Abstract
We characterized the full-length genomes of 16 distinct hepatitis C virus genotype 1 (HCV-1) isolates. Among them, four represented the first full-length genomes for subtypes 1d (QC103), 1i (QC181), 1j (QC329) and 1k (QC82), and another four corresponded to subtypes 1c (QC165), 1g (QC78), 1h (QC156) and 1e (QC172). Both QC196 and QC87 were assigned into a new subtype 1m, and QC113 and QC74 into another new subtype 1n. The remaining four (QC60, QC316, QC152 and QC180) did not classify among the established subtypes and corresponded to four new lineages. Subtypes 1j, 1k, 1m, 1n and the unclassified isolate QC60 were identified in Haitian immigrants. In the updated HCV nomenclature of 2005, a total of 12 subtypes of HCV-1 were designated. Including the data from the present study, all but subtype 1f now have their full-length genomes defined. Further analysis of partial NS5B sequences available in GenBank denoted a total of 21 unclassified lineages, indicating the taxonomic complexity of HCV-1. Among them, six have had their full-length genomes characterized. Based on the available full-length genome sequences, a timescale phylogenetic tree was reconstructed which estimated important time points in the evolution of HCV-1. It revealed that subtype 1a diverged from its nearest relatives 135 years ago and subtype 1b diverged from its nearest relatives 112 years ago. When subtypes 1a, 1j, 1k, 1m, 1n and six close relatives (all but one from Haitian immigrants) were considered as a whole, the divergence time was 176 years ago. This diversification was concurrent with the time period when the transatlantic slave trade was active. When taking all the HCV-1 isolates as a single lineage, the divergence time was 326 years ago. This analysis suggested the existence of a recent common ancestor for subtype 1a and the Haitian variants; a co-origin for subtypes 1b, 1i and 1d was also implied.
Introduction
Hepatitis C virus (HCV; genus Hepacivirus; family Flaviviridae) is a positive-sense ssRNA virus. As a blood-borne pathogen, HCV infects an estimated 170–200 million people worldwide (3 % of the world’s population) and poses a major threat to global public health (Alter, 2007). HCV infection is characterized by establishing chronic hepatitis in ~70–85 % of the infected individuals. Chronic infection leads to a major risk of developing liver cirrhosis and hepatocellular carcinoma, which are associated with substantial morbidity and mortality, and both are expected to increase over the next decades (CDC, 1998; WHO, 1997).
The analysis of HCV genetic sequences has resulted in the classification of the virus into seven genotypes (Smith et al., 2014). Except for genotypes 5 and 7, each of them is further divided into a number of subtypes. In terms of nucleotide identity, a genome-wide difference of 31–33 % is sufficient to distinguish a genotype and a difference of >15 % is sufficient to distinguish a subtype, given that at least three closely related but distinct isolates are identified. HCV genotypes have different geographical distribution patterns. Subtypes 1a, 1b, 2a, 2b and 3a are found worldwide. The other subtypes, however, are restricted primarily to indigenous regions (Simmonds et al., 2005). An example of the latter is seen for the numerous genotype 1 lineages. Taxonomically assigned or unassigned, these lineages are characterized by their endemic circulation in West and Central Africa. However, due to the scarcity in sampling of these variants, there is currently a lack of sufficient information about their genetic variation patterns over the full-length genome. This information is essential for a better understanding of HCV’s historical origin and epidemic potential.
As noted in the HCV nomenclature guidelines (Simmonds et al., 2005) and two recent studies (Bracho et al., 2008; Li et al., 2013), seven subtypes (1a, 1b, 1c, 1e, 1g, 1h and 1l) of HCV genotype 1 have now been confirmed with full-length genomic sequences. However, five subtypes (1d, 1f, 1i, 1j and 1k) remain assigned provisionally due to the availability of only partial sequences. In the present study, we aimed to help fill this information gap by characterizing the full-length genomes of 16 genotype 1 isolates, including the first full-length genome sequences for subtypes 1d, 1i, 1j and 1k, and others representing the unique variants.
Results
Full-length genome sequences and genomic organization
Full-length genome sequences were characterized for 16 HCV-1 isolates: QC165, QC103, QC172, QC78, QC156, QC181, QC329, QC82, QC60, QC74, QC87, QC113, QC152, QC180, QC196 and QC316, each with 19–22 overlapping fragments. These genomes were each 9398–9483 nt long, starting from the extreme 5′ UTR through to the variable region of the 3′ UTR. They each had a single ORF of 9033–9054 nt. The 5′ UTRs were of 341–342 nt, whilst the 3′ UTR varied from 21 to 106 nt. Excluding the E2 (363–369 aa) and NS5A (447–448 aa) regions, which were variable among isolates, the sizes of the other eight protein regions did not vary and were the same as those in the H77 genome.
Similarity plotting
To exclude the possibility of recent viral recombination events, pairwise nucleotide similarity curves were plotted along the HCV genome using rdp3 software. No evidence of recombination was detected when the 16 QC isolates were compared with each other and with 40 reference sequences representing various HCV genotypes and subtypes (data not shown). This analysis, however, revealed that isolate 1c.IN.Khaja1.AY651061 contained a recombination in its structural protein region.
Phylogenetic analysis and pairwise comparison of full-length genomes
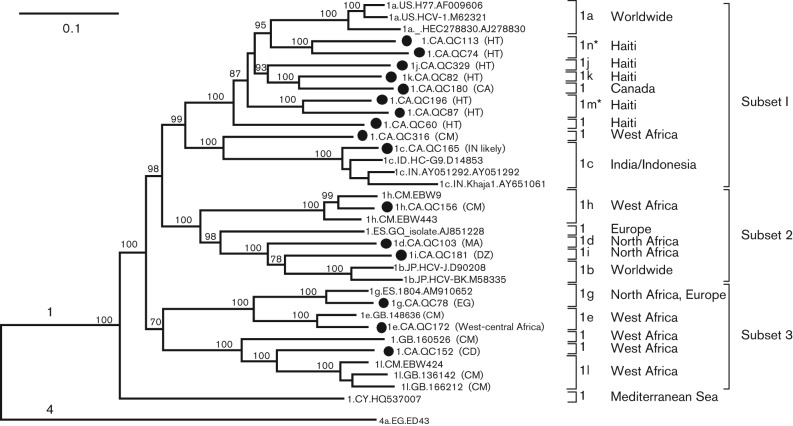
In a previous report, partial sequences were determined for the 16 isolates, which classified them into genotype 1. In more detail, QC165, QC103, QC172, QC78, QC156, QC181, QC329 and QC82 were classified into subtypes 1c, 1d, 1e, 1g, 1h, 1i, 1j and 1k, respectively, whilst QC60, QC74, QC87, QC113, QC152, QC180, QC196 and QC316 each represent an unclassified variant (Murphy et al., 2007). In this study, their full-length genomes were characterized. Based on the obtained sequences, a maximum-likelihood (ML) tree was reconstructed with the inclusion of a number of references (Fig. 1). For descriptive purposes, we arbitrarily divided the genotype 1 clade into three subsets in addition to an orphan branch. Ten, three and three sequences reported in this study fell into subsets 1, 2 and 3, respectively. In subset 1, QC82 and QC329 were the first full-length genomes of subtypes 1k and 1j, respectively, QC165 was the fourth full-length genome of subtype 1c, whilst QC60, QC74, QC113, QC87, QC196, QC180 and QC316 represented novel variants among which the former four could be assigned into two new subtypes, 1m and 1k. In subset 2, QC103 and QC181 were the first full-length genomes of subtypes 1d and 1i, respectively, and QC156 was the third full-length genome of subtype 1h. In subset 3, QC78 and QC172 were the second full-length genomes of subtypes 1g and 1e, respectively, and QC152 represented a novel variant. When a cluster was formed by several sequences, it was supported by a bootstrap value of 100 %.
Fig. 1.
ML tree estimated using 56 HCV full-length genome sequences. Reference sequences from confirmed subtypes of genotype 1 were analysed together with the 16 sequences characterized in this study (filled circles). Reference sequences from genotypes 2 to 7 were used as the outlier group; however, we only show the ED43 of subtype 4a in this tree. For genotype 1, all subtypes and unclassified lineages and the described subsets as well as the related larger geographical regions are indicated at the right-hand side of the tree. The two newly assigned subtypes 1m and 1n are indicated by asterisks. Isolates are named using the following format: subtype.sampling country (if available).isolate name.GenBank accession number. Bootstrap supports >70 % are shown at internal nodes. Bar, 0.10 nt substitutions per site. The patient’s country of origin or ancestral country (if available) is given in parentheses after the related taxa. The country codes refer to ISO 3166-1 alpha-2 (http://en.wikipedia.org/wiki/ISO_3166-1_alpha-2).
Subset 1 could be subdivided into two parts. The upper part was composed of subtype 1a sequences and the sequences isolated from Haitian immigrants and of QC180 obtained from a Caucasian. As these sequences formed a cluster that resembled a comb-like leaf suggesting active divergence processes, this may indicate a close phylogenetic link between subtype 1a and the Haitian sequences. In contrast, the lower part contained fewer sequences, including the single unassigned QC316 variant which was acquired from a Cameroonian immigrant and four 1c isolates which showed origins either in India or Indonesia. Subset 2 could also be subdivided into two parts when excluding the GQ isolate. The upper part was represented by subtype 1h, which was identified specifically in Cameroon, whilst the lower part was heterogeneous, consisting of subtypes 1d and 1i, and the globally distributed 1b strains. 1d_QC103 was obtained from a Moroccan immigrant, whilst 1i_QC181 was isolated from an Algerian immigrant. Morocco and Algeria are neighbouring countries in North Africa. Interestingly, subtype 1b is common in southern Europe, which is close to North Africa over the Mediterranean Sea. Such a phylogenetic and geographical closeness may suggest a co-origin for subtypes 1b, 1d and 1i. Subset 3 could also be subdivided into two parts. The upper part contained both subtypes 1e and 1g, whilst the lower part included the three 1l isolates and two unclassified variants. Excluding one of the two 1g isolates which was detected in Spain and the other which was from an Egyptian immigrant in Canada, all the remaining sequences in subset 3 had their origins in West-central Africa. Finally, the three subsets were joined by the orphan branch leading to the unclassified isolate of HQ537007 which was determined in Cyprus (Demetriou & Kostrikis, 2011).
Based on the 34 full-length genotype 1 sequences analysed in Fig. 1, pairwise p genetic distances were calculated (Table S1, available in the online Supplementary Material). This showed that 1c_QC165, 1e_QC172, 1g_QC78 and 1h_QC156 had p distances of 0.07, 0.084, 0.082 and 0.071, respectively, to reference isolates 1c_HC-G9, 1e_148636, 1g_ES.1804 and 1h_EBW9. These distances further supported the subtype assignments of these four QC isolates. The remaining 12 QC isolates showed p distances to their nearest relatives ranging from 0.13 to 0.195. As QC103, QC181, QC329 and QC82 represented the first full-length genomes of subtypes 1d, 1i, 1j and 1k, respectively, their p distances to established subtypes differed by >15 %. Likewise, QC316 and QC60 showed p distances of 0.195 and 0.175, respectively, to their closest relatives 1c_HC-G9 and 1a_H77, indicating that both QC316 and QC60 could correspond to new subtypes. QC152 showed p distances of 0.152, 0.154 and 0.149 to subtype 1l isolates 136142, 166212 and EBW424, respectively. Taking the mean p distance from these three isolates, QC152 could be also classified within a distinct subtype. Except for the above-mentioned 11 isolates, the other five isolates (QC113, QC196, QC180, QC74 and QC87) showed p distances to their nearest relatives that ranged from 0.13 to 0.149. Taken together, six sequences (QC152, QC113, QC196, QC180, QC74 and QC87) showed marginal genetic differences from the references and their implications for classification were considered.
Phylogenetic analysis of the partial NS5B sequences
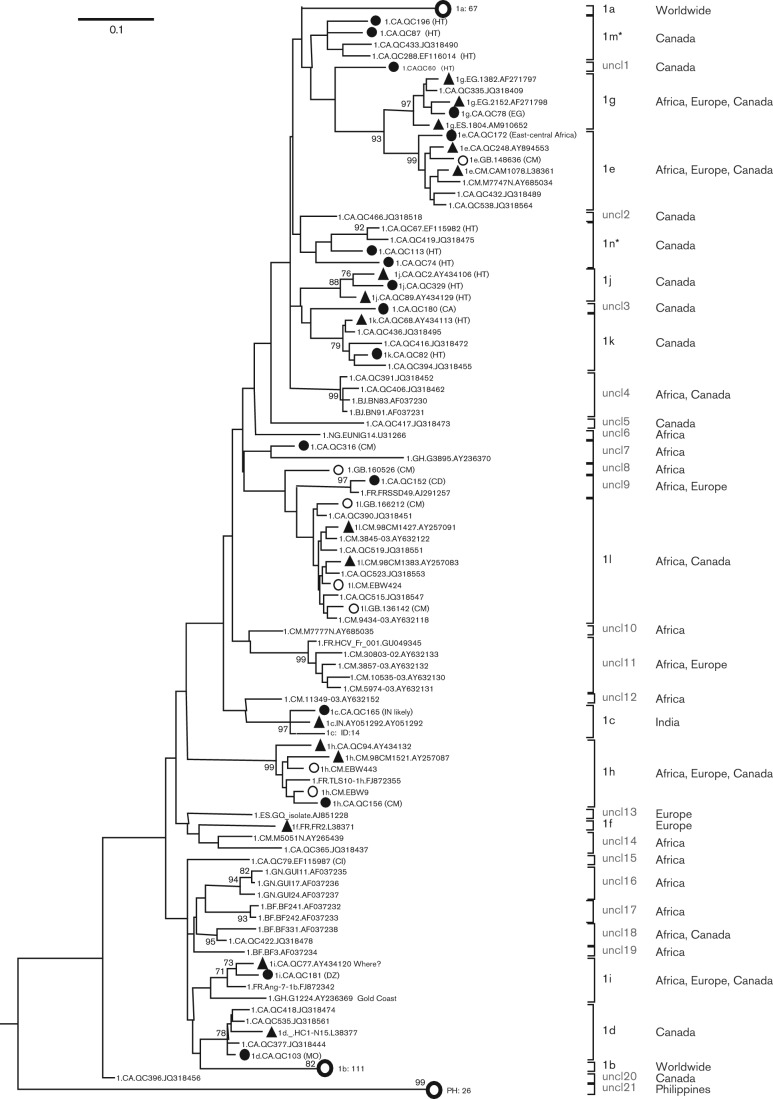
To further explore HCV-1 genetic diversity, 309 sequences in the NS5B region were analysed. Fig. 2 shows the ML tree reconstructed based on these 309 sequences. These included the 16 sequences that were determined in the present study (filled circles), seven that we have reported recently (open circles), 16 that represent subtypes 1c–1l (filled triangles) (Simmonds et al., 2005) and 270 that were retrieved from the Los Alamos HCV database because they were indicated as being unclassified HCV-1 isolates. The ML tree revealed that among the 270 unclassified HCV-1 isolates, 211 were grouped into subtypes 1a–1l (Jeannel et al., 1998; Simmonds et al., 2005), whilst 55 represented true novel variants. More precisely, 67 were classified into subtype 1a, 111 into 1b, 14 into 1c, three into each of 1d, 1e and 1k, one into each of 1g and 1h, six into 1l, and two into 1i. In addition to these 12 assigned subtypes, we designed two new subtypes, 1m and 1n, each containing four isolates. We further indicated 21 unclassified lineages (uncl) that may be new subtype candidates. For descriptive purposes, we temporarily numbered them uncl1–21. Of the 60 novel HCV-1 variants, one was grouped into each of uncl1, uncl2, uncl3, uncl5, uncl6, uncl8, uncl10, uncl12, uncl13, uncl15, uncl19 and uncl20, two into each of uncl7, uncl9, uncl14, uncl17 and uncl18, three into uncl16, four into uncl4, five into uncl11, and 26 into uncl21 (Fig. 2). The geographical sampling regions of these assigned subtypes and the unclassified lineages were identified (Table S2). Excluding subtypes 1a and 1b which were distributed worldwide, and 1c and 1g which were identified over several continents, all of the others appeared to be restricted geographically. However, most of these lineages have been detected in Canada, particularly among immigrants, with the exclusion of subtype 1f and 10 unclassified lineages. Thought to be the region of origin for HCV-1, only a fraction of its members have been identified in Africa, likely ascribed to undersampling.
Fig. 2.
ML tree based on 309 partial NS5B sequences of HCV-1, corresponding to nt 8316–8620 in the referenced H77 genome. The subtype is denoted at the right-hand side of the tree. The 16 isolates characterized in this study are labelled by filled circles. The seven isolates we reported recently are indicated by open circles (Li et al., 2013), whilst the 16 filled triangles mark the isolates that were assigned to the various HCV-1 subtypes in the consensus paper (Simmonds et al., 2005). To simplify the tree, isolates of subtype 1a, 1b and cluster PH-26 sequences are collapsed into single bold open circles. For other details, see legend for Fig. 1.
Unclassified HCV-1 isolates in the HCV database
Accessed on 1 April 2013, the Los Alamos HCV database had archived 1347 unclassified HCV-1 sequences corresponding to different genomic regions. Table 1 lists 347 out of the 1347 ‘unclassified’ HCV-1 sequences after excluding the 249 that were reclassified into subtype 1a, the 228 that were reclassified into subtype 1b and the 523 in the 5′ UTR that could not be assigned to any subtype. Subtypes 1c–1l were found to have a geographical distribution pattern similar to that we have described recently (Li et al., 2013). On the continental level, North America showed the largest genetic diversity of HCV-1 where 22 subtypes/lineages have been detected. This is followed by Africa where 17 subtypes/lineages have been identified and Europe where 10 subtypes/lineages have been found. In contrast, only one or two lineages have been reported in other continents. Although similar levels of HCV-1 genetic diversity were observed between North America and Africa, it is commonly believed that HCV-1 had its origin in Africa (Pybus et al., 2007). The latter is supported by the present study for which 15 of the 16 divergent HCV-1 isolates were isolated from African immigrants or descendants and 15 out of the 21 unclassified lineages in Table S2 showed such evidence.
Table 1. Distribution of genotype 1 subtypes/clusters in different continents.
| Location | No. of subtypes/unclassified lineages | Name of subtypes/unclassified lineages* | No. of isolates (n = 347) | Percentage of isolates |
| Africa | 6* | 1c, 1d, 1e, 1g, 1h, 1l | 109 | 36.6 |
| 11† (unclassified) | 4, 6, 7, 9, 11, 12, 14, 16, 17, 18, 19 | 18 | ||
| Asia | 2* | 1c, 1e | 101 | 36.6 |
| 1† (unclassified) | 21 | 26 | ||
| Eastern Mediterranean | 1* | 1g§ | 2 | 0.58 |
| Europe | 6* | 1c, 1d, 1f, 1g, 1h, 1i | 16 | 5.76 |
| 4† (unclassified) | 8||, 9, 11, 13|| | 4 | ||
| North America | 11* | 1c, 1d, 1e||, 1g||, 1h||, 1i||, 1j#, 1k, 1l, 1m#, 1n# | 42 | 15.56 |
| 11† (unclassified) | 1#, 2, 3, 4, 5, 7||, 9||, 14, 15||, 18, 20 | 12 | ||
| South America | 1* | 1g | 1 | 0.29 |
| Country unknown | 1* | 1l | 2 | 0.58 |
| Country unknown | Incomparable‡ | Core or E2 regions | 14 | 4.03 |
| Total (n = 347) | 11* | 1c–1n | 273 | 78.68 |
| 21† (unclassified) | 1–21 | 60 | 17.29 | |
| Incomparable‡ | 14 | 4.03 |
Subtypes 1a and 1b are excluded because they are distributed globally and therefore 10 assigned subtypes remain.
Sixty sequences could not be grouped into subtypes 1a–1n, based on which 21 unclassified lineages are numbered temporarily.
Incomparable.
From Lebanon.
||Origin in Africa (Murphy et al., 2007) or GenBank accession number AJ851228.
Origin in Haiti (Murphy et al., 2007).
Timescaled phylogenetic tree
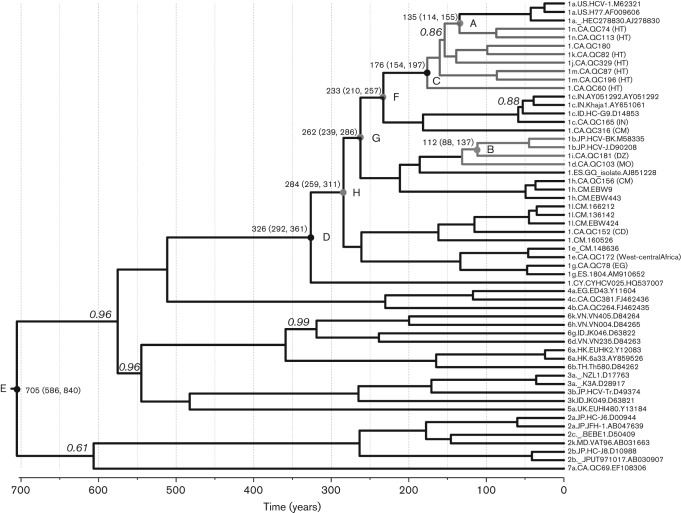
Based on 56 full-length HCV genome sequences, including the 16 determined in this study, a timescaled phylogenetic tree was reconstructed (Fig. 3). The tree showed the structure of the genotype 1 clade was highly similar to that of Fig. 1, except that all tips were aligned to the right ends and all branches were measured by grids corresponding to a timescale shown at the tree base. This scale indicated the year at which different branches or clusters had diverged. It was estimated that subtype 1a and two Haitian strains, QC74 and QC113, separated from each other 135 [95 % highest posterior density (HPD): 114, 155] years ago (point A), and subtype 1b and 1i (QC181) separated 112 (95 % HPD: 88, 137) years ago (point B). Considering subtypes 1a, 1j, 1k, 1m, 1n, and QC60 and QC180 as a whole, the most recent common ancestor was dated to 176 (95 % HPD: 154, 197) years ago (point C); taking all of the HCV-1 sequences as a single lineage, the most recent common ancestor was dated to 326 (95 % HPD: 292, 361) years ago (point D). However, when including all of the 56 sequences as a whole, the common ancestor was dated to 705 (95 % HPD: 586, 840) years ago (point E).
Fig. 3.
Timescaled phylogenetic tree estimated using the 56 sequences analysed in Fig. 1. Branch lengths represent the evolutionary time, which is measured by the grids corresponding to a timescale shown at the tree base. To simplify the tree, only those posterior probability scores <1 are shown in italics to the left of the related nodes, otherwise full scores of 1 are obtained. Eight time points are given after running the tMRCA function of beast and are indicated with circles labelled A–G in the related internal nodes, which measure the time and its 95 % HPD (shown above the circles) at which the related HCV lineages diverged. Branches corresponding to the Haitian sequences are shown in red. Similarly, the 1b branches and its most close relatives are shown in blue.
Discussion
In this study, full-length HCV genome sequences were characterized for 16 genotype 1 isolates. QC165, QC78, QC156 and QC172 correspond to subtypes 1c, 1g, 1h and 1e, and QC103, QC181, QC329 and QC82 correspond to subtypes 1d, 1i, 1j and 1k, respectively. Whilst the former four subtypes had their prototypic full-length genomes reported recently (Li et al., 2013), the latter four provisionally assigned subtypes are now confirmed for the first time. Similarly, the full-length genomes of QC113, QC74, QC196, QC87, QC60, QC316, QC152 and QC180 were described for the first time and represent six lineages. Of note, the classification of these subtypes and lineages was supported not only by phylogenetic and pairwise comparison of full-length genomes, but also by analysis of partial sequences in the NS5B region that included numerous additional sequences (Figs 1 and 2, Tables S1 and S2). Both QC196 and QC87 represent a new subtype, and both QC113 and QC74 represent another, as they fulfil two basic recommendations: (1) their full-length genomes are determined and (2) at least three closely related but independent isolates are identified (Fig. 2). Therefore, we propose to assign QC196 and QC87 into subtype 1m, and QC113 and QC74 into subtype 1n. As they each have two full-length genomes characterized, these two new subtypes can now be confirmed. However, the remaining four genomes, QC60, QC316, QC152 and QC152, each represent a new subtype candidate, but lack a sufficient number of closely related isolates to be identified. They remain categorized as unclassified lineages.
Among the pairwise nucleotide differences presented by the 16 QC genomes, six (QC152, QC113, QC196, QC180, QC74 and QC87) showed marginal percentages of 13–14.9 % from their nearest references, which fit into the gap of 13–15 % specified recently in a paper discussing the expanded HCV classification. This gap is a clear division to distinguish members of the same subtype that display differences <13 % from those of different subtypes that display differences >15 %. However, some exceptions have been noted for a few complete or nearly complete genomes that displayed pairwise nucleotide differences that fit into this gap, but whether such exceptions are due to technical problems or different epidemiological histories is unknown (Smith et al., 2014). As in a series of our recent studies, including the present one, we deliberately standardized all the experimental conditions to avoid possible cross-contamination, artificial recombination and other errors that may occur in full-length genome sequence assembly, such technical problems, are believed to have been minimized (Li et al., 2013). Therefore, the marginal distances presented by the six QC sequences are thought to reflect the natural viral evolution.
Analysis of unclassified HCV-1 sequences available in the Los Alamos HCV database revealed genotype 1 to be highly diverse and taxonomically complex. In addition to the 12 subtypes, 1a–1l, and two newly assigned subtypes, 1m and 1n, an additional 21 HCV-1 lineages that may represent new subtype candidates were also identified. Furthermore, there existed a number of untypable HCV-1 sequences in the core and E2 regions. Overall, at least 35 subtypes/lineages within genotype 1 could be identified. Excluding subtypes 1a and 1b which are distributed worldwide, the geographical sampling regions of the other 33 subtypes/lineages were found not to be as widespread. Although North America showed the largest genetic diversity where 22 HCV-1 subtypes/lineages have been detected, HCV-1 is believed to originate from Africa where 17 subtypes/lineages have been identified. This was also supported by the finding in the present study that the majority of the novel HCV-1 isolates detected in Canada were from African immigrants or descendants. Diverse HCV-1 isolates are also common in Europe where 10 subtypes/lineages have been found. This could result from the historical roles played by European explorers in Africa or other alternatives such as slave trading, recent immigration, geographical proximity etc.
There are two important findings in this study. The first finding is that subtype 1b is more closely related to subtypes 1d and 1i than to any other HCV-1 lineage. Both 1d and 1i are found not only in the French-speaking areas in North America and Europe, but also in North Africa (Algeria and Morocco) and West Africa (Ghana). Although there remains a lack of sufficient information to indicate the origin of 1b in an exact region of Africa, the limited data in this study seem to support its emergence later than 1a. The second finding is the close relationship observed between subtype 1a and the novel HCV-1 variants found in Haitian immigrants. This indicates an ancestral divergence correlating in time with a period when the transatlantic slave trade was active in Africa.
Currently, ~95 % of the Haitian population is descended directly from African slaves (CIA, 2008). The first wave of slaves came in 1502, but >75 % of them were imported during the late eighteenth century (Geggus, 2000, 2001). Although there is a lack of data on HCV molecular epidemiology in Haiti, a recent report regarding HBV genotype distribution may provide indirect information about the emergence of HCV in this country (Andernach et al., 2009). In that study, HBV subtypes A1, A5, D4 and D3 accounted for 43, 19.6, 16.2 and 3.9 % of strains, respectively, and were closely related to those identified in Africa. A1 represents the major HBV subtype in eastern Africa but it is essentially absent in West Africa. In contrast, A5 was only identified in the Bight of Benin, whilst D3 and D4 were only found in Rwanda. Analysis of the time of evolution indicated that the separation of the Haitian and African A5 strains was dated to 270 years ago, and that the separation of the Haitian and African A1 strains occurred in the past 100–190 years. The former timing was in agreement with the early phase of the slave trade which took place in West Africa (Geggus, 2001), whilst the latter timing was in accord with the late phase of slave importation which was mainly from eastern Africa (Morgan, 1998; Geggus, 2000, 2001). In this study, two important evolutionary times were estimated for HCV-1. Taking subtype 1a and all the Haitian HCV-1 variants as a whole, the common ancestor was dated to 176 (95 % HPD: 154, 197) years ago; considering all the HCV-1 subtypes/clusters as a single lineage, the common ancestor was dated to 326 (95 % HPD: 292, 361) years ago. The former timing is largely consistent with that of HBV A1, whilst the latter is congruent with that of HBV A5. The former timing seems to indicate that subtype 1a emerged only after the end of the transatlantic slave trade around the turn of the nineteenth century (Sepinwall, 2012). In contrast, the latter timing corresponds to the early phase of slave importation, by which the ancestral HCV-1 strains were likely brought outside Africa. The HCV-1 variants identified in Haitian immigrants were highly heterogeneous. This would suggest multiple early introductions of HCV strains into Haiti. Our understanding of the ancestral relationship that exists between subtype 1a and the Haitian variants characterized in this study is hampered by the lack of epidemiological data on HCV genotypes in Haiti. Closer and more recent 1a relatives may be identified if an extensive HCV survey is performed among the current Haitian population.
There exist a few uncertainties in this study with regard to the molecular rate and hence the evolutionary analysis. The major uncertainty is the possible higher rate we used and hence the underestimation of the ages of the HCV-1 common ancestors. We used the rate that was estimated previously based on the contemporarily sampled sequences from the globally epidemic subtype 1b strains (Yuan et al., 2013). However, the ancient rates may be lower than we estimate for modern HCV. Primarily, nucleotide substitutions of HCV are accumulated either within single infected individuals over time or upon the virus being transmitted into new hosts, likely with the latter being more significant. Compared with the ancient strains, modern HCV may have higher substitution rates because of more frequent and faster transmission to a wider range of individuals. In contrast, during the ancient era, most of the HCV strains may have been restrictively endemic with fewer transmissions, and hence lower substitution rates and a longer history of evolution for a given genetic distance. More importantly, the GORS (genome-scale ordered RNA structure) constriction on the viral genomic variations may lead to the possibility that many of the branches observed in the timescale trees actually occurred at more remote times in the past than we estimated (Simmonds, 2004). Thus, the ages of the HCV-1 common ancestors may in fact be much older. This would imply that all the HCV-1 diversity we observed among the Haitian immigrants could have been imported from Africa. This may help to explain why, when >75 % of the slaves arrived in Haiti in the late eighteenth century, the common ancestor we estimated for 1a and the subtypes from Haitian immigrants was dated to 1850.
The second uncertainty relates to possible sampling errors. Due to the limited number and the short period of time (only 20 years) for these contemporary HCV sequences to be sampled, sampling errors are unavoidable and a relatively small such error will have a very great impact on the estimated ages of the common ancestors. Other potential uncertainties may involve substitution saturation, different rates along branches and within genomic regions, etc. All evolutionary analyses of highly variable viruses such as HCV are subject to these uncertainties and therefore the results obtained need adjustments. However, currently we have little knowledge of these adjustments.
Methods
Subjects and specimens.
We first reviewed the data on patients who had been positive for HCV-1 in a previous report (Murphy et al., 2007). Serum samples had been collected from these patients for routine HCV genotyping during 2001–2006. Sixteen isolates were selected for entire genome sequencing because they represented HCV-1 subtypes or unassigned variants that lacked full-length genome sequences at the time when this project was initiated. The ethnic origins of these patients are seen in Fig. 1 and their ages ranged from 33 to 66 years.
Sequence amplification and analysis.
Each full-length HCV genome sequence was determined from 140 µl serum using the approaches we have described recently (Li et al., 2013) and the primers listed in Table S3. To reconstruct the ancestral relationship, a total of 40 full-length HCV genome sequences were retrieved from the Los Alamos HCV database and used as references. They represent not only HCV genotype 1, but also genotypes 2–7 (Kuiken et al., 2005). Based on this full-length sequence dataset, pairwise genetic distances were calculated using mega5 software (Tamura et al., 2011). To further comprehend the genetic complexity of HCV-1, a set of partial NS5B sequences representing all the assigned subtypes and unclassified variants was selected for analysis. The resulting dataset contained 309 sequences each having 305 nt corresponding to nt 8316–8620 in the H77 genome. For both sequence datasets, phylogenetic trees were reconstructed and for the full-length genome sequence dataset, virus recombination events were excluded, as described recently (Li et al., 2013). To better classify the HCV-1 strains, all the unassigned HCV-1 sequences in the Los Alamos HCV database were retrieved and analysed with the available full-length HCV-1 sequences. In total, 2689 sequences were identified that did not have a subtype assignment. After excluding recombinants, multiple clones and those from animals, 1347 sequences were found to represent individual isolates.
Evolutionary analysis.
Based on the full-length genome sequence dataset, a timescaled phylogenetic tree was estimated using the Bayesian Markov chain Monte Carlo (MCMC) algorithm implemented in the beast package (version 1.6.1) (Drummond & Rambaut, 2007). Recently, we analysed the full-length subtype 1a and 1b sequences, and identified that the log-normal model is better than the exponential and strict clock models (Yuan et al., 2013). Thus, we used this model in a combination with the GTR+I+Γ substitution model and the Bayesian skyline model for estimating the timescaled phylogenetic tree in this study. However, because the full-length sequence dataset we assembled lacked a sufficient temporal structure to allow for the direct estimation of an evolutionary rate, we had to use an outer rate of 1.13×10−3±6.66×10−6 substitutions per site per year which we estimated recently for subtype 1b (Yuan et al., 2013). Using these parameters, we ran the MCMC procedure for 100 million states and logged out a tree in every 10 000 states. After discarding the first 10 % burn-in, the output was examined for convergence by visual inspection of the chain length and comparing the statistics of the effective sample size (ESS) using the Tracer program (http://tree.bio.ed.ac.uk). Sufficient sampling was considered to have been achieved when all of the ESS numbers were >200. We used the TreeAnnotator program to summarize a tree from the resulting set of credible trees, which is called the MCC (maximum clade credibility) tree. As a molecular clock was incorporated, the branch lengths and the tree node height were marked in units of years. Phylogenetic structure was then displayed using the FigTree program, in which clades, lineages and internal node heights were indicated according to needs.
Acknowledgements
The study described was supported by a grant from the National Institute of Allergy and Infectious Diseases (5 R01 AI080734-03A).
Footnotes
Three supplementary tables are available with the online version of this paper.
References
- Alter M. J. (2007). Epidemiology of hepatitis C virus infection. World J Gastroenterol 13, 2436–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andernach I. E., Nolte C., Pape J. W., Muller C. P. (2009). Slave trade and hepatitis B virus genotypes and subgenotypes in Haiti and Africa. Emerg Infect Dis 15, 1222–1228 10.3201/eid1508.081642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracho M. A., Saludes V., Martró E., Bargalló A., González-Candelas F., Ausina V. (2008). Complete genome of a European hepatitis C virus subtype 1g isolate: phylogenetic and genetic analyses. Virol J 5, 72. 10.1186/1743-422X-5-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (1998). Recommendations for prevention and control of hepatitis C virus (HCV) infection and HCV-related chronic disease. MMWR Recomm Rep 47 (RR-19), 1–39 [PubMed] [Google Scholar]
- CIA (2008). World Factbook [accessed 11 May 2103]. https://www.cia.gov/library/publications/download/download-2008
- Demetriou V. L., Kostrikis L. G. (2011). Near-full genome characterization of unclassified hepatitis C virus strains relating to genotypes 1 and 4. J Med Virol 83, 2119–2127 10.1002/jmv.22237 [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A. (2007). beast: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7, 214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geggus D. (2000). La traite des esclaves aux Antilles francaises à la fin du 18me siècle: quelques aspects du marché local. In Négoce, ports et océans, XVIe–XXe siècles, pp. 235–245 Edited by Marzagalli S., Bonin H. Bordeaux: Presses Universitaires de Bordeaux [Google Scholar]
- Geggus D. (2001). The French slave trade: an overview. William Mary Q 58, 119–138 10.2307/2674421 [DOI] [PubMed] [Google Scholar]
- Jeannel D., Fretz C., Traore Y., Kohdjo N., Bigot A., Pê Gamy E., Jourdan G., Kourouma K., Maertens G. & other authors (1998). Evidence for high genetic diversity and long-term endemicity of hepatitis C virus genotypes 1 and 2 in West Africa. J Med Virol 55, 92–97 [DOI] [PubMed] [Google Scholar]
- Kuiken C., Yusim K., Boykin L., Richardson R. (2005). The Los Alamos hepatitis C sequence database. Bioinformatics 21, 379–384 10.1093/bioinformatics/bth485 [DOI] [PubMed] [Google Scholar]
- Li C., Njouom R., Pépin J., Nakano T., Bennett P., Pybus O. G., Lu L. (2013). Characterization of full-length hepatitis C virus sequences for subtypes 1e, 1h and 1l, and a novel variant revealed Cameroon as an area in origin for genotype 1. J Gen Virol 94, 1780–1790 10.1099/vir.0.048835-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan P. D. (1998). Slave trade: transatlantic. In Macmillan Encyclopedia of World Slavery, pp. 837–844 Edited by Finkelman P., Miller J. C. New York: Macmillan Reference [Google Scholar]
- Murphy D. G., Willems B., Deschênes M., Hilzenrat N., Mousseau R., Sabbah S. (2007). Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5′ untranslated region sequences. J Clin Microbiol 45, 1102–1112 10.1128/JCM.02366-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus O. G., Markov P. V., Wu A., Tatem A. J. (2007). Investigating the endemic transmission of the hepatitis C virus. Int J Parasitol 37, 839–849 10.1016/j.ijpara.2007.04.009 [DOI] [PubMed] [Google Scholar]
- Sepinwall A. G. (2012). Haitian History: New Perspectives. New York: Routledge [Google Scholar]
- Simmonds P. (2004). Genetic diversity and evolution of hepatitis C virus – 15 years on. J Gen Virol 85, 3173–3188 10.1099/vir.0.80401-0 [DOI] [PubMed] [Google Scholar]
- Simmonds P., Bukh J., Combet C., Deléage G., Enomoto N., Feinstone S., Halfon P., Inchauspé G., Kuiken C. & other authors (2005). Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42, 962–973 10.1002/hep.20819 [DOI] [PubMed] [Google Scholar]
- Smith D. B., Bukh J., Kuiken C., Muerhoff A. S., Rice C. M., Stapleton J. T., Simmonds P. (2014). Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59, 318–327 10.1002/hep.26744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). mega5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (1997). Hepatitis C. Wkly Epidemiol Rec 72, 65–69 [PubMed] [Google Scholar]
- Yuan M., Lu T., Li C., Lu L. (2013). The evolutionary rates of HCV estimated with subtype 1a and 1b sequences over the ORF length and in different genomic regions. PLoS ONE 8, e64698 10.1371/journal.pone.0064698 [DOI] [PMC free article] [PubMed] [Google Scholar]