Abstract
The overproduction and secretion of inositol (i.e., Opi−) phenotype is associated with defects in regulation of phospholipid biosynthesis in yeast. Here we report a screen of the essential yeast gene set using a conditional-expression library. This screen identified novel functions previously unknown to affect phospholipid synthesis.
Keywords: phospholipid synthesis, transcription, regulation, yeast, inositol
Transcription of the phospholipid biosynthetic structural genes in Saccharomyces cerevisiae is regulated by inositol and choline (Carman and Henry 1999; Greenberg and Lopes 1996; Henry et al. 2012; Henry and Patton-Vogt 1998; Jesch et al. 2005; Paltauf et al. 1992; Santiago and Mamoun 2003). Gene expression is maximally repressed in the presence of inositol and choline and derepressed when they are limiting. This regulation requires several transcription factors that when mutated display one of two phenotypes: inositol auxotrophy or overproduction and secretion of inositol (Opi−) (Carman and Han 2009; Greenberg and Lopes 1996; Henry et al. 2012). Some of these mutants were identified during the last three decades through traditional genetic screens. However, we previously reported a genomic screen of the viable yeast deletion set (VYDS) for Opi− mutants that identified 91 mutants (Hancock et al. 2006). Here we report a screen of the essential yeast gene set using a conditional-expression library (Mnaimneh et al. 2004).
Well studied regulators of phospholipid biosynthetic genes include the Ino2p:Ino4p activators, the Opi1p repressor, the Ume6p-Sin3p-Rpd3p histone deacetylase complex (HDAC), the SAGA histone acetyltransferase complex, the ISW2, INO80, SWI/SNF chromatin remodeling complexes, and Mot1p (Ambroziak and Henry 1994; Dasgupta et al. 2005; Elkhaimi et al. 2000; Fazzio et al. 2001; Ford et al. 2008; Jackson and Lopes 1996; Kadosh and Struhl 1997, 1998; Nikoloff and Henry 1994; Rundlett et al. 1996, 1998; Shen et al. 2000; White et al. 1991). Ino2p and Ino4p belong to a family of basic helix-loop-helix regulatory proteins, which form a heterodimer that binds to a UASINO sequence to activate transcription of most phospholipid biosynthetic genes (e.g., INO1, CHO2, and OPI3 in Figure 1) (Jesch et al. 2005; Santiago and Mamoun 2003). The Ume6p-Sin3p-Rpd3p HDAC, the ISW2 and INO80 chromatin remodeling complexes, and Mot1p are global regulators that play a negative role in phospholipid biosynthetic gene expression (Dasgupta et al. 2005; Elkhaimi et al. 2000; Fazzio et al. 2001; Grigat et al. 2012; Jackson and Lopes 1996; Kadosh and Struhl 1997, 1998; Rundlett et al. 1996, 1998; Shen et al. 2000). Opi1p was the first, and to date, the only repressor found that specifically regulates the phospholipid biosynthetic pathway.
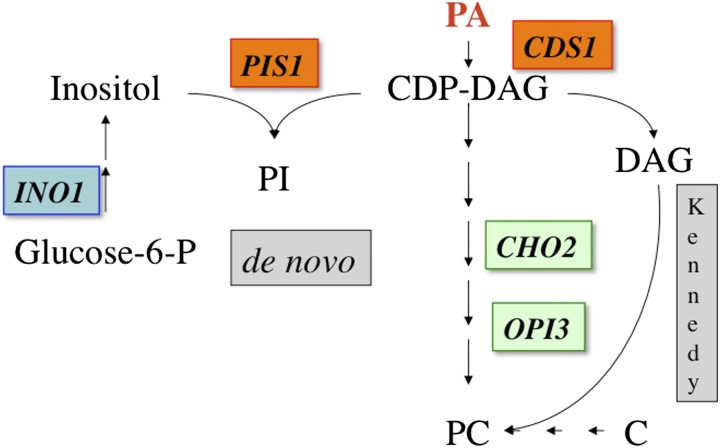
Figure 1.
Abridged yeast phospholipid biosynthetic de novo and Kennedy pathways. Genes encoding biosynthetic enzymes are italicized and boxed. Those genes noted in green and orange are nonessential and essential (respectively) and yield an Opi− phenotype when mutated. PA, phosphatidic acid; CDP-DAG, CDP-diacylglycerol; PI, phosphatidylinositol; PC, phosphatidylcholine; and C, choline
The OPI1 locus was first identified in a screen for mutants that overproduce and excrete inositol into the medium in the absence of inositol (Opi− phenotype) (Greenberg et al. 1982). The original opi1 mutant and a small set of similar mutants identified over the next two decades showed that the Opi− phenotype correlated with a defect in repression of the INO1 gene (Elkhaimi et al. 2000; Hirsch and Henry 1986; Hudak et al. 1994), which is required for inositol synthesis de novo (Figure 1) (Culbertson and Henry 1975). However, most of the 91 Opi− mutants identified in a more recent screen of the VYDS did not affect inositol-mediated repression of an INO1-lacZ reporter (Hancock et al. 2006).
Our current understanding of the mechanism for inositol-mediated repression of phospholipid biosynthetic gene expression is that it requires translocation of Opi1p from the endoplasmic reticulum (ER) to the nucleus. Repression in response to inositol is actually mediated by the level of phosphatidic acid (PA) (Figure 1). In the absence of inositol, PA levels are elevated and Opi1p binds PA (Loewen et al. 2004) and is tethered in the ER by Scs2p, an integral membrane protein (Gavin et al. 2002; Kagiwada and Zen 2003; Loewen et al. 2003, 2004; Loewen and Levine 2005). When inositol is added, phosphatidylinositol synthesis is increased, causing a decrease in PA levels, and Opi1p is released from the ER. Opi1p rapidly translocates to the nucleus, where it interacts with the Ino2p activator and recruits several HDACs to repress transcription. (Gardenour et al. 2004; Grigat et al. 2012; Heyken et al. 2005; Wagner et al. 2001). The addition of choline by itself has little effect on PA levels; however, in combination with inositol, choline further reduces PA levels, resulting in additional repression (Henry and Patton-Vogt 1998). Not surprisingly, blocks in de novo phosphatidylcholine (PC) biosynthesis that elevate PA levels also yield an Opi− phenotype (Klig et al. 1988; McGraw and Henry 1989; Shen and Dowhan 1996; Summers et al. 1988). Thus, cds1, cho2, and opi3 mutants all have the Opi− phenotype (Figure 1). The Opi− phenotype of these mutants is conditional and it can be suppressed by adding choline (i.e., C) to the medium. Choline restores PC synthesis through the Kennedy pathway, thereby alleviating the accumulation of PA caused by the block in the de novo PC pathway (Figure 1) (Henry and Patton-Vogt 1998).
Consistent with the role of PA as the signal, we reported that reduced expression of the PIS1 gene (Figure 1) yields an Opi− phenotype (Jani and Lopes 2009). Because PIS1 is an essential gene, we created a strain harboring a GAL1-PIS1 gene that allowed us to reduce PIS1 gene expression by growth in glucose or low galactose concentrations (Jani and Lopes 2009). These growth conditions reduced phosphatidylinositol levels and PA would therefore increase explaining the Opi− phenotype (Jani and Lopes 2009). These results are consistent with another study showing that GFP-Opi1p translocation into the nucleus is slow and impaired in a pis1 partial function mutant (Loewen et al. 2004).
Many studies have shown that screening the VYDS (Giaever et al. 2002; Winzeler et al. 1999) and an essential yeast mutant gene set (Mnaimneh et al. 2004) can yield valuable insight into well-studied processes such as regulation in response to phosphate concentration (Huang and O’Shea 2005). We previously reported the results of a VYDS screen for the Opi− phenotype to further understand repression of phospholipid biosynthesis (Hancock et al. 2006). That screen identified all seven of the Opi− mutants that had been identified by several labs over the previous 30 years but also identified 84 new Opi− mutants. Highly represented in this mutant set were the components of the Rpd3p HDAC complex and five of the six nonessential components of NuA4 KAT complex (EAF1, EAF3, EAF5, EAF7, and YAF9) (Hancock et al. 2006). The screen also identified the reg1 mutant (Hancock et al. 2006), which was known to regulate gene expression in response to changes in glucose. Early hypotheses suggested a coordination of glucose use and phospholipid synthesis; however, the mechanism for this coordination was unknown. More recently, it was found that the Opi− phenotype of a reg1 mutant is actually due to the altered protonation status of PA, as a function of cellular pH, which affects Opi1 translocation to the nucleus (Young et al. 2010).
It is well established that phospholipid biosynthesis is coordinated with the unfolded protein response (UPR) and that Opi1p plays a role in this coordination (Betz et al. 2002; Cox et al. 1997; Jesch et al. 2005). The UPR is initiated in the ER in response to accumulation of unfolded proteins (Schröder and Kaufman 2005) and is also induced by depleting inositol (Betz et al. 2002; Cox et al. 1997). Upon UPR induction, Ire1p is activated initiating splicing of HAC1 mRNA (Sidrauski and Walter 1997). The spliced HAC1 transcript produces the Hac1p basic leucine zipper transcription factor that binds to the UPR element of genes such as KAR2 but also regulates UASINO containing promoters by counteracting the function of Opi1p (Cox and Walter 1996). Thus, it was predictable that the VYDS Opi− screen identified genes that are known to affect the UPR (L. C. Hancock and J. M. Lopes, unpublished results). Screening the VYDS for the Opi− phenotype provided a wealth of information about other functions that affect regulation of phospholipid synthesis.
Materials and Methods
Strains and growth conditions
This study used the BY4742 (MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ1) wild-type and doxycycline (Dox) titratable strains (Giaever et al. 2002; Mnaimneh et al. 2004; Winzeler et al. 1999). The BRS1005 tester strain is a diploid homozygous for the ino1-13 and ade1 alleles (Hancock et al. 2006). Yeast cultures were grown at 30° in complete synthetic medium (Kelly and Greenberg 1990) containing 2% glucose (w/v) but lacking inositol and choline (I-C-). For the Opi− screen, agarose was reduced to 1.2%, and Dox was added to concentrations noted in the sections to follow.
Results and Discussion
Screen of an essential yeast gene library driven by a titratable promoter identifies 122 Opi− mutants
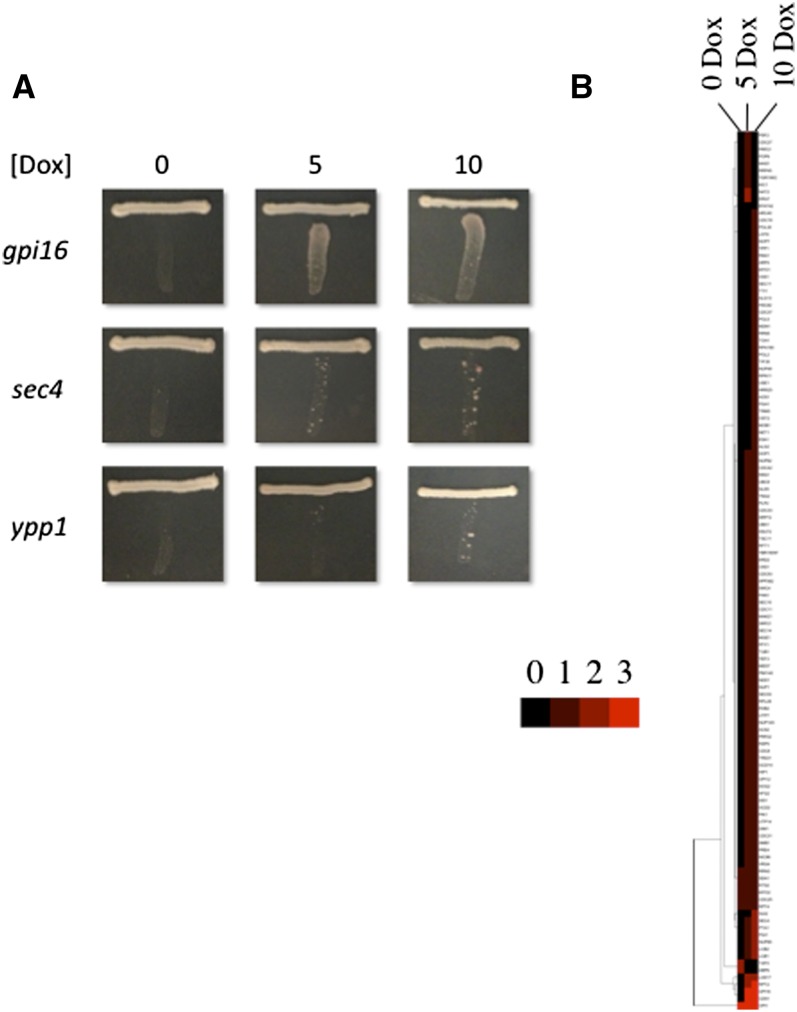
To date there had been no screen of the essential genes for defects in phospholipid synthesis, and it is clear that the essential gene set and VYDS are not identical with respect to the biological processes they affect (Winzeler et al. 1999). Motivated by this and the success of the VYDS Opi− screen, we conducted a screen of an essential gene library driven by a titratable promoter (Mnaimneh et al. 2004). The collection we used contains 838 essential yeast genes driven by a Tet-regulated promoter that is shut off by the addition of Dox. We tested a range of Dox concentrations because different strains have been shown to have differing growth sensitivities (Mnaimneh et al. 2004). Our screen of the VYDS for the Opi− phenotype used a pinning strategy (Hancock et al. 2006), but this strategy did not work for the essential gene collection. Thus, we used a more laborious but also more sensitive screening assay (Figure 2A) (McGee et al. 1994). Briefly, the Tet-driven strain was streaked at the top of plates containing various concentrations of Dox (0, 5, and 10 μg/mL), and lacking inositol and allowed to grow for 1−2 d. A tester strain was then streaked perpendicular to the Tet-driven strain. The tester strain is a diploid homozygous for ino1 and ade1 mutants (Swede et al. 1992). This strain does not normally grow on media lacking inositol because of the ino1 mutation. Thus, the Opi− phenotype is observed if the Tet-driven strain secretes inositol into the growth medium allowing the tester to grow. As inositol levels increase in the media, the tester grows more robustly as a red streak (ade1 phenotype). The tester strain was streaked 3x on each plate and each Tet-driven strain was analyzed in duplicate. The growth of the tester was scored as 0 (no growth), 1, 2, or 3 for progressively varying growth phenotypes. Three researchers independently scored each plate. The screen yielded 122 mutants that all three researchers agreed had a positive test in the two independent assays (Figure 2B and Supporting Information, Table S1). As a control, we included the BY4742 strain (wild type) and an opi1 mutant, which had an Opi− phenotype under all [Dox]. Sometimes the tester will show a papillar pattern rather than a uniform growth pattern (Figure 2A). These are not revertants or a result of rare mating since the tester is homozygous diploid. We have observed this pattern previously and shown that it correlates with a defect in transcription regulation (Elkhaimi et al. 2000; Hancock et al. 2006).
Figure 2.
Essential Opi− mutants. (A) Representative Opi− phenotype for the gpi16 (0,3,3), sec4 (0,1,2), and ypp1 (0,0,1) mutants grown under three Dox concentrations. (B) Mutants were clustered with respect to phenotype severity using Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) and displayed using Java Treeview (Saldanha 2004).
Most of the mutant strains did not display an Opi− phenotype in the absence of Dox but did have the phenotype with increasing [Dox] (Figure 2B). In a few cases the Opi− phenotype was observed at lower [Dox] but not at higher [Dox] (top of Figure 2B). This was because the mutant strains did not grow at the higher [Dox]. In a couple of cases the mutant strain yielded an Opi− phenotype in the absence of Dox and did not grow in the presence of Dox (bottom of Figure 2B). These may be false positives or they may result from reduced expression from the Tet promoter (in the absence of Dox) relative to the native promoter and lethality when expression is further reduced by the addition of Dox. As expected, the screen identified the cds1 mutant which is the only essential gene previously shown to yield an Opi− phenotype (the aforementioned pis1 allele was not present in the collection) (Klig et al. 1988; Shen and Dowhan 1996). The screen also identified five mutants that are duplicated in the collection (use1, cks1, rpn11, sec4, and vrg4). These results suggest that the screen was successful in identifying legitimate Opi− mutants. We should also note that four of the Opi− mutants (YNG2, HSC82, KIC1, and SMB1) are actually not classified as essential in the Saccharomyces Genome Database (http://www.yeastgenome.org/). Regardless of this fact, down-regulation did yield an Opi− phenotype so these mutants are retained in our dataset.
The essential gene and VYDS screens identified mutants in different sets of biological processes
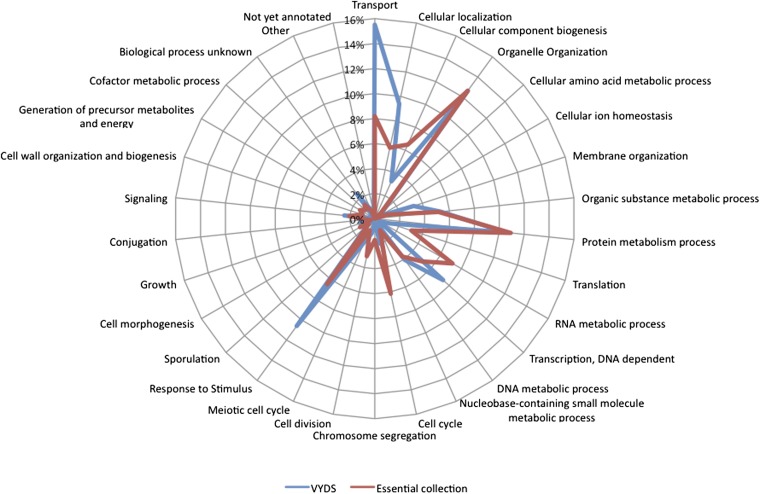
We predicted that the screen might reveal novel processes compared to the VYDS screen. To test this the mutants were grouped based on biological processes using the SGD Yeast Go Slim Mapper software (http://www.yeastgenome.org/cgi-bin/GO/goSlimMapper.pl). The results clearly showed that the two screens yielded different information with respect to biological processes (Figure 3). The essential mutant collection yielded significantly more mutants affecting RNA metabolic processes, the cell cycle, and cell division whereas the VYDS screen identified more mutants in transport, cellular localization, transcription, and response to stimulus.
Figure 3.
Radar chart comparing percentage of Opi− mutants in different biological processes for the VYDS (blue) and essential (red) mutant collections. Each point on the graph represents the percentage of mutants within each of the Opi− mutant sets in each functional category.
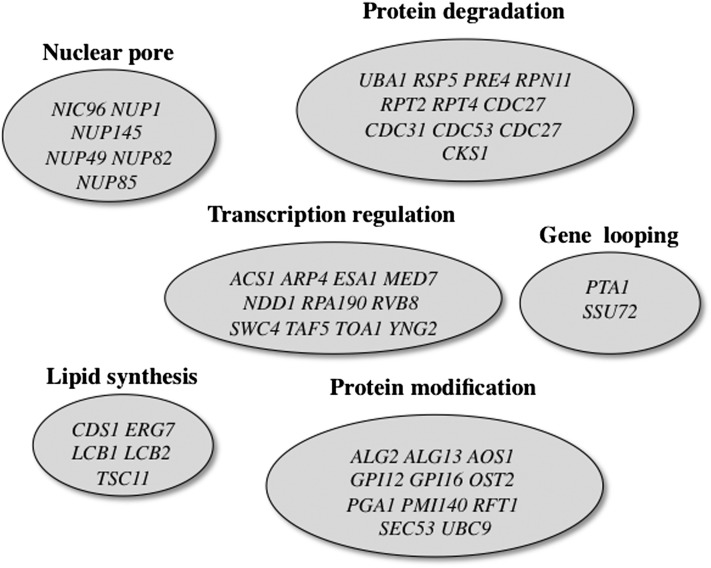
Consistent with the results from the VYDS screen and the coordination of phospholipid biosynthesis with the UPR, the current screen identified several mutants that affect protein modifications (Figure 4 and Table S1). These include several genes that glycosylate proteins in the ER (ALG2, ALG13, OST2, PMI40, RFT1, and SEC53). The screen also identified several genes required for synthesis of glycosylphosphatidylinositol anchors (GPI12, GPI12, and PGA1) and for sphingolipid synthesis (LCB1, LCB2, and TSC11) (Figure 4 and Table S1). This is the first report linking these two processes to phospholipid synthesis.
Figure 4.
Opi− mutants cluster by functional categories. Shown are those cases in which a significant set of mutants affected a biological function.
Expression of the INO1 gene is affected by a mechanism that involves both gene looping and association of the INO1 promoter with the nuclear pore complex (Brickner 2010; Kerr and Corbett 2010). Interestingly, mutants that affect both gene looping and nuclear pore complex were identified in the Opi− mutant screen (Figure 4 and Table S1). Both the pta1 and ssu72 mutants were identified in the essential gene screen. These proteins have been previously shown to be required for gene looping (promoter-terminator) of the INO1 gene (Ansari and Hampsey 2005). It is not immediately obvious why they should also have an Opi− phenotype but this does provide the first phenotype for gene looping. A significant number of nuclear pore complex mutants (Aitchison and Rout 2012) were identified in the two screens. The VYDS screen identified NUP84 whereas the essential gene screen identified NIC96, NUP1, NUP49, NUP82, NUP85, and NUP145. On activation, the INO1 promoter is recruited to the nuclear pore complex via cis sequences called DNA Zip Codes (GRS1 and II) within the INO1 promoter and the adjoining SNA3 ORF (Ahmed et al. 2010; Light et al. 2010). Upon transfer to repressing conditions, the INO1 promoter remains associated with the nuclear periphery for up to three to four generations (Brickner et al. 2007). This association is a mechanism for transcriptional memory of recently repressed INO1 transcription (Brickner et al. 2007; Light et al. 2010). This memory requires an 11-bp sequence, the memory recruitment sequence, within the INO1 promoter (Light et al. 2010). Importantly, both recruitment to the periphery and transcriptional memory involve distinct mechanisms with different cis elements and nuclear pore components, including the Nup1p, Nup84p, Nup145p, and Nic96p subunits (Light et al. 2010). Thus, identification of nuclear pore complex mutants in the Opi− screens is consistent with its role in recruiting and regulating the INO1 promoter.
The essential gene Opi− screen identified several interesting mutants in biological processes that were not identified in the VYDS screen. There was an overrepresentation of mutants in the ubiquitin-mediated degradation pathway (Figure 4 and Table S1). This included the UBA1 and RSP5 genes that encode E1 and E3 ubiquitinating enzymes (Kerscher et al. 2006). Interestingly, an rsp5 mutant has been shown to affect expression of an INO1-lacZ reporter under derepressing conditions (Kaliszewski et al. 2006). The screen also identified several genes required for proteasome function (Forster et al. 2010; Tomko and Hochstrasser 2011), including the PRE4 gene that is required for assembly of the 20S proteolytic core particle; the RPN11 gene that encodes a deubiquitylase present in the lid of the 19S regulatory particle (Guterman and Glickman 2004); and the RPT2 and RPT4 genes that are required for unfolding and translocating the protein substrates as well as opening of the proteasome gate (RPT2) (Forster et al. 2010; Tomko and Hochstrasser 2011). Another protein modification pathway that was illuminated by the screen is that of an ubiquitin-like modification, SUMO. The screen identified both E1 (AOS1) and E2 (UBC9) encoding genes (Figure 4 and Table S1) (Johnson 2004; Kerscher et al. 2006). This finding is consistent with recent published work showing that a mutation in a deubiquitylation enzyme (ULP2) affects INO1 expression under derepressing conditions by altering the sumoylation status of Scs2p, which normally retains Opi1p in the ER under derepressing conditions (Felberbaum et al. 2012).
Both Opi− screens identified subunits of the NuA4 HAT complex
We previously reported that the VYDS screen identified five of the six nonessential subunits of the NuA4 KAT complex (Hancock et al. 2006). The essential collection screen also identified three of the six essential subunits (ARP4, ESA1, and SWC4) (Note: YNG2 is included in the collection but is not essential.) (Figure 4). One of the essential subunits (ACT1) was not present in the collection. Our screen identified ESA1, which encodes the KAT activity and contains a chromodomain that interacts with methylated histones as well as YNG2, which contains a PHD domain that also interacts with methylated histones (Schulze et al. 2010). Thus, both screens collectively identified nine of the possible 12 NuA4 subunits.
It is possible that the proteasome and NuA4 complexes may regulate INO1 gene expression via a direct role since it has been shown that a 19S proteasome subcomplex works with NuA4 to regulate expression of ribosomal protein genes (Uprety et al. 2012). However, the finding that mutations in the 20S complex and the ubiquitin modification pathway yield an Opi− phenotype suggests that protein degradation is the more likely explanation for the phenotype. With respect to the NuA4 complex it is interesting that it functions in activation of gene expression while mutants in other transcription factors that also yield the Opi− phenotype (e.g., opi1, ume6, sin3, and rpd3) function in repression (Doyon and Cote 2004; Hancock et al. 2006; Schulze et al. 2010). In the case of the nonessential Opi− mutants, the mutants yielded elevated expression of the INO1 target gene under both repressing and derepressing growth conditions, that is, they had a defect in repression (Hancock et al. 2006). A trivial explanation for this would be that NuA4 affects repression of INO1 indirectly by controlling the activation of the OPI1 repressor gene. However, we found that these mutants did not affect activation of the OPI1 gene (Hancock et al. 2006). Moreover, there is evidence that NuA4 binds the INO1 promoter (Konarzewska et al. 2012). It is also important to note that some of the subunits of the NuA4 complex are shared with the SWR-C complex that is responsible for loading the modified H2A.Z into nucleosomes and H2A.Z is involved in regulation of INO1 (Lu et al. 2009). However, none of the SWR-C−specific components were identified in our screen suggesting that the Opi− phenotype is specific to the NuA4 complex. A more likely explanation is that NuA4 may be acetylating a non-histone regulatory protein that controls INO1 expression. Consistent with this, an in vitro protein acetylation microarray identified many non-histone targets of NuA4 (Lin et al. 2009). Along this line it is important that another HAT, Gcn5p, acetylates the Ume6p regulatory protein, which targets it for degradation via the anaphase-promoting complex/cyclosome ubiquitin ligase (Mallory et al. 2007, 2012). This occurs as cells are initiating the meiotic program. Consistent with this model the essential gene screen did identify the CDC27, which is a component of the anaphase-promoting complex/cyclosome (Figure 4 and Table S1). Although INO1 is not a meiotic gene, it is regulated by Ume6p and its associated Sin3p/Rpd3 complex (Eiznhamer et al. 2001; Elkhaimi et al. 2000; Hudak et al. 1994; Jackson and Lopes 1996; Kaadige and Lopes 2003; Kadosh and Struhl 1997, 1998). Thus, NuA4 could be regulating INO1 either through Opi1p or Ume6p via a mechanism that includes protein degradation. Future experiments will address this possibility.
Supplementary Material
Acknowledgments
We thank Aishwarya Swaminathan and Isabel Chang for discussions and comments on the manuscript. This work was supported by a National Science Foundation (NSF) Grant MCB-1020987 to J.M.L. B.S-S. was also supported by the Summer Program for Undergraduate Research (SPUR) as part of the Northeast Alliance for Graduate Education and the Professoriate Program (NEAGEP) (NSF grant 0450339) and a National Institutes of Health PREP grant R25GM086264.
Footnotes
Communicating editor: C. S. Hoffman
Literature Cited
- Ahmed S., Brickner D. G., Light W. H., Cajigas I., McDonough M., et al. , 2010. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol. 12: 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison J. D., Rout M. P., 2012. The yeast nuclear pore complex and transport through it. Genetics 190: 855–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambroziak J., Henry S. A., 1994. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J. Biol. Chem. 269: 15344–15349. [PubMed] [Google Scholar]
- Ansari A., Hampsey M., 2005. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 19: 2969–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz J. L., Chang M., Washburn T. M., Porter S. E., Mueller C. L., et al. , 2002. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics 268: 272–285. [DOI] [PubMed] [Google Scholar]
- Brickner D. G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P. C., et al. , 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5: e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner J. H., 2010. Transcriptional memory: staying in the loop. Curr. Biol. 20: R20–R21. [DOI] [PubMed] [Google Scholar]
- Carman G. M., Han G. S., 2009. Regulation of phospholipid synthesis in yeast. J. Lipid Res. 50(Suppl): S69–S73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G. M., Henry S. A., 1999. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid Res. 38: 361–399. [DOI] [PubMed] [Google Scholar]
- Cox J. S., Walter P., 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87: 391–404. [DOI] [PubMed] [Google Scholar]
- Cox J. S., Chapman R. E., Walter P., 1997. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 8: 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M. R., Henry S. A., 1975. Inositol-requiring mutants of Saccharomyces cerevisiae. Genetics 80: 23–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A., Juedes S. A., Sprouse R. O., Auble D. T., 2005. Mot1-mediated control of transcription complex assembly and activity. EMBO J. 24: 1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y., Cote J., 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 14: 147–154. [DOI] [PubMed] [Google Scholar]
- Eiznhamer D. A., Ashburner B. P., Jackson J. C., Gardenour K. R., Lopes J. M., 2001. Expression of the INO2 regulatory gene of Saccharomyces cerevisiae is controlled by positive and negative promoter elements and an upstream open reading frame. Mol. Microbiol. 39: 1395–1405. [PubMed] [Google Scholar]
- Elkhaimi M., Kaadige M. R., Kamath D., Jackson J. C., Biliran H., Jr, et al. , 2000. Combinatorial regulation of phospholipid biosynthetic gene expression by the UME6, SIN3 and RPD3 genes. Nucleic Acids Res. 28: 3160–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio T. G., Kooperberg C., Goldmark J. P., Neal C., Basom R., et al. , 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 21: 6450–6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felberbaum R., Wilson N. R., Cheng D., Peng J., Hochstrasser M., 2012. Desumoylation of the endoplasmic reticulum membrane VAP family protein Scs2 by Ulp1 and SUMO regulation of the inositol synthesis pathway. Mol. Cell. Biol. 32: 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J., Odeyale O., Shen C. H., 2008. Activator-dependent recruitment of SWI/SNF and INO80 during INO1 activation. Biochem. Biophys. Res. Commun. 373: 602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster F., Lasker K., Nickell S., Sali A., Baumeister W., 2010. Toward an integrated structural model of the 26S proteasome. Mol. Cell. Proteomics 9: 1666–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardenour K. R., Levy J., Lopes J. M., 2004. Identification of novel dominant INO2c mutants with an Opi-phenotype. Mol. Microbiol. 52: 1271–1280. [DOI] [PubMed] [Google Scholar]
- Gavin A. C., Bosche M., Krause R., Grandi P., Marzioch M., et al. , 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Greenberg M. L., Lopes J. M., 1996. Genetic regulation of phospholipid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev. 60: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M. L., Reiner B., Henry S. A., 1982. Regulatory mutations of inositol biosynthesis in yeast: isolation of inositol-excreting mutants. Genetics 100: 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigat M., Jaschke Y., Kliewe F., Pfeifer M., Walz S., et al. , 2012. Multiple histone deacetylases are recruited by corepressor Sin3 and contribute to gene repression mediated by Opi1 regulator of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Mol. Genet. Genomics 287: 461–472. [DOI] [PubMed] [Google Scholar]
- Guterman A., Glickman M. H., 2004. Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J. Biol. Chem. 279: 1729–1738. [DOI] [PubMed] [Google Scholar]
- Hancock L. C., Behta R. P., Lopes J. M., 2006. Genomic analysis of the Opi- phenotype. Genetics 173: 621–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry S. A., Patton-Vogt J. L., 1998. Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog. Nucleic Acid Res. Mol. Biol. 61: 133–179. [DOI] [PubMed] [Google Scholar]
- Henry S. A., Kohlwein S. D., Carman G. M., 2012. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190: 317–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyken W. T., Repenning A., Kumme J., Schuller H. J., 2005. Constitutive expression of yeast phospholipid biosynthetic genes by variants of Ino2 activator defective for interaction with Opi1 repressor. Mol. Microbiol. 56: 696–707. [DOI] [PubMed] [Google Scholar]
- Hirsch J. P., Henry S. A., 1986. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol. Cell. Biol. 6: 3320–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., O’Shea E. K., 2005. A systematic high-throughput screen of a yeast deletion collection for mutants defective in PHO5 regulation. Genetics 169: 1859–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudak K. A., Lopes J. M., Henry S. A., 1994. A pleiotropic phospholipid biosynthetic regulatory mutation in Saccharomyces cerevisiae is allelic to sin3 (sdi1, ume4, rpd1). Genetics 136: 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. C., Lopes J. M., 1996. The yeast UME6 gene is required for both negative and positive transcriptional regulation of phospholipid biosynthetic gene expression. Nucleic Acids Res. 24: 1322–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani N. M., Lopes J. M., 2009. Regulated transcription of the Saccharomyces cerevisiae phosphatidylinositol biosynthetic gene, PIS1, yields pleiotropic effects on phospholipid synthesis. FEMS Yeast Res. 9: 552–564. [DOI] [PubMed] [Google Scholar]
- Jesch S. A., Zhao X., Wells M. T., Henry S. A., 2005. Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J. Biol. Chem. 280: 9106–9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. S., 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73: 355–382. [DOI] [PubMed] [Google Scholar]
- Kaadige M. R., Lopes J. M., 2003. Opi1p, Ume6p and Sin3p control expression from the promoter of the INO2 regulatory gene via a novel regulatory cascade. Mol. Microbiol. 48: 823–832. [DOI] [PubMed] [Google Scholar]
- Kadosh D., Struhl K., 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89: 365–371. [DOI] [PubMed] [Google Scholar]
- Kadosh D., Struhl K., 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18: 5121–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiwada S., Zen R., 2003. Role of the yeast VAP homolog, Scs2p, in INO1 expression and phospholipid metabolism. J. Biochem. 133: 515–522. [DOI] [PubMed] [Google Scholar]
- Kaliszewski P., Ferreira T., Gajewska B., Szkopinska A., Berges T., et al. , 2006. Enhanced levels of Pis1p (phosphatidylinositol synthase) improve the growth of Saccharomyces cerevisiae cells deficient in Rsp5 ubiquitin ligase. Biochem. J. 395: 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly B. L., Greenberg M. L., 1990. Characterization and regulation of phosphatidylglycerolphosphate phosphatase in Saccharomyces cerevisiae. Biochim. Biophys. Acta 146: 144–150. [DOI] [PubMed] [Google Scholar]
- Kerr S. C., Corbett A. H., 2010. Should INO stay or should INO Go: a DNA “zip code” mediates gene retention at the nuclear pore. Mol. Cell 40: 3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O., Felberbaum R., Hochstrasser M., 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22: 159–180. [DOI] [PubMed] [Google Scholar]
- Klig L. S., Homann M. J., Kohlwein S. D., Kelley M. J., Henry S. A., et al. , 1988. Saccharomyces cerevisiae mutant with a partial defect in the synthesis of CDP-diacylglycerol and altered regulation of phospholipid biosynthesis. J. Bacteriol. 170: 1878–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarzewska P., Esposito M., Shen C. H., 2012. INO1 induction requires chromatin remodelers Ino80p and Snf2p but not the histone acetylases. Biochem. Biophys. Res. Commun. 418: 483–488. [DOI] [PubMed] [Google Scholar]
- Light W. H., Brickner D. G., Brand V. R., Brickner J. H., 2010. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol. Cell 40: 112–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. Y., Lu J. Y., Zhang J., Walter W., Dang W., et al. , 2009. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell 136: 1073–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C. J., Levine T. P., 2005. A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J. Biol. Chem. 280: 14097–14104. [DOI] [PubMed] [Google Scholar]
- Loewen C. J., Roy A., Levine T. P., 2003. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22: 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C. J., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., et al. , 2004. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304: 1644–1647. [DOI] [PubMed] [Google Scholar]
- Lu P. Y., Levesque N., Kobor M. S., 2009. NuA4 and SWR1-C: two chromatin-modifying complexes with overlapping functions and components. Biochem. Cell Biol. 87: 799–815. [DOI] [PubMed] [Google Scholar]
- Mallory M. J., Cooper K. F., Strich R., 2007. Meiosis-specific destruction of the Ume6p repressor by the Cdc20-directed APC/C. Mol. Cell 27: 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory M. J., Law M. J., Sterner D. E., Berger S. L., Strich R., 2012. Gcn5p-dependent acetylation induces degradation of the meiotic transcriptional repressor Ume6p. Mol. Biol. Cell 23: 1609–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee T. P., Skinner H. B., Bankaitis V. A., 1994. Functional redundancy of CDP-ethanolamine and CDP-choline pathway enzymes in phospholipid biosynthesis: ethanolamine-dependent effects on steady-state membrane phospholipid composition in Saccharomyces cerevisiae. J. Bacteriol. 176: 6861–6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw P., Henry S. A., 1989. Mutations in the Saccharomyces cerevisiae opi3 gene: effects on phospholipid methylation, growth and cross-pathway regulation of inositol synthesis. Genetics 122: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S., Davierwala A. P., Haynes J., Moffat J., Peng W. T., et al. , 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118: 31–44. [DOI] [PubMed] [Google Scholar]
- Nikoloff D. M., Henry S. A., 1994. Functional characterization of the INO2 gene of Saccharomyces cerevisiae. A positive regulator of phospholipid biosynthesis. J. Biol. Chem. 269: 7402–7411. [PubMed] [Google Scholar]
- Paltauf F., Kohlwein S. D., Henry S. A., 1992. Regulation and compartmentalization of lipid synthesis in yeast, pp. 415–499 in The Molecular Biology of the Yeast Saccharomyces cerevisiae: Gene Expression, edited by Jones E. W., Pringle J. R., Broach J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Rundlett S. E., Carmen A. A., Kobayashi R., Bavykin S., Turner B. M., et al. , 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93: 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett S. E., Carmen A. A., Suka N., Turner B. M., Grunstein M., 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392: 831–835. [DOI] [PubMed] [Google Scholar]
- Saldanha A. J., 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- Santiago T. C., Mamoun C. B., 2003. Genome expression analysis in yeast reveals novel transcriptional regulation by inositol and choline and new regulatory functions for Opi1p, Ino2p, and Ino4p. J. Biol. Chem. 278: 38723–38730. [DOI] [PubMed] [Google Scholar]
- Schröder M., Kaufman R. J., 2005. ER stress and the unfolded protein response. Mutat. Res. 569: 29–63. [DOI] [PubMed] [Google Scholar]
- Schulze J. M., Wang A. Y., Kobor M. S., 2010. Reading chromatin: insights from yeast into YEATS domain structure and function. Epigenetics 5: 573–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Dowhan W., 1996. Reduction of CDP-diacylglycerol synthase activity results in the excretion of inositol by Saccharomyces cerevisiae. J. Biol. Chem. 271: 29043–29048. [DOI] [PubMed] [Google Scholar]
- Shen X., Mizuguchi G., Hamiche A., Wu C., 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406: 541–544. [DOI] [PubMed] [Google Scholar]
- Sidrauski C., Walter P., 1997. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90: 1031–1039. [DOI] [PubMed] [Google Scholar]
- Summers E. F., Letts V. A., McGraw P., Henry S. A., 1988. Saccharomyces cerevisiae cho2 mutants are deficient in phospholipid methylation and cross-pathway regulation of inositol synthesis. Genetics 120: 909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swede M. J., Hudak K. A., Lopes J. M., Henry S. A., 1992. Strategies for generating phospholipid synthesis mutants in yeast. Methods Enzymol. 209: 21–34. [DOI] [PubMed] [Google Scholar]
- Tomko R. J., Jr, Hochstrasser M., 2011. Order of the proteasomal ATPases and eukaryotic proteasome assembly. Cell Biochem. Biophys. 60: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uprety B., Lahudkar S., Malik S., Bhaumik S. R., 2012. The 19S proteasome subcomplex promotes the targeting of NuA4 HAT to the promoters of ribosomal protein genes to facilitate the recruitment of TFIID for transcriptional initiation in vivo. Nucleic Acids Res. 40: 1969–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C., Dietz M., Wittmann J., Albrecht A., Schuller H. J., 2001. The negative regulator Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol. Microbiol. 41: 155–166. [DOI] [PubMed] [Google Scholar]
- White M. J., Hirsch J. P., Henry S. A., 1991. The OPI1 gene of Saccharomyces cerevisiae, a negative regulator of phospholipid biosynthesis, encodes a protein containing polyglutamine tracts and a leucine zipper. J. Biol. Chem. 266: 863–872. [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., et al. , 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Young B. P., Shin J. J., Orij R., Chao J. T., Li S. C., et al. , 2010. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science 329: 1085–1088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.