Abstract
We have screened chromosome arm 3L for ethyl methanesulfonate−induced mutations that disrupt localization of fluorescently labeled gurken (grk) messenger (m)RNA, whose transport along microtubules establishes both major body axes of the developing Drosophila oocyte. Rapid identification of causative mutations by single-nucleotide polymorphism recombinational mapping and whole-genomic sequencing allowed us to define nine complementation groups affecting grk mRNA localization and other aspects of oogenesis, including alleles of elg1, scaf6, quemao, nudE, Tsc2/gigas, rasp, and Chd5/Wrb, and several null alleles of the armitage Piwi-pathway gene. Analysis of a newly induced kinesin light chain allele shows that kinesin motor activity is required for both efficient grk mRNA localization and oocyte centrosome integrity. We also show that initiation of the dorsoanterior localization of grk mRNA precedes centrosome localization, suggesting that microtubule self-organization contributes to breaking axial symmetry to generate a unique dorsoventral axis.
Keywords: oogenesis, RNA localization, microtubules, whole-genome sequencing, mutant mapping
Asymmetric messenger (m)RNA localization in the cytoplasm acts to restrict the sites of protein synthesis, particularly in large or polarized cells (Martin and Ephrussi 2009). For example, specification and maintenance of Drosophila oocyte fate depends on the transport of selected mRNAs along microtubules (MTs) from accessory nurse cells to the adjacent and interconnected oocyte (St. Johnston 2005). The minus-end-directed motor dynein and its cofactors Bicaudal-D (BicD) and Egalitarian (Egl) are required for selective RNA transport into the oocyte and for the localization of certain transcripts in later-stage oocytes (Bullock and Ish-Horowicz 2001; MacDougall et al. 2003; Navarro et al. 2004). Posterior localization of the mRNA encoding the germline determinant Oskar depends on the plus-end-directed motor kinesin-1 (Brendza et al. 2000). Genetic and biochemical experiments have shown that the ultimate destinations of transported RNAs depend on recognition of cargo RNAs by appropriate MT motors and on the organizational architecture of the MT cytoskeleton (MacDougall et al. 2003; Dienstbier et al. 2009; Parton et al. 2011).
Dynein-dependent RNA transport in Drosophila eggs and oocytes relies on short RNA signals that are presumably recognized by motor components and adapter proteins. However, the basis for the signals’ specificity and recognition is unclear. One such signal forms a novel helical RNA structure (Bullock et al. 2010), but its generality in directing RNA transport is not currently known. There is strong in vitro evidence that the Egl protein acts as an adapter between dynein and cargo mRNA (Dienstbier et al. 2009), but some signals may have different structures and operate via other adapters.
A particularly significant target of dynein-mediated transport is gurken (grk) whose transcript localization is key to establishing the prospective body axes of the future embryo. grk mRNA localizes posteriorly in early oocytes and is translated during stage 5 into a transforming growth factor-α−like protein that signals to overlying, somatic follicle cells to specify their posterior character (Gonzalez-Reyes et al. 1995). During this stage, the minus-ends of MTs are orientated predominantly toward the oocyte posterior.
During stages 7−8, grk transcripts delocalize to a dorsoanterior corner, allowing localized Grk signaling to establish the dorsoventral axis of the oocyte (Neuman-Silberberg and Schüpbach 1993). At this time, the nucleus and the oocyte centrosome also migrate from the oocyte posterior to its dorsoanterior corner (Januschke et al. 2006), and the cytoskeleton is remodeled so that MTs with anteriorly orientated minus-ends predominate (Theurkauf et al. 1992). How MTs are reorganized at this stage remains controversial, but a recent study has suggested that anterior migration of the oocyte nucleus during stage 7 is due to its being pushed by the posterior-lying centrosome (Zhao et al. 2012).
Several studies indicate that MTs can nucleate from the lateral and anterior cortex of the oocyte and from the centrosome and the nuclear envelope (Cha et al. 2002; Januschke et al. 2006; Parton et al. 2011). It is unclear whether the nucleus and the centrosome localize first or whether cortical MTs prefigure organelle localization, nor is it understood how different classes of MTs might contribute to the asymmetric localization of grk mRNA.
In this paper, we report a novel genetic screen for maternal factors needed to localize fluorescently labeled endogenous grk transcripts during oogenesis. We also describe the combined use of whole-genome sequencing (WGS) and single-nucleotide polymorphism (SNP)-marked recombination to rapidly identify new genes required for grk localization, egg-chamber morphogenesis, and correct organization of the MT cytoskeleton. Finally, we present novel analysis of wild-type and kinesin light chain (klc) mutant oocytes that reveals roles for centrosome-dependent and -independent MTs in grk mRNA localization and axial patterning.
Materials and Methods
Genetic screen
Details of fly stocks, mutagenesis, and the screen are described in Figure 1 and the Supporting Information, File S1. In summary, novel mutations were identified by dissecting one to three females of the genotype hsFLP, nanos::MCP-mCherry/w; grk(MS2)12/w; *FRT2A/ P{ovoD1} FRT2A (“*FRT2A” representing the mutagenized chromosome 3L) 8−10 d after heat-shock to induce homozygous mutant germlines and stage 8−9 oocytes screened for the distribution of fluorescently marked grk mRNA (grk*mCherry). Only germline cysts that are homozygous for the mutagenized chromosome lack the dominant female-sterile mutation ovoD1 and can develop to later stages.
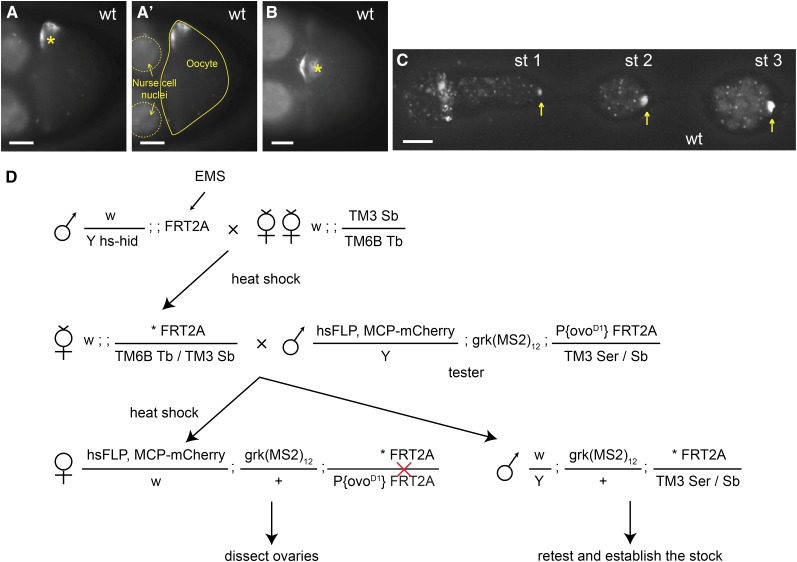
Figure 1.
Genetic screen based on in vivo RNA imaging. (A−C) grk transcript localization in wild-type egg-chambers. (A, B) Localization of grk*mCherry (MCP-labeled grk transcripts) as visualized in differently orientated stage 8−9 wild-type oocytes: the nucleus (asterisk) is at the side or at the top center of the anterior end in A and B, respectively. (A′) Same image as in (A) outlining the oocyte (solid line) and two nurse cell nuclei (dashed lines). (C) grk*mCherry in the germarium and stage 1−3 egg-chambers. grk*mCherry localizes to the wild-type oocyte, which is at the posterior of the egg-chambers (yellow arrows). (D) Crossing scheme for generating mosaic females with homozygous mutant germline. Asterisk indicates the mutagenized chromosome. hs-hid males are selectively eliminated by heat shock during larval stages. The red cross indicates a FLP-induced mitotic recombination event between the two FRT sequences. All developing egg-chambers are homozygous for induced mutations on 3L, because they lack the dominant female-sterile ovoD1 gene. Scale bars = 20 μm.
Approximately one-third of the mutagenized lines lacked egg-chambers that reached stage 8−9, perhaps reflecting mutation that cause early defects in oogenesis. To detect such mutations, ovaries larger than those of ovoD1 flies were scored for the presence and position of the grk*mCherry spot that marks the early oocyte (Figure 1C).
WGS and SNP mapping
Parental and ru h th st cu st e ca (rucuca) chromosomes were sequenced at 168- and 28-fold coverage, respectively. These levels of coverage allowed us to reassemble ~99% of the euchromatic coding genome for each stock and provided a high density of euchromatic SNP markers (averaging 1 per 1.5 kb) for recombination mapping. Individual mutant chromosomes were sequenced at about 20-fold coverage.
The basis for SNP-based mapping of causative mutations is presented in Figure 2. Recombinant females (*?FRT2A/TM3, Sb or TM6, Tb F2 virgin recombinant females; “*?” representing the possibly mutant recombinant chromosome) were individually mated with tester males carrying TM3, Ser for phenotypic retesting, SNP mapping, and to make stocks of informative recombinants. DNA was extracted from individual *?FRT2A/TM3, Ser F3 males and used for high-throughput allelic-discrimination polymerase chain reaction (KASPAr; LGC Genomics, http://www.lgcgenomics.com/), which provide a two-color fluorescence assay for SNP genotyping in microtiter plates (File S2). F3 females showing recombination between proximal and distal SNPs were tested phenotypically as in the primary screen. Additional details are presented in the section Results and File S1.
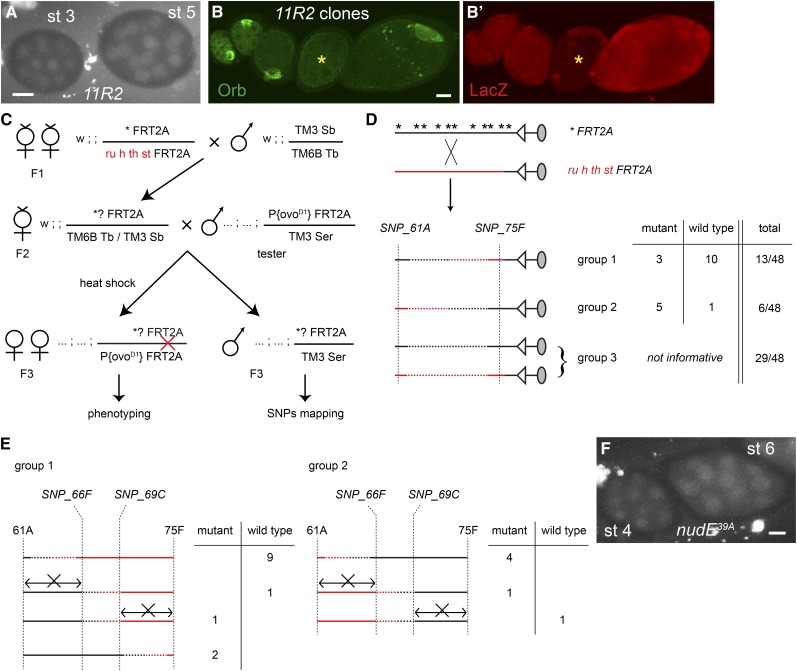
Figure 2.
11R2 is an allele of nudE: overview of the single-nucleotide polymorphism (SNP) mapping. (A and F) grk*mCherry in 11R2 (A) and nudE39A (F) stage 3−6 egg chambers. Lack of localized grk*mCherry indicates the absence of oocyte. (B) Lack of Orb staining in a 11R2 germline clone (*, marked by the absence of LacZ expression; B′) confirms the failure to form an oocyte. Shown is a projection of a series of z-stack confocal sections that cover the whole egg chamber. (C) Crossing scheme for the SNP mapping. Individual F2 females are mated to the tester males (as in the primary screen), and F3 progeny are used for phenotyping (in germline clones) and genotyping. (D, E) summary of SNP genotyping. (D) 48 putative recombinants were genotyped for the SNPs at 61A and at 75F. The ratio of mutant and wild-type recombinants in each group yields an approximate map position. Nonrecombinant lines (group 3) are excluded from the subsequent assays. (E) Genotyping by exclusion shows that the causative lesion lies between 66F and 69C. If necessary, further mapping could have been performed using the four remaining recombinants. Scale bars = 20 μm.
Other methods
Plasmid construction, RNA quantification, antibody staining, in situ hybridization, and X-gal staining were performed as described in File S1.
Results
Genetic screen based on sensitive in vivo fluorescent labeling of grk mRNA
grk transcripts have previously been visualized in vivo by labeling with a fluorescent MS2 Coat Protein (MCP) fusion protein, which binds to endogenous grk transcripts that include 12 repeats of the MCP binding-site (grk-(MS2)12; Jaramillo et al. 2008). To sensitize the system for large-scale screening, we used MCP fused to the more photostable mCherry (Shaner et al. 2004) and excluded fluorescence from the follicle cells by driving MCP-mCherry expression selectively in the germline using the nanos promoter and a shortened 3′UTR from fs(1)K10 that both stabilizes germline transcripts and restricts them to the nurse cells (Serano et al. 1994).
Together, these changes allow rapid and sensitive visualization of the tagged grk transcript (which we refer to as grk*mCherry) in live ovaries. grk*mCherry and endogenous grk transcripts both localize tightly around the nucleus at the dorsoanterior corner of the oocyte from stage 8 and appear as a dorsoanterior crescent or perinuclear halo depending on whether viewed from the top or side, respectively (Figure 1, A, A′, and B). In younger oocytes, grk*mCherry shows a distinctive spot at the posterior of the early egg-chamber due to transport of grk mRNA into the developing oocyte (Neuman-Silberberg and Schüpbach 1993).
We used ethyl methanesulfonate (EMS) mutagenesis and the FLP-ovoD1-dominant-female-sterile system to generate germline clones of newly- induced mutations on chromosome arm 3L and screened for those affecting grk mRNA localization (Figure 1D; Materials and Methods; Chou et al. 1993). We scored 4911 independently mutagenized lines and found 39 lines with apparent defects in grk*mCherry localization, 11 of whose phenotypes repeated on rescreening. We also identified several mutations affecting egg-chamber and oocyte morphology.
Identification of newly induced mutations by WGS and SNP-based recombinational mapping
A standard EMS dose in Drosophila melanogaster (25 mM) induces an average of one mutation per 400 kb (Cooper et al. 2008), corresponding to about 10 mutations per chromosome arm that affect protein-coding regions (Misra et al. 2002). These were easily identified by deep genomic sequencing of the parental and mutated chromosomes (Materials and Methods; File S1). To determine which mutation was causative, we first tested for allelism. If present, we compared the genome sequences of two alleles to identify a gene on 3L that is mutated in each allele. For mutants represented by single alleles, we used high-throughput SNP markers and genetic recombination to map the mutation to a 2- to 3-Mb region (~10% of a chromosome arm), which is usually sufficient to restrict the number of candidate causative coding mutations to one or two. The phenotype and the molecular lesions of all mutants are summarized in Table 1, Table 2, and Table 3.
Table 1.
Mutants identified from the screen
| gene | alleles | grk mRNA localization in stage 6 | grk mRNA localization in stage 9 | other phenotypes | |
|---|---|---|---|---|---|
| mislocalised grk mRNA in late stages | armitage (armi) | 3R13, 9R1, 10M12, 11R3, 11S9, 13P2, 13P5, 14P4 | diffuses internally | weakly accumulates around the nucleus, spreads along anterior periphery | defective follicle cell development |
| saturn*: kinesin light chain (klc) & maelstrom (mael) | 5R12 | diffuses internally | no longer localizes to the anterior periphery nor to the nucleus | mispositioned nucleus | |
| scaf6 + unmapped mutation | 12M9 | diffuses internally | weakly accumulates around the nucleus, spreads along anterior periphery | defective reproduction of germline stem cells | |
| elg1 | 3R7 | localizes posteriorly as in wild type | spreads along anterior periphery | defective nurse cell DNA endoreplication |
| Phenotypes | |||||
| other mutants | nudE | 11R2 | Oocyte is misspecified. Most egg chambers contain 16 nurse cells. | ||
| quemao (qm) | 5R4, 11S5 | Plasma membranes of nurse cells and the oocyte collapse. Released ring canals cluster. grk mRNA colocalizes to the ring canal cluster. | |||
| Tsc2/gigas | 1R8, 2R5, 11R10 | Oocyte is frequently mispositioned. | |||
| rasp | 11R5, 13P2, 13P10, 13P13 | Mature eggs have fused appendages. Gurken proteins diffuse in the cytoplasm. | |||
| Chd5/Wrb | 9R9, 13P2 | Disorganized nurse cells in late stage 10. | |||
saturn (5R12) is a synthetic phenotype caused by mutations in klc and mael (see Results).
Table 2. Molecular lesions identified by whole-genome sequencing.
| Allele | Affected Gene(s) | Nucleotide Change | Translational Change |
|---|---|---|---|
| 5R12 | klc | AG>GG | Truncation due to mutated intron 2 acceptor |
| mael | GAG>AAG | E131 > R | |
| 12M9 | scaf6 | CAG>TAG | Q560 > stop |
| 3R7 | elg1 | TGG>TGA | W212 > stop |
| 11R2 | nudE | CAG>TAG | Q33 > stop |
| 5R4 | qm | CAG>TAG | Q116 > stop |
| 11S5 | qm | GCC > GTC | A104 > V |
| 1R8 | Tsc2/gigas | AG>AA | Truncation due to mutated intron 11 acceptor |
| 2R5 | Tsc2/gigas | GTG > GAG | V1726 > E |
| 9R9 | Chd/Wrb | ATAaccgtgtttaTCA > ATA—TCA | Truncation after 107 aa due to 10 bp deletion |
| 13P2 | Chd/Wrb | TGG>TGA | W158 > stop |
| 11R5 | rasp | TAT>TAA | Y210 > stop |
| 13P10 | rasp | ATT|gtcacaat|GTC > ATTgtcacaat-HOBO-gtcacaatGTC | Insertion of HOBO transposon at 177 aa |
The mutated bases are underlined.
Table 3. Molecular lesions of armitage alleles.
| Allele Name | Amino Acid Change | Domain | grk Mislocalization | Eggs Laid by Hemizygous Females |
|---|---|---|---|---|
| 3R13, 9R1 | Q315 > stop | C-terminal truncation | + | – |
| 11R3 | G583 > D | Unknown domain | + | – |
| 14P4 | Q674 > stop | C-terminal truncation | + | – |
| 13P2 | G728 > E | Helicase domain | + | – |
| 10M12 | Splicing donor of intron 7 (GT to AT) | Exon 8 skipped, which truncates after aa 1082 | + | – |
| 11S9 | E1082 > K | Helicase domain | + | + |
| 13P5 | R1121 > stop | C-terminus of helicase domain truncated | + | – |
The helicase domain lies between amino acids 698−1132 of the 1188 amino acids Genbank accession no ABX00729.1. Df(3L)E1 was used to make flies hemizygous for each allele.
To illustrate the strategy, we present the mapping of 11R2, a mutation that causes oocyte misspecification and that was not allelic to other mutations from our screen. Germline clones of 11R2 lack both the early, oocyte-specific spot of grk*mCherry expression (Figure 2A). They also fail to express another oocyte marker, Orb (Figure 2B), confirming that no oocyte has been specified.
The principle and crosses for the mapping are illustrated in Figure 2. The mutated chromosome *FRT2A was mated with a presequenced ru h th st FRT2A chromosome. F2-recombinant females were mated with tester males and, prior to phenotyping, F3 animals were SNP-genotyped using a rapid single-fly polymerase chain reaction assay based on two-color fluorescence that can be assayed in microtiter plates (Figure 1D, Figure 2C, File S1, File S2; and Materials and Methods). SNP genotyping was performed in two stages over a single generation: individual F3-recombinant males were identified first by genotyping for SNPs at each end of the chromosome arm (Figure 2D; Materials and Methods), allowing early discard of nonrecombinant flies and crosses. Additional SNPs were then used for “digital” fine mapping to define a minimal causative interval by exclusion (Figure 2E). Further technical details are in the section Materials and Methods and File S1.
For 11R2, 19 of 48 F2 females had recombined between tester distal and proximal SNPs (SNP_61A and SNP_75F, respectively in Figure 2D); 3 of 13 recombinants with distal FRT2A homology (group 1) and 5 of 6 recombinants with proximal FRT2A homology (group 2) were mutant, mapping the mutation closer to 75F than to 61A. Internal SNPs localized the mutation between 66F and 69C, based on a mutant recombinant that excludes the 69C−75F interval and a wild-type recombinant that excludes the region distal to 66F (Figure 2E).
WGS of the original mutant chromosome revealed a nonsense mutation in the region of interest: Q33 > stop in the nudE gene (at 67D). NudE is a component of the dynein motor complex (Shu et al. 2004; McKenney et al. 2010) that is implicated in several transport processes linked to oocyte specification: fusome organization, centrosome migration, and RNA transport into the pro-oocyte (Theurkauf et al. 1993; Bolivar et al. 2001; Navarro et al. 2004). We confirmed that the nudE nonsense mutation explains the 11R2 phenotype by showing that germline clones of nudE39A, a previously described null allele (Wainman et al. 2009), also fail to specify an oocyte (Figure 2F).
Mutations affecting egg-chamber morphology
The background cytoplasmic fluorescence of MCP-mCherry allowed us to identify a number of mutations with defects in oocyte positioning or the morphology of egg chamber. Two allelic mutations (5R4 and 11S5) are associated with lesions in the quemao (qm) gene (Table 2) encoding geranylgeranyl pyrophosphate synthase, which synthesizes a lipid substrate for protein prenylation. Most mutant egg-chambers older than stage 5 have unevenly spaced nurse cell nuclei (Figure 3A). They also lack clear boundaries between the nurse cells and the oocyte, suggesting that nurse-cell plasma membranes have collapsed (Figure 3B). The qm mutations are indeed responsible for the oocyte phenotypes because germline clones of the previously described null allele (Lai et al. 1998) have the same maternal phenotype (Figure 3C).
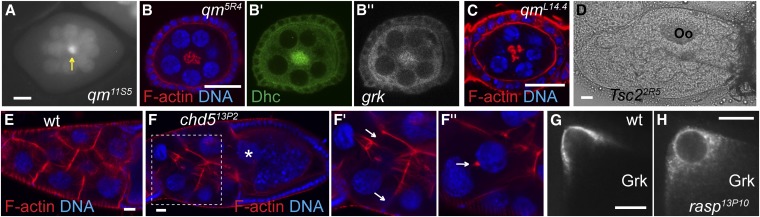
Figure 3.
Phenotypes of other mutants identified in the screen. (A) grk*mCherry localizes to the center of the qm11S5 egg-chamber (arrow). (B−B′′) Clustering of ring canals in center of qm5R4 oocytes as revealed by coincident central staining of mutant oocyte for F-actin (phalloidin, B), Dynein heavy chain (Dhc, B′), and grk*mCherry (grk, B′′). The lack of F-actin staining between nurse cell nuclei suggests that the cell membranes have collapsed. (C) Staining of F-actin (phalloidin) in qmL14.4 germline clone. (D) Laterally mispositioned oocyte (Oo) in a Tsc22R5 mutant stage 7−9 egg-chamber. (E, F) Phalloidin staining of wild-type (E) and Chd513P2 (F) stage 10B egg-chambers showing discontinuous actin filaments (arrows in F′) and a nurse cell nucleus (asterisk) going beyond the border of nurse cells and the oocyte. A different z-section of the same egg-chamber shows a collapsed ring canal (arrow in F′′). (G, H) Grk protein is more diffuse in the cytoplasm of rasp13P10 (H) than wild-type (G) stage 8−9 egg-chambers. scale bars = 20 μm.
In qm mutant egg-chambers, the ring canals are clustered together centrally (Figure 3, B and C), and Dynein heavy chain (Dhc), which accumulates at MT minus-ends, is enriched adjacent to the ring canal aggregates (Figure 3B′), supporting the idea that ring canals can nucleate nurse cell MTs to facilitate the transcript transport from nurse cells to the oocyte (Clark et al. 2007; Mische et al. 2007). grk mRNA and Dhc still colocalize, indicating that the transcripts are still being transported to MT minus-ends (Figure 3, B′ and B′′).
We also recovered three complementation groups affecting egg-chamber or egg-shell morphology in which grk localization appears normal (Table 1). One set of three alleles (1R8, 2R5, and 11R10) showed frequent oocyte mispositioning (Figure 3D) and mutant egg chambers that deteriorate before stage 10. Sequencing two alleles revealed mutations in Tsc2/gigas (Table 2), which encodes a negative regulator of cell cycle and tissue growth through the target of rapamycin pathway (Ito and Rubin 1999; Potter et al. 2001; Tapon et al. 2001). All three alleles fail to complement each other or Tsc2192.1, a previously described null allele.
Two allelic mutations (9R9 and 13P2) affect CG32022 (Table 2), a gene that encodes a homolog of human Congenital Heart Disease 5 (Chd5)/Tryptophan-rich basic protein (Wrb), which is required for protein insertion into the endoplasmic reticulum membrane (Favaloro et al. 2010). The arrangement of nurse cells is disorganized in late stage 10 Chd5 mutant oocytes, and their nuclei often invade the oocyte (Figure 3, F and F′′). Several aspects of the mutant phenotype are suggestive of defective actin polymerization during the “dumping” of nurse cell contents into the late oocyte: ring canals have collapsed, actin filaments along the nurse cell plasma membrane are discontinuous (Figure 3, F′ and F′′), and the radial arrays of actin cables emanating from the nurse cell plasma membrane are less abundant than the wild type (Cooley et al. 1992; Murphy and Montell 1996; Li et al. 1999).
One complementation group (11R5, 13P2, 13P10, and 13P13) has lesions in the rasp gene (Table 2), which encodes a Protein-cysteine N-palmitoyltransferase (Chamoun et al. 2001). All alleles generate eggs with fused appendages, indicative of reduced Grk signaling, although levels and localization of grk mRNA appear normal. Rasp is required for the palmitoylation and activation of Spitz, an epidermal growth factor receptor ligand, and has been proposed to be also required for Grk activity (Miura et al. 2006). Grk protein is diffusely localized in the cytoplasm of mutant oocytes, rather than adjacent to the cortex as in the wild type (Figure 3, G and H). This result suggests that lack of posttranslational modification causes Grk to be mistrafficked and not fully activated.
elg1 is required for grk transcript localization and nurse cell endoreplication
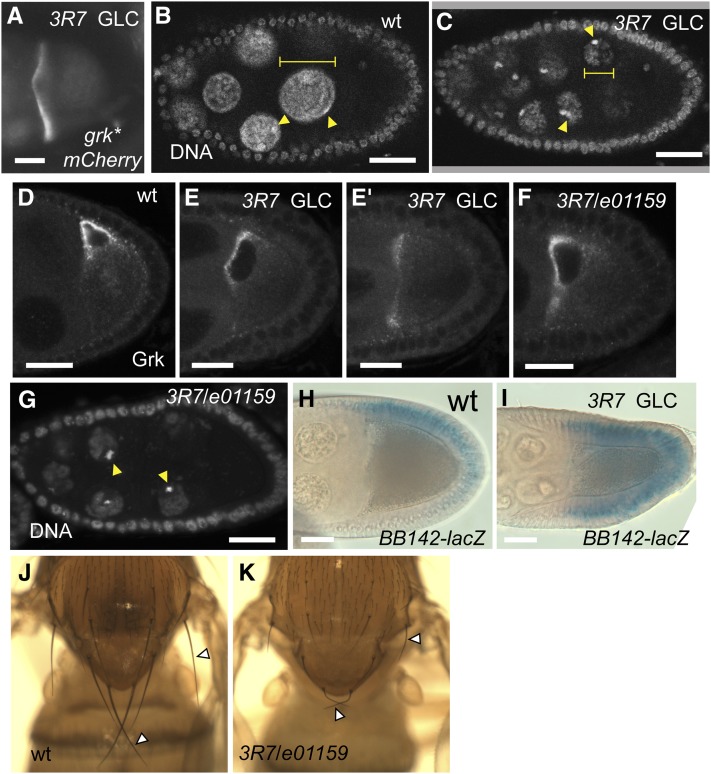
grk transcripts mislocalize along the entire anterior margin of 3R7 oocytes (Figure 4A). 3R7 also affects nurse cell DNA endoreplication: mutant nurse cell nuclei are smaller and apparently underreplicated (Figure 4, B and C). Translation of the mislocalized transcripts is not repressed, and so Grk protein accumulates anteriorly (Figure 4, D, E, and E′), where it drives ectopic expression in follicle cells of a Grk-responsive lacZ enhancer-trap (BB142; Figure 4, H and I; Schüpbach and Roth 1994). In these and other respects, the 3R7 oocyte phenotype differs from that associated with a DNA-damage response (see following section).
Figure 4.
elg1 is required for grk mRNA localization and nurse cell DNA endoreplication. (A) grk*mCherry localization, spreading along the anterior periphery in elg13R7 stage 8−9 oocytes. (B, C, and G) DAPI staining of stage 8 egg-chambers showing the elg1 nurse cell nuclei [(B, G) are smaller than wild-type (diameters; yellow bars]. Mutant nurse cell nuclei include an increased proportion of condensed heterochromatin (arrowheads), which is usually relatively underreplicated during endoreplication (Dej and Spradling 1999). (C) elg13R7 germline clone (GLC) and (G) 3R7/e01159 egg-chamber. (D−F) Anti-Grk staining of the wild-type (D) and elg1 mutant stage 8−9 egg-chambers, showing that Grk protein localizes across the entire anterior end in elg13R7 germline clone oocytes (E and E′; two different planes of germline clone) and 3R7/e01159 egg-chamber (F). (H, I) X-Gal staining of BB142-lacZ enhancer trap in wild-type (H) and 3R7 germline clone egg-chambers (I), showing ectopic (ventral; bottom) staining in the latter. (J, K) Macrochaetae (arrowhead) are shorter in 3R7/e01159 scutellum (J) than in the wild type (I). Scale bars = 20 μm.
3R7, which is represented by a single allele, maps by SNP-recombination between 70E and 73E, within which WGS identified a W212 > stop nonsense mutation that would truncate most of the 1162 amino acid protein in CG16838, a Drosophila homolog of yeast elg1 (Table 2). The mutation is likely to eliminate Elg1 activity since 3R7 is not complemented for female fertility, Grk protein localization, or nurse cell DNA replication by e01159, a piggybac insertion into the coding region of CG16838 (Figure 4, F and G), confirming that the causative mutation indeed lies in elg1.
Elg1 is a factor that binds to and loads proliferating cell nuclear antigen onto replicating DNA (reviewed in Aroya and Kupiec 2005) and is required for maintaining genome stability in yeast and human cells (Kanellis et al. 2003; Sikdar et al. 2009). Homozygous 3R7 flies as well as 3R7/e01159 mutant flies are viable (albeit female sterile) and develop without significant difference in adult body size. Also, their nota display shortened macrochaetae (Figure 4, J and K), whose lengths also depend on endoreplication (Weng et al. 2003). This phenotype suggests that elg1 is also required for the growth of other cell types that undergo endoreplication.
armi is required for somatic ovarian development
Other newly induced mutations appear to affect transposon silencing. Complementation analysis and DNA sequencing showed that 8 of the 11 mutants affecting grk mRNA localization are allelic to armitage (armi), which encodes a putative RNA helicase required for the transposon silencing by Piwi-interacting RNAs (piRNAs), small RNAs that associate with the Piwi protein (Cook et al. 2004; Malone et al. 2009). Like other piRNA pathway mutations, armi oocytes show phenotypes associated with the response to DNA damage caused by excessive transposon expression, including failure to restrict anterior grk mRNA localization to a dorsal corner (Figure 5A) due to a misorganized MT cytoskeleton, silencing of Grk translation, and disruption of the karyosome (not shown; Klattenhoff et al. 2007; Pane et al. 2007).
Figure 5.
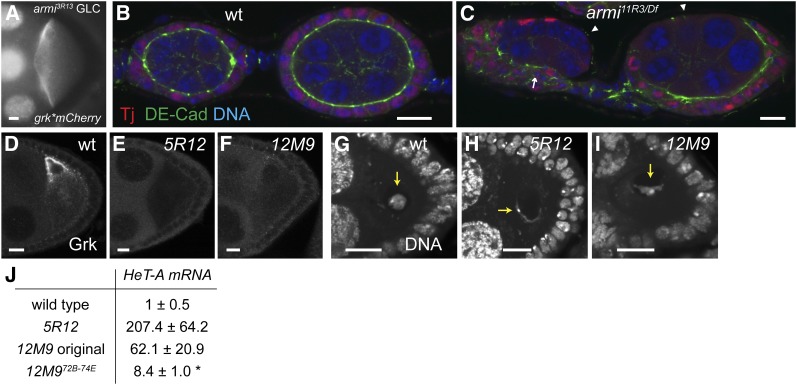
Mutants affecting transposon silencing. (A) grk*mCherry localization, spreading along the anterior periphery in armi3R13 stage 8−9 oocytes. (B) wild-type and (C) armi11R3 / Df(3L)E1 ovarioles. Costainings of Traffic jam (Tj, marker for follicle cells), DE-Cadherin (DE-Cad), and DNA (DAPI). Follicle cells in armi mutant are disorganized (arrow) and fail to encapsulate the germline cyst (arrowheads). (D−F) lack of Grk protein expression in mutant stage 8−9 egg-chambers. (G-I) DAPI staining in stage 3-5 egg-chambers showing distorted oocyte karyosomes (arrows). (D and G) wild type. (E and H) 5R12 germline clone. (F and I) 12M9 germline clone. (J) Mean levels of HeT-A transcripts (± SD) are elevated in mutant egg-chambers compared with the wild type. *P < 0.001. Scale bars = 10 μm.
Armi has been shown to function together with Piwi and Fs(1)Yb (Saito et al. 2010; Qi et al. 2011). However, previous armi alleles (armi72.1; armi1) did not show the abnormal follicle cell development and defective germline stem cell reproduction seen in piwi mutant females (Cox et al. 1998; Cook et al. 2004). Our results show that this is probably due to residual somatic armi activity. Females hemizygous for our novel, strong armi alleles produce no mature eggs (Table 3). Ovaries of those females display a variety of defects in follicle cell organization, including a failure to encapsulate the germline and formation of a multilayered epithelium (Figure 5, B and C). Similar phenotypes are seen in another piRNA pathway mutant, vreteno (Zamparini et al. 2011).
CG33522 (Scaf6/Cherp) is required for transposon silencing
The nonallelic grk mRNA mislocalization mutants 5R12 and 12M9 both show grk mistranslation and a defective karyosome indicative of a DNA damage response (Figure 5, E, F, H, and I). To confirm that the novel mutations affect transposon silencing, we measured transcription of telomeric transposon HeT-A, whose expression is sensitive to piRNA-mediated silencing (Pane et al. 2007; Li et al. 2009). For both mutations, HeT-A mRNA levels in the ovaries are greatly increased (Figure 5J), indicating that piRNA silencing is indeed disrupted.
SNP-recombination analysis showed that 12M9 is due to at least two interacting lesions. In summary, a distal mutation between 72B−74E causes weaker transposon activation (Figure 5J), which is enhanced by a proximal mutation between 74E−80B that also reduces germ cell maintenance such that no developing egg-chambers remain 10 d after clone induction. The enhancer mutation had no other apparent maternal phenotype in the absence of the distal mutation and so was not investigated further.
Sequencing 12M9 revealed the induction of a nonsense mutation (Q560 > stop) in CG33522 (at 73E5), the Drosophila homolog of human SR-related CTD associated factor 6 (scaf6)/calcium homeostasis endoplasmic reticulum protein 6 (cherp) (Will et al. 2002; Jurica and Moore 2003). Animals transheterozygous for the 12M9 chromosome and Df(3L)ED4674, a deficiency lacking 73B5−73E5, live until the late pupal stage but rarely eclose. Viability and fertility are rescued by a 20-kb transgene covering scaf6 (Pacman genomic clone CH322-104B15; Venken et al. 2009), implying that the scaf6 mutation is indeed causative.
saturn is due to combined inactivation of klc and the piRNA pathway
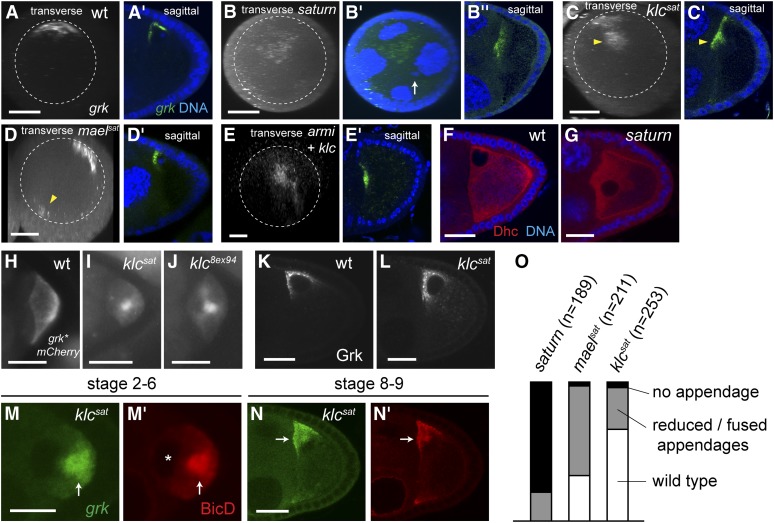
The final mutation (5R12, which we named “saturn”), also affects transposon expression but differs phenotypically from other piRNA pathway mutants in that grk mRNA and the nucleus localize anteriorly but not in a corner (Figure 6, A−G). SNP-recombination mapping combined with WGS showed that saturn is a synthetic mutation: klc (AG > GG in the splice-acceptor of the second intron, which probably inactivates the gene due to skipping of the 121 base exon 3 and a translational frame shift), combined with maelstrom (mael; E131 > R), which encodes a piRNA pathway component that is required for grk mRNA localization and MT polarization (Clegg et al. 1997; Findley et al. 2003). We call these individual alleles klcsat and maelsat, respectively (see Figure S2 for mapping details).
Figure 6.
Anterior-central grk mRNA localization in saturn is due to joint inactivation of klc and piRNA pathway. (A−E) Sagittal views as well as transverse projections of a series of z-stack confocal images of endogenous grk mRNA visualized by in situ hybridization in wildtype (A), saturn (B), klcsat (C), maelsat (D), and armi3R13 klc8ex94 double mutant (E) stage 8−10 egg-chambers. Nuclear DNA in the saturn oocyte is marked by arrow (B′), showing that the majority of grk mRNA is not associated to the nucleus. Approximately one-half the grk mRNA (arrowhead, C) is internalized in klcsat germline clones. grk mRNA is also seen in the opposite side of the anterior periphery (arrowhead, D) in maelsat germline clones. (F, G) Anti-Dhc staining in wild-type (F) and in saturn (G) stage 8−9 egg chambers. Dhc is excluded from the nucleus, thus showing the mispositioned nucleus. (H, I, J) grk*mCherry localization in stage 6 oocytes. (H) wild-type (wt), (I) klcsat, and (J) klc8ex94 showing internally localized grk mRNA in klc mutants. (K, L) Anti-Grk staining in stage 8−9 egg-chambers. (K) wildtype (wt). (L) klcsat, showing that Grk translation is not disrupted in klc mutant. (M, N) Localization (arrows) of grk*mCherry (grk, M and N) and the dynein cofactor, BicD (M′ and N′) indicates that dynein-dependent grk mRNA transport is not affected in klcsat germline clone. BicD and grk mRNA are excluded from the oocyte nucleus, showing a posteriorly mislocalized nucleus before its migration to the dorsoanterior corner. (O) Frequencies of abnormal or absent dorsal appendages in mature eggs of indicated genotypes. Scale bars = 5 μm in (M) and 20 μm in the rest.
To check whether combined reduction of piRNA and klc activities explains the saturn phenotype, we combined a different null klc allele with a mutation in a different piRNA pathway gene. klc8ex94 armi3R13 double-mutant oocytes show a saturn-like phenotype with a complete lack of cortical localization of grk mRNA in stage 10 oocytes (Figure 6E), indicating that the saturn phenotype is indeed caused by the klcsat maelsat mutations.
MT organization is altered in klc mutant oocytes
grk mRNA is normally tightly associated with the oocyte cortex but, in stage 9 klcsat oocytes, approximately one-half of the grk mRNA is somewhat internally localized (Figure 6C), a phenotype that is evident from as early as stage 2 (Figure 6I). A similar phenotype is seen in klc8ex94 germline clones (Figure 6J; Gindhart et al. 1998).
Levels of Grk protein appear unaffected in klcsat oocytes (cf. Figure 6, K and L), indicating that a DNA damage checkpoint has not been triggered. Nevertheless, the mutant eggs are somewhat ventralized, showing that Grk signaling activity is reduced (Figure 6O). Presumably, grk mRNA mislocalization causes Grk to be less efficiently processed or secreted.
Internally mislocalized grk mRNA in klc mutant oocytes could be due either to impaired RNA transport or to MT misorganization. Dynein cofactors Egl and BicD have been previously shown to be in the grk mRNA transport complex (Delanoue et al. 2007), and most grk mRNA colocalizes with BicD from early stages until at least stage 9 (File S4). BicD and grk transcripts still colocalize in klcsat mutant oocytes (Figure 6, M, M′, N, and N′), indicating that dynein-mediated transport of grk mRNA is still active in klc mutant oocytes and the grk mRNA mislocalization is due to an altered MT cytoskeleton.
klc and piRNA pathways are required for the clustering of oocyte centrosomes
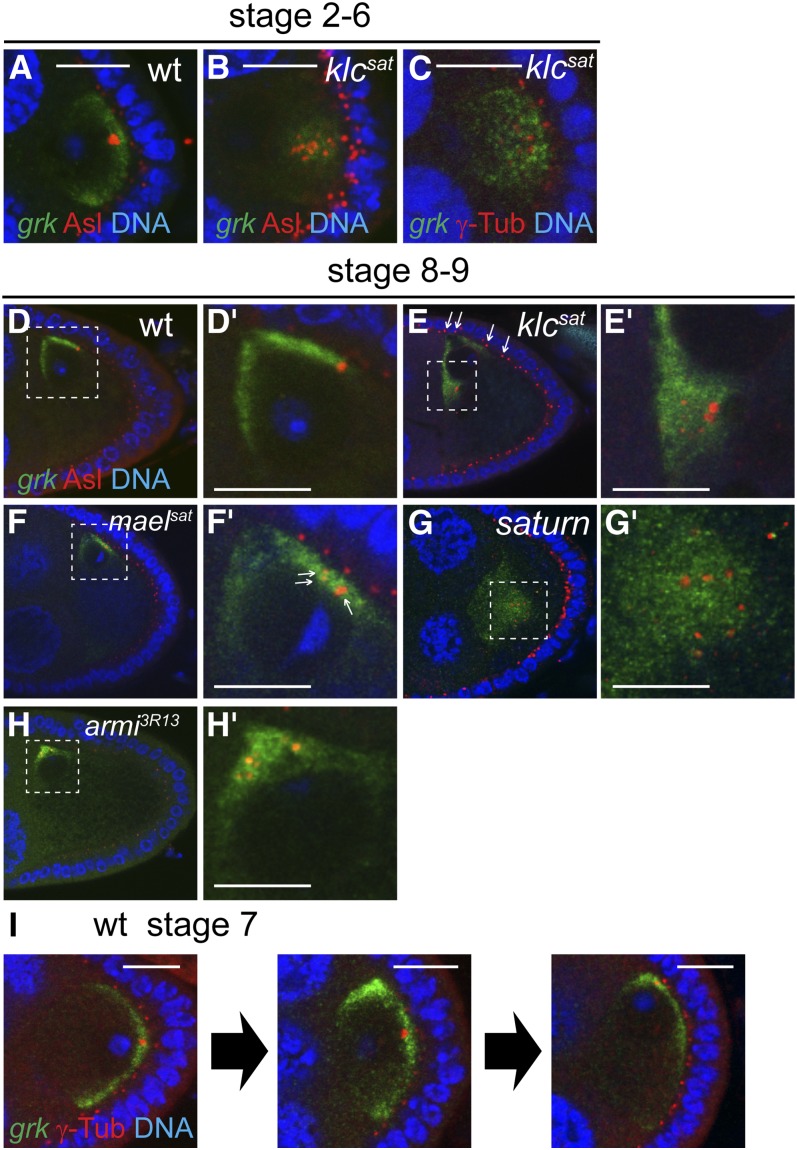
During early oogenesis, nurse cell centrosomes are exported to the prospective oocyte where they form a single, large cluster (Bolivar et al. 2001). Our results show that efficient formation of this aggregate is dependent on kinesin activity. Staining klc mutant oocytes for the pericentriolar proteins Asterless and γ-tubulin reveals 10−20 foci clustered posteriorly at stage 2 and anteriorly but somewhat internally from stage 8 (Figure 7, A−E and E′). Costaining for Asl. and grk*mCherry shows that internally localized grk mRNA in klc mutant oocytes follows the dispersed centrosomes (Figure 7, B, C, and E), either as a consequence of displacing the centrosomes, or because related cues drive grk RNA and centrosomal localization.
Figure 7.
Disrupted centrosome clustering in klc and piRNA pathway mutant oocytes reveals two modes of microtubule organizations involved in grk mRNA localization. Costaining of grk*mCherry (grk) and centrosome components Asterless (Asl, A, B, D-H) or γ-Tubulin (γ-Tub, C, I) in wild-type (A, D, I), and klcsat (B, C, E), maelsat (F), saturn (G), and armi3R13 (H) germline clones. D′-H′ are the enlarged images of selected square in D−H, respectively. Centrosome components are dispersed both in klc (B, C, and E) and piRNA pathway mutants (mael in F′ and armi in H′) and are internally misplaced in klc mutant, like grk mRNA (E). Arrows indicate faint centrosomes deriving from follicle cells (E). Centrosome components are more dispersed in saturn (G′) oocyte than in klc (E’) and mael (F′) mutant oocytes alone. (I) Snapshots of stage 7 wild-type egg-chambers, showing that dorsal-anterior localization of grk mRNA precedes the centrosome migration. Scale bars = 10 μm.
Centrosomes are also less tightly clustered in maelsat oocytes (Figure 7, F and F′), and in armi3R13 mutant oocytes (Figure 7, H and H′), suggesting that defects in the piRNA pathway also lead to this phenotype. Centrosomes are even more dispersed, and grk mRNA is even less focused in saturn mutant oocytes (Figure 7G′), illustrating that loss of function of klc and defective piRNA pathway have additive effects.
Breakage of dorsoventral asymmetry is associated with centrosome-independent transport
Although most grk transcripts localize around the centrosomes, a significant proportion accumulates in a centrosome-free domain adjacent to the nucleus. In stage 8−9 wild-type oocytes, the centrosome lies dorsal to the nucleus, yet some grk RNA lies anterior to the nucleus (Figure 7D). In klc oocytes, a population of grk mRNA still localizes to the dorsal cortex, away from the centrosomes, which now lie ventral to the nucleus (Figure 7E). Together, these results suggest that a population of grk mRNA is transported on MTs that nucleate from the cortex.
To test the relationship between grk mRNA localization and centrosomes, we focused on stage 7, when the site of grk localization changes from posterior to dorsoanterior (Neuman-Silberberg and Schüpbach 1993). Many wild-type stage 7 oocytes show an intermediate pattern of grk RNA localization in which transcripts lie both dorsoanteriorly and posteriorly (Figure 7I, middle image). We find that the centrosome is still at the posterior in 60% (19/31) of such oocytes, indicating that grk relocalization usually precedes that of the centrosome. Together, these results argue that breakage of radial symmetry is not driven by centrosome migration.
Early, centrosome-independent grk localization reflects asymmetric RNA transport rather than differential anchoring of RNA because BicD, which is involved in transport but not anchoring of grk mRNA (Delanoue et al. 2007), also accumulates dorsoanteriorly (Figure S3B). These results suggest that breakage of radial symmetry and formation of a dorsoventral axis arises from centrosome-independent reorganization of the MT cytoskeleton (see Discussion).
Discussion
Rapid gene identification by SNP mapping combined with WGS
WGS has revolutionized methods for identifying mutational lesions and has been applied to many organisms, including humans (Sarin et al. 2008; Blumenstiel et al. 2009; Irvine et al. 2009; Berger et al. 2012). Such sequencing is readily affordable for Drosophila. A single multiplexed pool of 5−10 mutants yields ~3.3 Gb of mutant sequence data with which we could reassemble >99% of the euchromatic region of chromosome 3L, reducing the cost to about $150 per mutant (unpublished data R. Hayashi and S. Horswell).
By using two-stage recombinational mapping with SNP markers in a single generation, we were also able to map causative molecular lesions very rapidly, thereby rendering unbiased EMS-based genetic screens practical, even for complex phenotypes (Figure 2 and File S3). The first stage uses telomeric and centromeric SNPs to identify recombinants, and to generate linkage data that map the causative locus approximately. A second stage of “digital” mapping with internal SNPs, performed without additional crosses, locates the lesion to a minimal region. A related recombinational strategy using dominant markers and WGS has been proposed by Sapiro et al. (2013). Our strategy would seem to be at least as rapid, requires fewer recombinants, and maps the mutations more accurately. It is also more flexible as it is not prescriptive as to the tester chromosome, which needs only to be unrelated to the mutagenized one (particularly important for phenotypes that are sensitive to genetic background).
SNP mapping also makes it worthwhile following up synthetic phenotypes, as demonstrated by our mapping and analysis of the saturn double-mutant (Figure S2). Such occurrences are surprisingly frequent due to the efficiency of EMS mutagenesis; the phenotypes of two of the four single-allele mutants in our 3L screen result from two interacting mutations, as do approximately one-third of single-allele mutants in the equivalent 3R screen (unpublished data).
Kinesin-1 is required for centrosome clustering and cortical localization
By analyzing klcsat null oocytes, we have shown that klc activity is required for cortical grk mRNA localization and, later, also for efficient dorsoanterior localization of the RNA (Figure 6, C, I, L, and O). BicD is similarly affected in the mutant oocytes, demonstrating that the grk mislocalization is due to alterations of MT cytoarchitecture (Figure 6, M and N). Kinesin-1 is also needed for the clustering that follows the migration of nurse cell centrosomes into the pro-oocyte. Centrosome aggregation is never seen in klc mutant oocytes (Figure 7, B, C, and E), suggesting that klc is required to initiate a cluster rather than only for its stabilization.
The latter phenotype is probably due to reduced dynein activity, which can mediate the sliding of antiparallel MTs that is required for centromere clustering (Goshima et al. 2005; Braun et al. 2009; Fink et al. 2009; Gatlin and Bloom 2010). Evidence for the linkage of kinesin and dynein activities comes from studies showing that mid-stage Khc mutant oocytes have reduced Dhc levels (Loiseau et al. 2010) and that klc embryos are sensitive to reduced dynein activity (Duncan and Warrior 2002). Dynein activity is also altered in piRNA pathway mutants (Navarro et al. 2009), in which centromeric clustering is also impaired (Figure 7, F and H).
Clustering may also depend on mutual tension between centrosomes, the cortex and the nucleus, which is likely to be weakened in klc mutants, as evidenced by the relative displacement of the dispersed centrosomes away from the cortex (Figure 7, B, C, and E). Such tension, generated from MT attachment to the cortex and the kinetochore, is thought to be responsible for the clustering of supernumerary centrosomes in cancer cells (Kwon et al. 2008; Leber et al. 2010).
Centrosome-independent transport of grk mRNA suggests breakage of axial symmetry via MT self-organization
The dorsoanterior localization of grk mRNA and the oocyte nucleus marks the formation of a single organizing center and represents a key breakage of radial symmetry. This center is clearly a major site of MT minus-end nucleation, as marked by BicD staining (Figure S3).
The association between centrosomes and the major domain of grk localization in both wild-type and klc oocytes (Figures 7, A−E) indicates that centrosomes promote nucleation of many of the MTs used to transport grk mRNA. However, a significant proportion of grk transcripts localizes in a distinct domain that is not associated with the centrosomes (anteriorly in wild-type oocytes; dorsally in klc oocytes; Figure 7, D and E), implying that centrosome-independent MTs are also involved in grk mRNA localization. Such acentrosomal MTs could be nucleated by the oocyte cortex and the nucleus. Injection of bicoid transcripts into the oocyte and MT regrowth experiments have suggested that MTs can grow from the anterior and lateral oocyte cortex and from subnuclear regions (Cha et al. 2001; Januschke et al. 2006; Parton et al. 2011) and the dynein component Dynamitin localizes around the oocyte nucleus (Januschke et al. 2002).
These two domains of grk transcript localization largely overlap throughout oogenesis in wild-type oocytes, except during stage 7 when MT reorganization and nuclear migration take place (Figure 7I). The trigger for this repolarization is an unknown signal from the posterior follicle cells (Gonzalez-Reyes and St. Johnston 1998), but it is clearly accompanied by extensive MT remodeling. Zhao et al. (2012) have suggested that the posterior-lying oocyte nucleus is “pushed” anteriorly by centrosomally nucleated MTs. This mechanism accounts for anterior movement of the nucleus but does not explain why it ends up in a corner rather than at the center of the anterior surface or the formation of a single dorsoventral axis. The oocyte centrosome cannot be the basis of such uniqueness because dorsoanterior grk mRNA localization usually precedes anterior centrosome migration (Figure 7I), suggesting that the behavior of noncentrosomal MTs are key to defining the unique axis.
How might noncentrosomal MTs generate a unique dorsoventral axis? Jaramillo et al. (2008) have described a transient phase of dorsoventral patterning in which grk mRNA is partly localized in an anterior ring. To resolve such radial symmetry requires selective stabilization of a single dorsoventral organizer site at the expense of others which, in turn, implies a form of positive feedback that favors the final site.
Such feedback might be driven by MT self-organization, which has been observed in several other contexts (Wasteneys and Ambrose 2009; Mogilner and Craig 2010; Sumino et al. 2012). The oocyte itself is not radially symmetric (due to distortions imposed on its anterior surface by the nurse cells; File S5), and this spatial constraint may restrict the number of dorsoventral MT foci, and might explain why the reduced nurse cell size of elg1 egg-chambers affects grk mRNA localization (Figure 4). Further genetic approaches, as well as direct in vivo visualization of the MT and motor dynamics, should lead to a better understanding of how MT reorganization and anterior nuclear migration are achieved during oogenesis.
Supplementary Material
Acknowledgments
We thank Sally Leevers, Nic Tapon, and Barry Thompson for comments on the manuscript and Nik Matthews and the Advanced Sequencing Facility for the DNA sequencing. This research was supported by Cancer Research UK.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Aroya S. B., Kupiec M., 2005. The Elg1 replication factor C-like complex: a novel guardian of genome stability. DNA Repair (Amst.) 4: 409–417. [DOI] [PubMed] [Google Scholar]
- Berger M. F., Hodis E., Heffernan T. P., Deribe Y. L., Lawrence M. S., et al. , 2012. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 485: 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstiel J. P., Noll A. C., Griffiths J. A., Perera A. G., Walton K. N., et al. , 2009. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics 182: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar J., Huynh J. R., Lopez-Schier H., Gonzalez C., St Johnston D., et al. , 2001. Centrosome migration into the Drosophila oocyte is independent of BicD and egl, and of the organisation of the microtubule cytoskeleton. Development 128: 1889–1897. [DOI] [PubMed] [Google Scholar]
- Braun M., Drummond D. R., Cross R. A., McAinsh A. D., 2009. The kinesin-14 Klp2 organizes microtubules into parallel bundles by an ATP-dependent sorting mechanism. Nat. Cell Biol. 11: 724–730. [DOI] [PubMed] [Google Scholar]
- Brendza R. P., Serbus L. R., Duffy J. B., Saxton W. M., 2000. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science 289: 2120–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock S. L., Ish-Horowicz D., 2001. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature 414: 611–616. [DOI] [PubMed] [Google Scholar]
- Bullock S. L., Ringel I., Ish-Horowicz D., Lukavsky P. J., 2010. A’-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat. Struct. Mol. Biol. 17: 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha B. J., Koppetsch B. S., Theurkauf W. E., 2001. In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell 106: 35–46. [DOI] [PubMed] [Google Scholar]
- Cha B. J., Serbus L. R., Koppetsch B. S., Theurkauf W. E., 2002. Kinesin I-dependent cortical exclusion restricts pole plasm to the oocyte posterior. Nat. Cell Biol. 4: 592–598. [DOI] [PubMed] [Google Scholar]
- Chamoun Z., Mann R. K., Nellen D., von Kessler D. P., Bellotto M., et al. , 2001. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science 293: 2080–2084. [DOI] [PubMed] [Google Scholar]
- Chou T. B., Noll E., Perrimon N., 1993. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development 119: 1359–1369. [DOI] [PubMed] [Google Scholar]
- Clark A., Meignin C., Davis I., 2007. A Dynein-dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte. Development 134: 1955–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg N. J., Frost D. M., Larkin M. K., Subrahmanyan L., Bryant Z., et al. , 1997. maelstrom is required for an early step in the establishment of Drosophila oocyte polarity: posterior localization of grk mRNA. Development 124: 4661–4671. [DOI] [PubMed] [Google Scholar]
- Cook H. A., Koppetsch B. S., Wu J., Theurkauf W. E., 2004. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 116: 817–829. [DOI] [PubMed] [Google Scholar]
- Cooley L., Verheyen E., Ayers K., 1992. chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophila oogenesis. Cell 69: 173–184. [DOI] [PubMed] [Google Scholar]
- Cooper J. L., Greene E. A., Till B. J., Codomo C. A., Wakimoto B. T., et al. , 2008. Retention of induced mutations in a Drosophila reverse-genetic resource. Genetics 180: 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D. N., Chao A., Baker J., Chang L., Qiao D., et al. , 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12: 3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dej K. J., Spradling A. C., 1999. The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development 126: 293–303. [DOI] [PubMed] [Google Scholar]
- Delanoue R., Herpers B., Soetaert J., Davis I., Rabouille C., 2007. Drosophila Squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev. Cell 13: 523–538. [DOI] [PubMed] [Google Scholar]
- Dienstbier M., Boehl F., Li X., Bullock S. L., 2009. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 23: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan J. E., Warrior R., 2002. The cytoplasmic dynein and kinesin motors have interdependent roles in patterning the Drosophila oocyte. Curr. Biol. 12: 1982–1991. [DOI] [PubMed] [Google Scholar]
- Favaloro V., Vilardi F., Schlecht R., Mayer M. P., Dobberstein B., 2010. Asna1/TRC40-mediated membrane insertion of tail-anchored proteins. J. Cell Sci. 123: 1522–1530. [DOI] [PubMed] [Google Scholar]
- Findley S. D., Tamanaha M., Clegg N. J., Ruohola-Baker H., 2003. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development 130: 859–871. [DOI] [PubMed] [Google Scholar]
- Fink G., Hajdo L., Skowronek K. J., Reuther C., Kasprzak A. A., et al. , 2009. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat. Cell Biol. 11: 717–723. [DOI] [PubMed] [Google Scholar]
- Gatlin J. C., Bloom K., 2010. Microtubule motors in eukaryotic spindle assembly and maintenance. Semin. Cell Dev. Biol. 21: 248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindhart J. G., Jr, Desai C. J., Beushausen S., Zinn K., Goldstein L. S., 1998. Kinesin light chains are essential for axonal transport in Drosophila. J. Cell Biol. 141: 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reyes A., Elliott H., St. Johnston D., 1995. Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature 375: 654–658. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes A., St. Johnston D., 1998. Patterning of the follicle cell epithelium along the anterior-posterior axis during Drosophila oogenesis. Development 125: 2837–2846. [DOI] [PubMed] [Google Scholar]
- Goshima G., Nedelec F., Vale R. D., 2005. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol. 171: 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine D. V., Goto D. B., Vaughn M. W., Nakaseko Y., McCombie W. R., et al. , 2009. Mapping epigenetic mutations in fission yeast using whole-genome next-generation sequencing. Genome Res. 19: 1077–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N., Rubin G. M., 1999. gigas, a Drosophila homolog of tuberous sclerosis gene product-2, regulates the cell cycle. Cell 96: 529–539. [DOI] [PubMed] [Google Scholar]
- Januschke J., Gervais L., Dass S., Kaltschmidt J. A., Lopez-Schier H., et al. , 2002. Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr. Biol. 12: 1971–1981. [DOI] [PubMed] [Google Scholar]
- Januschke J., Gervais L., Gillet L., Keryer G., Bornens M., et al. , 2006. The centrosome-nucleus complex and microtubule organization in the Drosophila oocyte. Development 133: 129–139. [DOI] [PubMed] [Google Scholar]
- Jaramillo A. M., Weil T. T., Goodhouse J., Gavis E. R., Schüpbach T., 2008. The dynamics of fluorescently labeled endogenous gurken mRNA in Drosophila. J. Cell Sci. 121: 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica M. S., Moore M. J., 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12: 5–14. [DOI] [PubMed] [Google Scholar]
- Kanellis P., Agyei R., Durocher D., 2003. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr. Biol. 13: 1583–1595. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C., Bratu D. P., McGinnis-Schultz N., Koppetsch B. S., Cook H. A., et al. , 2007. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 12: 45–55. [DOI] [PubMed] [Google Scholar]
- Kwon M., Godinho S. A., Chandhok N. S., Ganem N. J., Azioune A., et al. , 2008. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 22: 2189–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C., McMahon R., Young C., Mackay T. F., Langley C. H., 1998. quemao, a Drosophila bristle locus, encodes geranylgeranyl pyrophosphate synthase. Genetics 149: 1051–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber B., Maier B., Fuchs F., Chi J., Riffel P., et al. , 2010. Proteins required for centrosome clustering in cancer cells. Sci. Transl. Med. 2: 33ra38. [DOI] [PubMed] [Google Scholar]
- Li C., Vagin V. V., Lee S., Xu J., Ma S., et al. , 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. G., Serr M., Edwards K., Ludmann S., Yamamoto D., et al. , 1999. Filamin is required for ring canal assembly and actin organization during Drosophila oogenesis. J. Cell Biol. 146: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau P., Davies T., Williams L. S., Mishima M., Palacios I. M., 2010. Drosophila PAT1 is required for Kinesin-1 to transport cargo and to maximize its motility. Development 137: 2763–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall N., Clark A., MacDougall E., Davis I., 2003. Drosophila gurken (TGFalpha) mRNA localizes as particles that move within the oocyte in two dynein-dependent steps. Dev. Cell 4: 307–319. [DOI] [PubMed] [Google Scholar]
- Malone C. D., Brennecke J., Dus M., Stark A., McCombie W. R., et al. , 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K. C., Ephrussi A., 2009. mRNA localization: gene expression in the spatial dimension. Cell 136: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney R. J., Vershinin M., Kunwar A., Vallee R. B., Gross S. P., 2010. LIS1 and NudE induce a persistent dynein force-producing state. Cell 141: 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mische S., Li M., Serr M., Hays T. S., 2007. Direct observation of regulated ribonucleoprotein transport across the nurse cell/oocyte boundary. Mol. Biol. Cell 18: 2254–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S., Crosby M. A., Mungall C. J., Matthews B. B., Campbell K. S., et al. , 2002. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 3: RESEARCH0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura G. I., Buglino J., Alvarado D., Lemmon M. A., Resh M. D., et al. , 2006. Palmitoylation of the EGFR ligand Spitz by Rasp increases Spitz activity by restricting its diffusion. Dev. Cell 10: 167–176. [DOI] [PubMed] [Google Scholar]
- Mogilner A., Craig E., 2010. Towards a quantitative understanding of mitotic spindle assembly and mechanics. J. Cell Sci. 123: 3435–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A. M., Montell D. J., 1996. Cell type-specific roles for Cdc42, Rac, and RhoL in Drosophila oogenesis. J. Cell Biol. 133: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C., Puthalakath H., Adams J. M., Strasser A., Lehmann R., 2004. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat. Cell Biol. 6: 427–435. [DOI] [PubMed] [Google Scholar]
- Navarro C., Bullock S., Lehmann R., 2009. Altered dynein-dependent transport in piRNA pathway mutants. Proc. Natl. Acad. Sci. USA 106: 9691–9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman-Silberberg F. S., Schüpbach T., 1993. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell 75: 165–174. [PubMed] [Google Scholar]
- Pane A., Wehr K., Schüpbach T., 2007. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell 12: 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R. M., Hamilton R. S., Ball G., Yang L., Cullen C. F., et al. , 2011. A PAR-1-dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte. J. Cell Biol. 194: 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter C. J., Huang H., Xu T., 2001. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 105: 357–368. [DOI] [PubMed] [Google Scholar]
- Qi H., Watanabe T., Ku H. Y., Liu N., Zhong M., et al. , 2011. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J. Biol. Chem. 286: 3789–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Ishizu H., Komai M., Kotani H., Kawamura Y., et al. , 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 24: 2493–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapiro A. L., Ihry R. J., Buhr D. L., Konieczko K. M., Ives S. M., et al. , 2013. Rapid recombination mapping for high-throughput genetic screens in Drosophila. G3 (Bethesda) 3: 2313–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S., Prabhu S., O’Meara M. M., Pe’er I., Hobert O., 2008. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat. Methods 5: 865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T., Roth S., 1994. Dorsoventral patterning in Drosophila oogenesis. Curr. Opin. Genet. Dev. 4: 502–507. [DOI] [PubMed] [Google Scholar]
- Serano T. L., Cheung H. K., Frank L. H., Cohen R. S., 1994. P element transformation vectors for studying Drosophila melanogaster oogenesis and early embryogenesis. Gene 138: 181–186. [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., et al. , 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572. [DOI] [PubMed] [Google Scholar]
- Shu T., Ayala R., Nguyen M. D., Xie Z., Gleeson J. G., et al. , 2004. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron 44: 263–277. [DOI] [PubMed] [Google Scholar]
- Sikdar N., Banerjee S., Lee K. Y., Wincovitch S., Pak E., et al. , 2009. DNA damage responses by human ELG1 in S phase are important to maintain genomic integrity. Cell Cycle 8: 3199–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Johnston D., 2005. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 6: 363–375. [DOI] [PubMed] [Google Scholar]
- Sumino Y., Nagai K. H., Shitaka Y., Tanaka D., Yoshikawa K., et al. , 2012. Large-scale vortex lattice emerging from collectively moving microtubules. Nature 483: 448–452. [DOI] [PubMed] [Google Scholar]
- Tapon N., Ito N., Dickson B. J., Treisman J. E., Hariharan I. K., 2001. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105: 345–355. [DOI] [PubMed] [Google Scholar]
- Theurkauf W. E., Smiley S., Wong M. L., Alberts B. M., 1992. Reorganization of the cytoskeleton during Drosophila oogenesis: implications for axis specification and intercellular transport. Development 115: 923–936. [DOI] [PubMed] [Google Scholar]
- Theurkauf W. E., Alberts B. M., Jan Y. N., Jongens T. A., 1993. A central role for microtubules in the differentiation of Drosophila oocytes. Development 118: 1169–1180. [DOI] [PubMed] [Google Scholar]
- Venken K. J., Carlson J. W., Schulze K. L., Pan H., He Y., et al. , 2009. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6: 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainman A., Creque J., Williams B., Williams E. V., Bonaccorsi S., et al. , 2009. Roles of the Drosophila NudE protein in kinetochore function and centrosome migration. J. Cell Sci. 122: 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys G. O., Ambrose J. C., 2009. Spatial organization of plant cortical microtubules: close encounters of the 2D kind. Trends Cell Biol. 19: 62–71. [DOI] [PubMed] [Google Scholar]
- Weng L., Zhu C., Xu J., Du W., 2003. Critical role of active repression by E2F and Rb proteins in endoreplication during Drosophila development. EMBO J. 22: 3865–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C. L., Urlaub H., Achsel T., Gentzel M., Wilm M., et al. , 2002. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 21: 4978–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamparini A. L., Davis M. Y., Malone C. D., Vieira E., Zavadil J., et al. , 2011. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development 138: 4039–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Graham O. S., Raposo A., St. Johnston D., 2012. Growing microtubules push the oocyte nucleus to polarize the Drosophila dorsal-ventral axis. Science 336: 999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.