Term Three-Dimensional Neural Tissue Cultures in Functionalized Self-Assembling Peptide Hydrogels, Matrigel, and Collagen I. Acta Biomater. 2013, 9, 5162–5169. [DOI] [PubMed] [Google Scholar]
- Davis M. E.; Motion J. P. M.; Narmoneva D.A.; Takahashi T.; Hakuno D.; Kamm R. D.; Zhang S.; Lee R.T. Injectable Self-Assembling Peptide Nanofibers Create Intramyocardial Microenvironments for Endothelial Cells. Circulation 2005, 111, 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsopoulos S.; Unsworth L.D.; Nagai Y.; Zhang S. Controlled Release of Functional Proteins through Designer Self-Assembling Peptide Nanofiber Hydrogel Scaffold. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 4623–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai Y.; Unsworth L.D.; Koutsopoulos S.; Zhang S. Slow Release of Molecules in Self-Assembling Peptide Nanofiber Scaffold. J. Controlled Release 2006, 115, 18–25. [DOI] [PubMed] [Google Scholar]
- Koutsopoulos S.; Zhang S. Two-Layered Injectable Self-Assembling Peptide Scaffold Hydrogels for Long-Term Sustained Release of Human Antibodies. J. Controlled Release 2012, 160, 451–458. [DOI] [PubMed] [Google Scholar]
- Vauthey S.; Santoso S.; Gong H.; Watson N.; Zhang S. Molecular Self-Assembly of Surfactant-like Peptides to Form Nanotubes and Nanovesicles. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 5355–5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso S.; Hwang W.; Hartman H.; Zhang S. Self-Assembly of Surfactant-like Peptides with Variable Glycine Tails to Form Nanotubes and Nanovesicles. Nano Lett. 2002, 2, 687–691. [Google Scholar]
- Koutsopoulos S.; Kaiser L.; Eriksson H. M.; Zhang S. Designer Peptide Surfactants Stabilize Diverse Functional Membrane Proteins. Chem. Soc. Rev. 2012, 41, 1721–1728. [DOI] [PubMed] [Google Scholar]
- Meng Q. B.; Kou Y. Y.; Ma X.; Liang Y. J.; Guo L.; Ni C. H.; Liu K. L. Tunable Self-Assembled Peptide Amphiphile Nanostructures. Langmuir 2012, 28, 5017–5022. [DOI] [PubMed] [Google Scholar]
- Wang Q. R.; Yu J.; Zhang X.; Liu D. J.; Zheng J. H.; Pan Y.; Lin Y. J. Controlled Biosilification Using Self-Assembled Short Peptides A(6)K and V6K. RSC Adv. 2013, 3, 2784–2793. [Google Scholar]
- Castelletto V.; Nutt D. R.; Hamley I. W.; Bucak S.; Cenker C.; Olsson U. Structure of Single-Wall Peptide Nanotubes: In Situ Flow Aligning X-ray Diffraction. Chem. Commun. 2010, 46, 6270–6272. [DOI] [PubMed] [Google Scholar]
- Qiu F.; Chen Y. Z.; Zhao X. J. Comparative Studies on the Self-Assembling Behaviors of Cationic and Catanionic Surfactant-like Peptides. J. Colloid Interface Sci. 2009, 336, 477–484. [DOI] [PubMed] [Google Scholar]
- Xu H.; Wang J.; Han S. Y.; Wang J. Q.; Yu D. Y.; Zhang H. Y.; Xia D. H.; Zhao X. B.; Waigh T. A.; Lu J. R. Hydrophobic-Region-Induced Transitions in Self-Assembled Peptide Nanostructures. Langmuir 2009, 25, 4115–4123. [DOI] [PubMed] [Google Scholar]
- Matsumoto K.; Vaughn M.; Bruce B. D.; Koutsopoulos S.; Zhang S. Designer Peptide Surfactants Stabilize Functional Photosystem-I Membrane Complex in Aqueous Solution for Extended Time. J. Phys. Chem. B 2009, 113, 75–83. [DOI] [PubMed] [Google Scholar]
- Wang X. Q.; Corin K.; Baaske P.; Wienken C. J.; Jerabek-Willemsen M.; Duhr S.; Braun D.; Zhang S. G. Peptide Surfactants for Cell-Free Production of Functional G Protein-Coupled Receptors. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 9049–9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R.; Kiley P. J.; Segal M.; Norville J.; Yu A. A.; Wang L.; Trammell S. A.; Reddick L. E.; Kumar R.; Stellacci F.; Lebedev N.; Schnur J.; Bruce B. D.; Zhang S.; Baldo M. Integration of Photosynthetic Protein Molecular Complexes in Solid-State Electronic Devices. Nano Lett. 2004, 4, 1079–1083. [Google Scholar]
- Berne B. J.; Pecora R.. Dynamic Light Scattering. Wiley: New York, 1976. [Google Scholar]
- Provencher S. W. A Constrained Regularization Method for Inverting Data Represented by Linear Algebraic or Integral Equations. Comput. Phys. Commun. 1982, 27, 213–227. [Google Scholar]
- Denkov N. D.; Velev O. D.; Kralchevsky P. A.; Ivanov I. B.; Yoshimura H.; Nagayama K. 2-Dimensional Crystallization. Nature 1993, 361, 26–26.8421493 [Google Scholar]
- Deegan R. D.; Bakajin O.; Dupont T. F.; Huber G.; Nagel S. R.; Witten T. A. Capillary Flow as the Cause of Ring Stains from Dried Liquid Drops. Nature 1997, 389, 827–829. [Google Scholar]
- Birgin E. G.; Sobral F. N. C. Minimizing the Object Dimensions in Circle and Sphere Packing Problems. Comput. Oper. Res. 2008, 35, 2357–2375. [Google Scholar]
- Greenspan P.; Fowler S. D. Spectrofluorometric Studies of the Lipid Probe, Nile Red. J. Lipid Res. 1985, 26, 781–789. [PubMed] [Google Scholar]
- Yaghmur A.; Laggner P.; Zhang S.; Rappolt M. Tuning Curvature and Stability of Monoolein Bilayers by Designer Lipid-like Peptide Surfactants. PLoS One 2007, 5, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]


