Abstract
We show that coating of decellularized extracellular matrix (DC-ECM) on substrate surfaces is an efficient way to generate a platform mimicking the native ECM environment. Moreover, the DC-ECM can be modified with a peptide (QK) mimicking vascular endothelial growth factor without apparently compromising its integrity. The modification was achieved through metabolic incorporation of a “clickable” handle to DC-ECM followed by rapid attachment of the QK peptide with an azido tag using copper-catalyzed click reaction. The attachment of the QK peptide on to DC-ECM in this way further enhanced the angiogenic responses (formation of branched tubular networks) of endothelial cells.
Keywords: angiogenesis, decellularization, extracellular matrix, copper-catalyzed click chemistry, QK peptide
1. Introduction
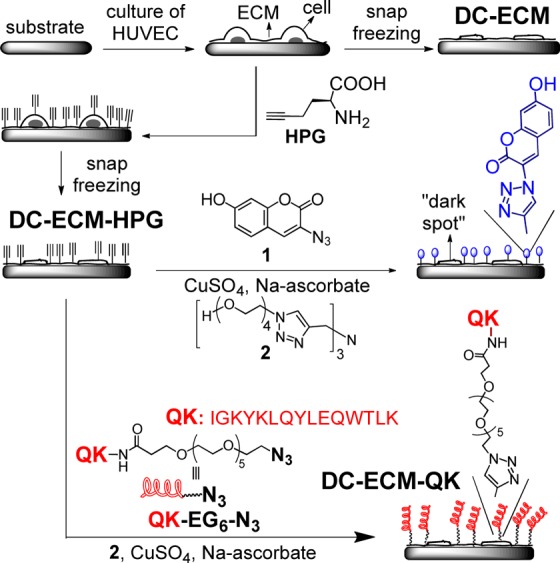
The bottleneck in development of tissue engineering scaffolds often lies on the formation of vascular networks through angiogenesis and vasculougenesis.1−3 The vascularization and other processes for tissue regeneration are regulated through the extracellular matrix (ECM) that dynamically presents a large collection of cues as part of the ECM molecules, growth factors, and physical features at macro- to nanoscales.1,4 For the development of synthetic scaffolds, the ultimate goal is to mimic the essentials of these molecular and physical cues in a cost-effective way.1,4 Recent advances in this field include nanostructured scaffolds,5,6 degradable hydrogel scaffolds,7,8 spatial and dynamic control of ligand, and/or growth factor presentation and delivery,9−12 polymers for engineering cell surfaces13 and covalent ligation of cells.14 Despite the great progress,1,4 current synthetic scaffolds can present only one or a handful of cues.15 In comparison, natural scaffolds prepared by removal of the cellular contents (decellularization) of tissues or organs may ideally retain the major structural integrity and present much more molecular cues than synthetic scaffolds.16−19 Nevertheless, decellularization is inherently accompanied by substantial loss of ECM molecules and growth factors and altering the structures and mechanical properties.20 Few methods are available to recapitulate the loss of functions and to enhance other desired functions without further deteriorating the delicate decellularized ECM (DC-ECM). Herein, we present a versatile and mild method for rapid attachment of biomolecules, such as the QK peptide (Scheme 1), via click chemistry onto DC-ECM coatings on substrates to enhance the desired functions (e.g., angiogenesis).
Scheme 1. Preparation of DC-ECM, DC-ECM-HPG, and DC-ECM-QK.
A wide variety of synthetic substrates with excellent shape and physical properties have been developed.1,5−8 Coating of DC-ECM on to the substrate is easier to perform than a typical multistep covalent modification of the substrate. It simply involves in vitro growth of the cells of interest in the substrates followed by decellularization.21−23 Unlike ECM components that are generally conserved among species, intracellular materials contain antigens that elicit host inflammatory response and, hence, should be removed. Decellularization of thick tissues and organs usually requires strong chemicals (acid/base, detergents, and enzymes), leading to substantial deformation of the scaffold and loss of ECM molecules.20 In contrast, a monolayer of cells on the substrate can be readily decellularized under mild conditions. Nevertheless, this approach still cannot avoid partial removal of ECM molecules, particularly the weakly bound growth factors. We envisioned that some of the lost functions might be recapitulated by attaching proper biomolecules to the DC-ECM platforms, which might also enhance other desired functions, especially angiogenesis. However, covalent modification on the complex, delicate DC-ECM without compromising its integrity is challenging. Few methods have been reported so far, which were mostly based on carbodiimide chemistry and only on decellularized organs consisting of collagens.24,25 When applied to DC-ECM with a wide variety of functional proteins, this method will suffer from a lack of selectivity and a high susceptibility to cross-link the proteins.
Our approach for functionalization of DC-ECMs is based on efficient incorporation of a biocompatible handle (ethynyl or azido group) to allow subsequent bioconjugation via copper-catalyzed alkyne–azide cyclization (CuAAC, a click reaction).26−29 This reaction is rapid, specific and compatible with live cells,26 and the resulted triazole linkages hardly affect the integrity of the DC-ECM. We demonstrate this strategy with a peptide (QK, Scheme 1) mimicking vascular endothelial growth factor (VEGF) to enhance angiogenesis.30
VEGF is a major growth factor specific for vascular endothelial cells to initiate angiogenesis by binding to the transmembrane VEGF receptors (VEGFRs).31 The 15-amino-acid QK peptide (Scheme 1) mimics the helical structures at the binding site of VEGF and exhibits a high binding affinity to VEGFRs, and in vitro and in vivo angiogenic activities.30 Recently, West and co-workers attached acetyl-protected QK onto succinimidyl ester functionalized PEG hydrogel and showed that the immobilized peptide also promoted angiogenesis.32
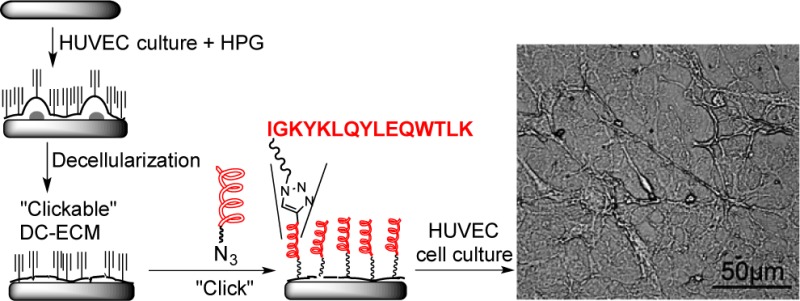
In our click chemistry based strategy, the first step is to introduce a handle to DC-ECM rendering it “clickable”. We chose to biosynthetically incorporate l-homopropargylglycine (HPG, an alkynyl surrogate of methionine28,29) to the ECM proteins of HUVECs. Since the functional ECM proteins are generally synthesized and degraded in a high turnover rate, we anticipate that the incorporation of HPG to ECM will be efficient.
2. Experimental Section
2.1. Reagents
Rat collagen I (Sigma), BD growth factor-reduced MatrigelTM basement membrane matrix (BD Bioscience, Bedford, MA), endothelial cell growth medium EGM-2 (Lonza, Walkersiville), trypsin/EDTA, and fetal bovine serum (HyClone, Logan, UT), rabbit IgG against VEGF antibody, and FITC conjugated goat antirabbit IgG antibody (Syd Laboratories, Inc., Malden, MA), and CellTiter 96 aqueous assay kit for MTS assay (Promega) were purchased and used directly.
2.2. Synthesis of Azido-Terminated QK Peptide
The C-amide-terminated QK peptide with an azido-hexa(ethylene glycol) tag at the N-terminus (N3-EG6-COO-KLTWQELYQLKYKGI-CONH2, QK-EG6-N3) was prepared on Rink Amide MBHA resin (Novabiochem, Darmstadt, Germany) by standard Fmoc chemistry using a manual peptide synthesizer. After the protected peptide sequence was assembled, the Fmoc group of the last amino acid (K) was removed and a solution of 0.3 mmol of N3-EG6-COOH (Quanta Biodesign Ltd., Powell, OH), 0.3 mmol of N-Hydroxybenzotriazole (HOBt) and 0.3 mmol of N,N′-diisopropylcarbodiimde (DIC) in THF was added. The coupling reaction was allowed to proceed for 2 h at 50 °C. The coupling step was repeated with a freshly prepared solution of HOBt and DIC before the peptide was deprotected and cleaved from the solid support by treatment with a mixture of trifluoacetic acid, triisopropylsilane and water (95:2.5:2.5) to provide QK-EG6-N3 with 78% purity shown by HPLC. The HPLC purified peptide was characterized with a Voyager DE-STR MALDI-TOF mass spectrometry (Applied Biosystems) using sinipanic acid as matrix, operated in positive ion linear mode with acceleration voltage at 20 kV, extraction delay time from 100 to 200 ns and grid voltage at 94%. m/z: [M + H]+ calcd for C107H171N25O29, 2271.27; found: 2271.87 (Supporting Information Figure S1).
2.3. Collagen I and BD Matrigel Coatings on Glass Substrates
Clean glass slides were cut into pieces of 1 × 1 cm2, and sterilized with 75% ethanol for 2 h. After sterilization, the slides were placed individually into a 24-well plate and washed three times with phosphate buffered saline (PBS). Rat tail collagen I (Sigma-Aldrich) was diluted to 50 μg/mL with 0.01 N HCl. An aliquot of 300 μL of the solution was added to cover each glass slide. After incubation for 1 h, the glass slides were rinsed with PBS and used immediately. Coating of BD Matrigel basement membrane matrix growth factor on the glass slides was preformed following the manufacturer’s instruction. Briefly, a 24-well plate housing the glass slides were placed on ice. An aliquot of 50 μL of Matrigel per cm2 was applied evenly onto each prechilled glass slide. The gel was allowed to polymerize at 37 °C for 30 min. The samples were rinsed with PBS and used immediately.
2.4. Culture Human Umbilical Vein Endothelial Cells (HUVECs)
HUVECs were grown in endothelial cell growth medium EGM-2 with 10% fetal bovine serum (FBS) at 37 °C and 5% CO2. The medium was refreshed every 2 days, and the cells were passaged at a ratio of 1:3 with routine trypsinization every 4 days when the confluence reached about 90%.
2.5. Culture HUVECs on glass substrates for the preparation of DC-ECM-HPG and DC-ECM
To prepare the ECM coating by decellularization of HUVECs, passage 5 or 6 HUVECs were seeded directly onto the coated glass slides in a 24-well plate at a density of 2 × 104 cells/well and cultured until reaching 90% confluence. To prepare the DC-ECM-HPG samples, HUVECs-covered glass slides were immersed in methionine-free Dulbecco’s modified eagle medium (DMEM) (Invitrogen, Carlsbad, CA) for 30 min to deplete the residual methionine, and then immersed in methionine-free DMEM with 50 μM HPG (Invitrogen) for 1 h to biosynthetically incorporate HPG as a surrogate of methionine into the newly synthesized proteins.29 This group of samples was then subjected to the following decellularization process to provide the group of samples named DC-ECM-HPG (Table 1). On the other hand, the samples cultured in EGM-2 with 10% FBS followed by the decellularization are named DC-ECM. All samples were then washed with PBS before decellularization.
Table 1. Summary of Sample Groups.
| name | descriptiona |
|---|---|
| glass | clean glass slides (1 × 1 cm2) |
| collagen I | collagen I coated on glass slides |
| Matrigel | Matrigel coated on glass slides |
| DC-ECM | decelluarized ECM of HUVECs on glass slides |
| DC-ECM-HPG | decelluarized ECM of HPG incorporated HUVECs on glass slides |
| DC-ECM-QK | DC-ECM-HPG samples (with HPG) treated with the “click solution”b containing QK-EG6-N3 |
| DC-ECM-Ctrl | DC-ECM samples (without HPG) treated with the “click solution”b containing QK-EG6-N3 |
See text for detailed preparation procedures.
“Click solution”: a freshly prepared solution of reagents for the CuAAC reaction (10 min at 37 °C), containing 0.1 mM CuSO4, 0.2 mM ligand 2 and 2.5 mM l-ascorbic acid sodium salt in 10% (v/v) DMEM/PBS.
2.6. Decellularization
Decellularization was performed by snap freezing and thawing as previously described.33 Briefly, the HUVECs covered glass slides was sealed in a small vial and placed into liquid nitrogen for 20 min, followed by thawing in a 37 °C water bath for 10 min. This freezing-thawing circle was repeated three times and the substrates were rinsed with PBS.
2.7. Bright Field and Fluorescence Images of HUVECs, DC-ECM, and DC-ECM-HPG
HUVECs adhered on glass substrates were fixed with 100% isopropyl alcohol for 10 min and then treated with 10 μL of 1 μg/mL propidium iodide (PI, Calbiochem, Darmstadt, Germany). Decellularized samples (DC-ECM and DC-ECM-HPG) were directly stained with 10 μL of 1 μg/mL PI. The samples were imaged with Nikon eclipse 80i fluorescence microscope in TRITC channel.
2.8. Attachment of Coumarin-Azide 1 on DC-ECM-HPG
To DC-ECM and DC-ECM-HPG samples in a 24-well plate were added a solution of coumarin-azide 1 (0.1 mM, Figure 1) and a freshly prepared “click solution” containing 0.1 mM CuSO4, 0.2 mM of ligand 2 and 2.5 mM l-ascorbic acid sodium salt in 10% (v/v) DMEM/PBS. The samples were incubated for 10 min at 37 °C. The solution was removed, and the sample was washed 3 times with PBS. The samples were then examined with a Nikon eclipse 80i fluorescence microscope at bright field and DAPI channel using a 20 × objective. A CoolSnap HQ2 camera (Photometrics, Tuscon, AZ) and NIS Elements software (Version 3.0, Nikon Instruments) were used for image acquisition and analysis. The images are shown in Figure 2a and Supporting Information Figure S4. The mean fluorescence intensities (MFI) of DC-ECM and DC-ECM-HPG surfaces after treatment with the coumarin-azide 1 under CuAAC reaction conditions were 1800 ± 500 and 6000 ± 1200 au, respectively. Result was obtained by measurement on three random areas on each of three samples.
Figure 1.
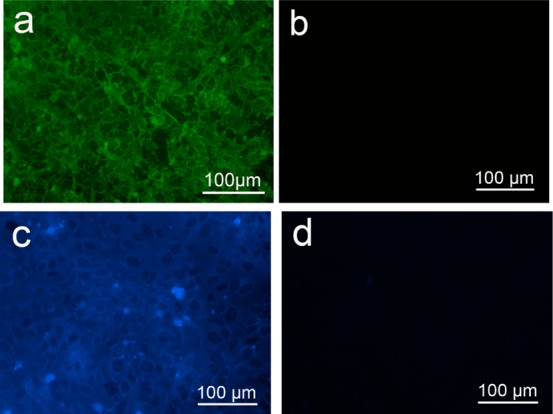
Immunofluorescence image of VEGF in DC-ECM after treatment first with VEGF pAb followed by FITC-labeled goat antirabbit IgG antibody (a) and its control only treated with FITC-labeled goat antirabbit IgG antibody (b), and a DAPI fluorescence image of the coumarin-triazole in DC-ECM-HPG after CuAAC reaction with the coumarin-azide 1 (c), and its control as DC-ECM after CuAAC reaction with the coumarin-azide 1 (d).
Figure 2.
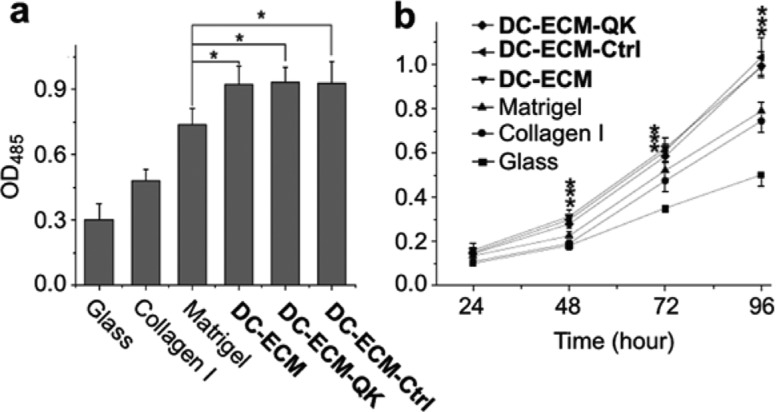
Adhesion (a) and proliferation (b) of HUVECs on different surfaces. Cell numbers are plotted as OD485 measured by MTS assay at 3 h (a) and during 96 h (b) post seeding. Data represent mean ± SD (n = 4; * denotes p < 0.05 compared to the Matrigel group).
2.9. Attachment of QK Peptide on DC-ECM-HPG
To attach QK peptide on the ECM surfaces via CuAAC reaction, DC-ECM-HPG samples were immersed in the above-mentioned “click solution” containing 0.1 mM QK-EG6-N3. After incubation for 10 min at 37 °C, the samples were washed 3 times with PBS. The resultant samples are named DC-ECM-QK. As a control, DC-ECM substrates were subjected to the same conditions to provide the samples named DC-ECM-Ctrl. All experimental and control groups prepared in this work are listed in Table 1.
2.10. Immunofluorescence Assay to Evaluate VEGF Retained on Surfaces
The presence of VEGF on DC-ECM and DC-ECM-QK samples was visualized with fluorescent secondary antibody that bounded to the primary antibody of VEGF. Specifically, the samples were incubated overnight at 4 °C with VEGF polyclonal antibody (pAb, 1 μg/mL, PA000214-PA1080, Syd Laboratories, Malden, MA) in PBS. The samples were rinsed 3 times with PBS to remove the excess antibody before incubation with goat antirabbit IgG labeled with FITC (1 μg/mL, Syd Laboratories, Malden, MA) at room temperature for another 40 min. The samples were then washed with PBS for 3 times and observed with a fluorescence microscope at bright field and FITC channel. Meanwhile, the DC-ECM and DC-ECM-QK samples treated without the primary antibody (VEGF pAb) but only with FITC-labeled goat antirabbit IgG in the same way were used as the negative controls.
2.11. Adhesion and Proliferation of HUVECs on DC-ECM Surfaces
The cell adhesion and proliferation assays were performed using CellTiter 96 aqueous assay kit following the protocols recommended by the manufacturer. Briefly, HUVECs (30 000 cells/well for adhesion assay and 5000 cells/well for proliferation assay) were seeded on the substrates in 24-well plates. After 3 h of incubation for adhesion assay or 1–4 days of incubation for proliferation assay, the medium was removed and the cells were washed 3 times with PBS. Then, MTS solution (MTS/PMS = 20:1) with DMEM (MTS solution/DMEM = 1:5) was added into each well (400 μL/well), and the samples were incubated at 37 °C for 2 h. The absorbance of the culture was then measured at 485 nm with HTS 700 Bio Assay microplate reader (Perkin Elemer, MA).
2.12. HUVECs Tubule Formation on DC-ECM Surfaces
HUVECs were seeded at a density of 20 000 cells/well on substrates in a 24-well plate and incubated at 37 °C with 5% CO2 for 6 h. The tube formation was examined using phase-contrast microscopy with 400× magnification. The length of each tubule was calculated using ImageJ software (http://rsbweb.nih.gov/ij/). The tubules with a length exceeding 200 μm were selected and summed up to provide the total length of tubules in each field of view on 5 randomly selected locations on the sample. ANOVA with Tukey’s LSD was performed to determine significant differences between groups, with p < 0.05 considered statistically significant. All data in Figure 3c are presented as mean ± standard deviation. While round cells were randomly distributed on glass (Supporting Information Figure S6a), a small fraction of the cells on the collagen I surfaces began to polarize and associate with each other (Figure S6b).
Figure 3.
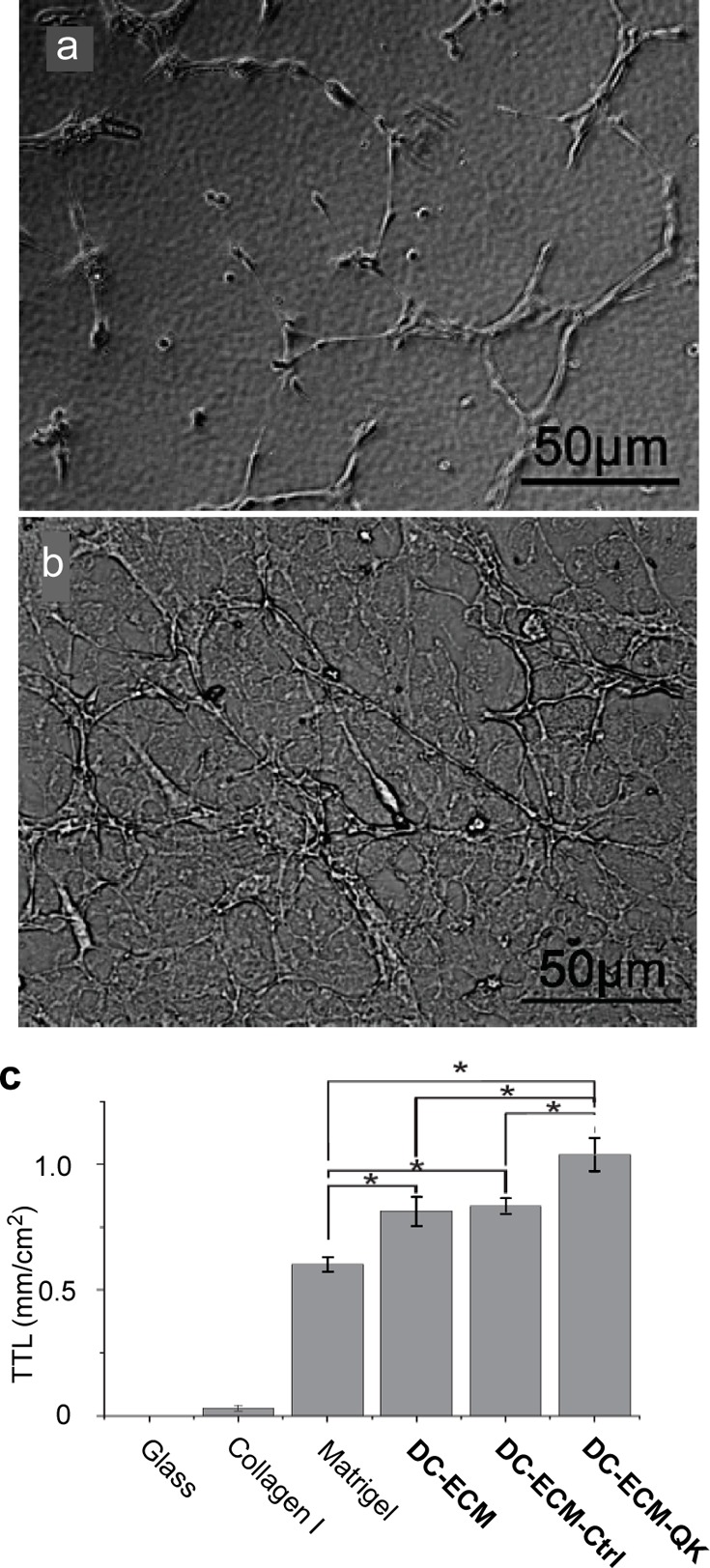
Bright field images of HUVECs on Matrigel (a) and DC-HPG-QK (b) at 6 h post seeding, and plot of the mean total tube length in mm/cm2 (c) on various surfaces. Only the lengths of tubules exceeding 200 μm were summed up using ImageJ software on 5 random locations of the sample. Data represent mean ± standard deviation. * denotes p < 0.05. For images of HUVECs on all surfaces, see Figure S6 in Supporting Information.
2.13. Statistics
The fluorescence intensity and MTS measurement were repeated at least three times and the results were expressed as mean ± standard deviation. Statistical significance was calculated using the SPSS 17.0 statistical software. Statistical significance was defined as p < 0.05.
3. Results and Discussion
After HUVECs were grown to >90% confluence on the substrate, we replaced methionine in the culture medium with HPG. After culture for 1 h to incorporate HPG into the newly synthesized ECM, the adhered cells were then decellularized by repeated freezing/thawing to disrupt the cell membrane followed by washing off the cell contents with PBS.23 The retaining of ECM on substrate was confirmed by the N 1s region scanning by X-ray photoelectron spectroscopy (XPS) (Supporting Information Figure 3S). The resultant substrate is designated as DC-ECM-HPG, while the substrate without HPG incorporation is named DC-ECM. The decellularization process effectively removed the nucleic acids, as shown by the absence of fluorescence from all DC-ECMs stained with PI (Supporting Information Figure S2). Bright field and fluorescence images of HUVECs before decellularization are shown in Supporting Information Figure S2a and b. Strong fluorescence was found in the nucleus of HUVECs prior to decellularization (Supporting Information Figure S2b), but completely absent after decellularization (Supporting Information Figure S2d and f). Upon decellularization the major cell bodies shrank and flattened to form thin patches found on both DC-ECM and DC-ECM-HPG samples (Supporting Information Figure S2c and e). These results support that the nucleic acids and most intracellular contents were effectively removed by the decellularization and washing process. While the decellularization of the adhered HUVECs was effective, the process was mild enough to retain some VEGF, as shown by immunofluorescence images after staining with VEGF pAb followed by FITC-labeled secondary antibody (Figures 1a). Supporting Information Figure S5a is the FITC fluorescence images of DC-ECM-QK sample, indicating the presence of VEGF in the ECM. In a control experiment, no fluorescence was detected on both samples that were not treated with the primary antibody before treatment with the FITC-labeled secondary antibody (Supporting Information Figures S1b and S5b). The result indicates that the biosynthetic incorporation of HPG and the attachment of QK peptide via click reaction did not apparently affect the amount of VEGF on the ECM and the binding to the VEGF antibody.
To probe the presence of HPG on DC-ECM-HPG, we treated both DC-ECM and DC-ECM-HPG surfaces with the fluorogenic coumarin-azide 1 under the CuAAC reaction conditions (Scheme 1, 0.1 mM CuSO4, 0.2 mM 2, 2.5 mM sodium ascorbate in 10% (v/v) DMEM/PBS for 10 min 37 °C).27 The CuAAC reaction generated the fluorescent triazole-coumarin product (λem = 465 nm) on DC-ECM-HPG, as indicated by the fluorescence image in Figure 1b, while the fluorescence intensity on the control (DC-ECM) was three times lower. The “dark spots” in Figure 1b colocalized with the cell patches formed after decellularization (Supporting Information Figure S4, see also Scheme 1 for an illustration). The weaker fluorescence on the patches suggests that these regions contain less newly synthesized proteins than the ECM bound on the substrate.
After confirming the existence of the HPG residues on DC-ECM-HPG, we then attached the QK peptide QK-EG6-N3 (C-amide-terminated QK peptide with an azido-hexa(ethylene glycol) tag at the N-terminus) to the surface under the same CuAAC reaction conditions to generate the QK-modified DC-ECM-QK (Scheme 1). As a control, DC-ECM was subjected to the same conditions to provide the group designated as DC-ECM-Ctrl.
The adhesion and proliferation of HUVECs on DC-ECMs with and without QK peptide (DC-ECM-QK, DC-ECM, and DC-ECM-Ctrl) were compared with those on the isolated ECMs (Collagen I and Matrigel). Collagen is a major structural component of ECM, which facilitates cell adhesion initiated by binding of its RGD motifs with the transmembrane integrin receptors. However, its ability to enhance cell proliferation is limited. In comparison, BD Matrigel contains many ECM proteins and growth factors, and has been widely used for cell proliferation and angiogenesis assays.34 The MTS assay shows that all DC-ECM samples were more effective in promoting cell adhesion (Figure 2a) and proliferation (Figure 2b) than the Collagen I and Matrigel surfaces (p < 0.05). Thus, at 3 h post seeding, the number of endothelial cells adhered on DC-ECMs were about 3, 2, and 1.3 times of those on bare glass, Collagen I and Matrigel surfaces, respectively. A similar ratio was found for cell proliferation on the DC-ECMs compared to the other surfaces after 4 days.
The CuAAC modification of the DC-ECM surfaces did not affect the adhesion and proliferation of HUVECs. Nearly identical growth curves were obtained for HUVECs on all three DC-ECM samples (Figure 2), regardless of the presence or absence of the immobilized QK peptide (DC-ECM-QK, DC-ECM, and DC-ECM-Ctrl). This result indicates that the CuAAC modification did not affect the function of the ECM and growth factors for promoting endothelial cell proliferation. Furthermore, the biosynthetic incorporation of HPG and the attachment of QK peptide via CuAAC reaction did not apparently affect the amounts of VEGF on the DC-ECM, as shown by the immunofluorescence images of DC-ECM-QK (Supporting Information Figure S5c).
Attachment of QK peptide onto the DC-ECM surfaces significantly enhanced the angiogenic response, as indicated by the increased formation of branched tubular networks.34 At 6 h post seeding, tubular structures were found on Matrigel coated surfaces (Figure 3a). However, these tubular structures appeared to be lesser branching than those found on all DC-ECM coated surfaces (Supporting Information Figure S6c–f), in particular on DC-ECM-QK (Figure 3b). This result is consistent with the fact that Matrigel reduced the amount of growth factors promoting vasculogenesis (formation of new blood vessels) more than angiogenesis (branching and spouting from existing blood vessels).12,33 Not only all three DC-ECM coatings induced more tube branching than Matrigel, they also increased the mean total tube length per unit area (TTL), which can be readily quantified (Figure3c). Thus, the TTL of the DC-ECM, DC-ECM-Ctrl, and DC-ECM-QK groups were about 1.30, 1.33, and 1.66 fold of that on Matrigel. Remarkably, DC-ECM-QK significantly enhanced the TTL by about 25% (Figure 3c).
4. Conclusion
In conclusion, as compared to Matrigel, all DC-ECM coatings promote far greater endothelial cell responses desired for vascularization, including adhesion, proliferation, migration, and formation of branched tubular networks. These results indicate that DC-ECMs present a more complete collection of molecular and physical cues for guiding vascularization. To modify the delicate DC-ECM, we took advantage of the high turnover rates of most ECM proteins for efficient incorporation of HPG, followed by conjugating to azido-tagged biomolecules via the rapid CuAAC reaction. Although the DC-ECM retained some VEGF which is desirable for angiogenesis, attaching QK peptide in this way to some extent supplemented the VEGF lost during DC-ECM preparation, thus enhancing the angiogenic responses without apparent effect on the cell adhesion and proliferation. Therefore, we have developed a general method to present many molecular cues on artificial scaffolds that can be further covalently modified to enhance a desired function with minimum deterioration of the system. To achieve specificity of modification, the “clickable” handle can be genetically encoded into specific ECM proteins.35
Acknowledgments
This work was supported by National Institutes of Health (NIH) (R21HD058985), National Science Foundation (NSF) (DMR-0706627 and DMR-1207583), and The Alliance for NanoHealth (W81XWH-09-02-0139). L.W. thanks support from the China Scholarship Council (2010615034).
Supporting Information Available
MALDI spectra of QK-EG6-N3, microscopic images of PI stained DC-ECMs, XPS spctra of DC-ECM substrates, microscopic images of coumarin-triazole on DC-ECM-HPG, immunofluorescence images of VEGF on DC-ECMs and bright field images of HUVECs tubule formation on different surfaces. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Place E. S.; Evans N. D.; Stevens M. M. Complexity in Biomaterials for Tissue Engineering. Nat. Mater. 2009, 8, 457–470. [DOI] [PubMed] [Google Scholar]
- Novosel E. C.; Kleinhans C.; Kluger P. J. Vascularization Is the Key Challenge in Tissue Engineering. Adv. Drug Delivery Rev. 2011, 63, 300–311. [DOI] [PubMed] [Google Scholar]
- Moon J. J.; West J. L. Vascularization of Engineered Tissues: Approaches to Promote Angiogenesis in Biomaterials. Curr. Top. Med. Chem. 2008, 8, 300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutolf M. P.; Hubbell J. A. Synthetic Biomaterials as Instructive Extracellular Microenvironments for Morphogenesis in Tissue Engineering. Nat. Biotechnol. 2005, 23, 47–55. [DOI] [PubMed] [Google Scholar]
- Dvir T.; Timko B. P.; Kohane D. S.; Langer R. Nanotechnological Strategies for Engineering Complex Tissues. Nat. Nanotechnol. 2011, 6, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed L. E.; Engelmayr G. C.; Borenstein J. T.; Moutos F. T.; Guilak F. Advanced Material Strategies for Tissue Engineering Scaffolds. Adv. Mater. 2009, 21, 3410–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S. W.; Yu T. B.; Guan Z. De Novo Design of Saccharide-Peptide Hydrogels as Synthetic Scaffolds for Tailored Cell Responses. J. Am. Chem. Soc. 2009, 131, 17638–17646. [DOI] [PubMed] [Google Scholar]
- Galler K. M.; Aulisa L.; Regan K. R.; D’Souza R. N.; Hartgerink J. D. Self-Assembling Multidomain Peptide Hydrogels: Designed Susceptibility to Enzymatic Cleavage Allows Enhanced Cell Migration and Spreading. J. Am. Chem. Soc. 2010, 132, 3217–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J. L. Protein-Patterned Hydrogels: Customized Cell Microenvironments. Nat. Mater. 2011, 10, 727–729. [DOI] [PubMed] [Google Scholar]
- Wylie R. G.; Ahsan S.; Aizawa Y.; Maxwell K. L.; Morshead C. M.; Shoichet M. S. Spatially Controlled Simultaneous Patterning of Multiple Growth Factors in Three-Dimensional Hydrogels. Nat. Mater. 2011, 10, 799–806. [DOI] [PubMed] [Google Scholar]
- Liu B.; Liu Y.; Riesberg J. J.; Shen W. Dynamic Presentation of Immobilized Ligands Regulated through Biomolecular Recognition. J. Am. Chem. Soc. 2010, 132, 13630–13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S. M.; Siegman S. N.; Segura T. The Effect of Vascular Endothelial Growth Factor (VEGF) Presentation within Fibrin Matrices on Endothelial Cell Branching. Biomaterials 2011, 32, 7432–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. T.; Krishnamurthy V. R.; Cui W.; Qu Z.; Chaikof E. L. Noncovalent Cell Surface Engineering with Cationic Graft Copolymers. J. Am. Chem. Soc. 2009, 131, 18228–18229. [DOI] [PubMed] [Google Scholar]
- Dutta D.; Pulsipher A.; Luo W.; Yousaf M. N. Synthetic Chemoselective Rewiring of Cell Surfaces: Generation of Three-Dimensional Tissue Structures. J. Am. Chem. Soc. 2011, 133, 8704–8713. [DOI] [PubMed] [Google Scholar]
- Derda R.; Musah S.; Orner B. P.; Klim J. R.; Li L.; Kiessling L. L. High-Throughput Discovery of Synthetic Surfaces That Support Proliferation of Pluripotent Cells. J. Am. Chem. Soc. 2010, 132, 1289–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Castillejos R. Replication of the 3D Architecture of Tissues. Mater. Today 2010, 13, 32–41. [Google Scholar]
- Ott H. C.; Clippinger B.; Conrad C.; Schuetz C.; Pomerantseva I.; Ikonomou L.; Kotton D.; Vacanti J. P. Regeneration and Orthotopic Transplantation of a Bioartificial Lung. Nat. Med. 2010, 16, 927–933. [DOI] [PubMed] [Google Scholar]
- Petersen T. H.; Calle E. A.; Zhao L.; Lee E. J.; Gui L.; Raredon M. B.; Gavrilov K.; Yi T.; Zhuang Z. W.; Breuer C.; Herzog E.; Niklason L. E. Tissue-Engineered Lungs for In Vivo Implantation. Science 2010, 329, 538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner W. R.; Griffith B. P. Medicine Reconstructing the Lung. Science 2010, 329, 520–522. [DOI] [PubMed] [Google Scholar]
- Crapo P. M.; Gilbert T. W.; Badylak S. F. An Overview of Tissue and Whole Organ Decellularization Processes. Biomaterials 2011, 32, 3233–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N.; Pham Q. P.; Sharma U.; Sikavitsas V. I.; Jansen J. A.; Mikos A. G. In Vitro Generated Extracellular Matrix and Fluid Shear Stress Synergistically Enhance 3D Osteoblastic Differentiation. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 2488–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham M. T.; Reuther H.; Maitz M. F. Native Extracellular Matrix Coating on Ti Surfaces. J. Biomed. Mater. Res., Part A 2003, 66, 310–316. [DOI] [PubMed] [Google Scholar]
- Datta N.; Holtorf H. L.; Sikavitsas V. I.; Jansen J. A.; Mikos A. G. Effect of Bone Extracellular Matrix Synthesized In Vitro on the Osteoblastic Differentiation of Marrow Stromal Cells. Biomaterials 2005, 26, 971–977. [DOI] [PubMed] [Google Scholar]
- Liao D.; Wang X.; Lin P. H.; Yao Q.; Chen C. Covalent Linkage of Heparin Provides a Stable Anti-coagulation Surface of Decellularized Porcine Arteries. J. Cell. Mol. Med. 2009, 13, 2736–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu L. L. Y.; Radisic M. Scaffolds with Covalently Immobilized VEGF and Angiopoietin-1 for Vascularization of Engineered Tissues. Biomaterials 2010, 31, 226–241. [DOI] [PubMed] [Google Scholar]
- Del Amo D.; Wang W.; Jiang H.; Besanceney C.; Yan A.; Levy M.; Liu Y.; Marlow F.; Wu P. Biocompatible Copper(I) Catalysts for In Vivo Imaging of Glycans. J. Am. Chem. Soc. 2010, 132, 16893–16899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong V.; Presolski S. I.; Ma C.; Finn M. G. Analysis and Optimization of Copper-Catalyzed Azide-Alkyne Cycloaddition for Bioconjugation. Angew. Chem., Int. Ed. 2009, 48, 9879–9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hest J. C. M.; Kiick K. L.; Tirrell D. A. Efficient Incorporation of Unsaturated Methionine Analogues into Proteins In Vivo. J. Am. Chem. Soc. 2000, 122, 1282–1288. [Google Scholar]
- Beatty K. E.; Liu J. C.; Xie F.; Dieterich D. C.; Schuman E. M.; Wang Q.; Tirrell D. A. Fluorescence Visualization of Newly Synthesized Proteins in Mammalian Cells. Angew. Chem., Int. Ed. 2006, 45, 7364–7367. [DOI] [PubMed] [Google Scholar]
- D’Andrea L. D.; Iaccarino G.; Fattorusso R.; Sorriento D.; Carannante C.; Capasso D.; Trimarco B.; Pedone C. Targeting Angiogenesis: Structural Characterization and Biological Properties of a De Novo Engineered VEGF Mimicking Peptide. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 14215–14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos G. D.; Davis S.; Gale N. W.; Rudge J. S.; Wiegand S. J.; Holash J. Vascular-Specific Growth Factors and Blood Vessel Formation. Nature 2000, 407, 242–248. [DOI] [PubMed] [Google Scholar]
- Leslie-Barbick J. E.; Saik J. E.; Gould D. J.; Dickinson M. E.; West J. L. The Promotion of Microvasculature Formation in Poly(ethylene glycol) Diacrylate Hydrogels by an Immobilized VEGF-Mimetic Peptide. Biomaterials 2011, 32, 5782–5789. [DOI] [PubMed] [Google Scholar]
- Francis M. E.; Uriel S.; Brey E. M. Endothelial Cell-Matrix Interactions in Neovascularization. Tissue Eng., Part B 2008, 14, 19–32. [DOI] [PubMed] [Google Scholar]
- Arnaoutova I.; George J.; Kleinman H. K.; Benton G. The Endothelial Cell Tube Formation Assay on Basement Membrane Turns 20: State of the Science and the Art. Angiogenesis 2009, 12, 267–274. [DOI] [PubMed] [Google Scholar]
- Wang L.; Schultz P. G. Expanding the Genetic Code. Angew. Chem., Int. Ed. 2005, 44, 34–66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.