Abstract
In Colombia, knowledge of the yeast and yeast-like fungi community is limited because most studies have focused on species with clinical importance. Sediments and water represent important habitats for the study of yeast diversity, especially for yeast species with industrial, biotechnological, and bioremediation potential. The main purpose of this study was to identify and compare the diversity of yeast species associated with sediment and water samples from two artificial lakes in Universidad del Valle (Cali-Colombia). Yeast samplings were performed from fifteen sediment samples and ten water samples. Grouping of similar isolates was initially based on colony and cell morphology, which was then complemented by micro/mini satellite primed PCR banding pattern analysis by using GTG5 as single primer. A representative isolate for each group established was chosen for D1/D2 domain sequencing and identification. In general, the following yeast species were identified: Candida albicans, Candida diversa, Candida glabrata, Candida pseudolambica, Cryptococcus podzolicus, Cryptococcus rajasthanensis, Cryptococcus laurentii, Williopsis saturnus, Hanseniaspora thailandica, Hanseniaspora uvarum, Rhodotorula mucilaginosa, Saccharomyces cerevisiae, Torulaspora delbrueckii, Torulaspora pretoriensis, Tricosporon jirovecii, Trichosporon laibachii and Yarrowia lypolitica. Two possible new species were also found, belonging to the Issatchenkia sp. and Bullera sp. genera. In conclusion, the lakes at the Universidad del Valle campus have significant differences in yeast diversity and species composition between them.
Keywords: yeast diversity, artificial lake, molecular identification, MSP-PCR fingerprinting
Introduction
Unlike other fungi, yeasts and yeast-like organisms can be potentially found everywhere in an aquatic ecosystem. They present an ecological flexibility that allows them to tolerate a wide range of salinity, environmental temperatures, oxygen saturation, and acidity in the surrounding medium (Boguslawska-Was and Dabrowski, 2001). They have been isolated from lake and sea sediments (Boguslawska-Was and Dabrowski, 2001; MacGillivray and Shiaris, 1993) and from all the levels of the water column (Wurzbacher et al., 2010).
A wide variety of yeasts have been isolated from lakes with different organic loads. They have been found in oligotrophic lakes in Patagonia, Argentina (Brandão et al., 2011), mesotrophic lakes in the United States (van Uden and Ahearn, 1963), in lakes receiving sewage discharges (Meyers et al., 1970), and lakes for recreational tourism in Brazil (Medeiros et al., 2008). In most cases, the predominant species found belonged to the Rhodotorula, Candida, and Cryptococcus genera. Pathogenic species of these genera were found in lakes contaminated by human activities and can be used as biological indicators to establish the contamination levels of these water bodies (Hagler, 2006).
Besides their function as water quality indicators, the diversity and identification of yeast species can be useful as a first approach to evaluate potential characteristics useful in industry, biotechnology, or bioremediation; a potential that comes from the ability of yeasts and yeast-like fungi to live in variable environments, which is not only due to their quick growth but also to the adaptation of their enzymatic apparatus to unpredictable food sources (Boguslawska-Was and Dabrowski, 2001). For these reasons, the purpose of this research was to isolate, identify, and compare the diversity of the yeasts species between the lakes at Universidad del Valle in Cali, Colombia, by employing Mini/Micro-satellite primed-PCR fingerprinting (MSP-PCR) and sequencing of D1/D2 domains.
Materials and Methods
Sampling sites
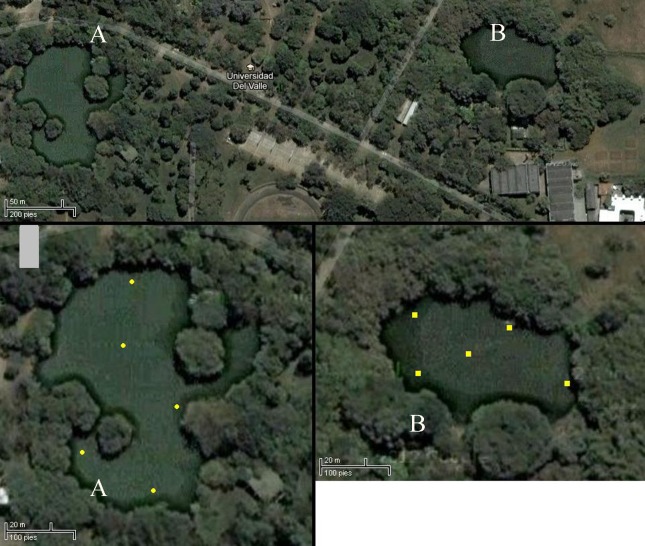
Yeast samplings were collected from two artificial lakes located inside the main campus of the Universidad del Valle (Figure 1). These lakes were built approximately 30 years ago and are nourished by the Rio Melendez’s waters which enter first to Central Lake and overflows through a water pipeline to Estación Lake. Both lakes have an average depth of 3 m at their centers and are completely surrounded by 3–15 m trees. Central Lake mainly has fruiting trees like Mango (Mangifera indica) and Pomarroso (Syzygium malaccense) and Estación Lake is predominantly surrounded by ornamental trees like Samán (Samanea saman) and Chiminango (Pithecellobium dulce).
Figure 1.
Satellite image of the lakes and location of the sampling sites. Central Lake (A); Estación Lake (B). (Image obtained from Google Maps).
Yeast sampling
From March to May 2011, five independent 100 g sediment samples per month were collected from each artificial lake with a lab-manufactured scoop consisting of a metal glass attached to a 3 m extendable pole stick. The samples were deposited in sterile screw-capped glass bottles. The sampling site locations are shown in Figure 1. Five samples of water, from the same sampling sites in each lake, were collected in June at a depth of c. 30 cm using sterile screw-capped glass bottles. All the samples were immediately processed in the “Laboratorio de Biología Molecular de Microorganismos” form the Universidad del Valle.
Samplings were performed at midday (12:00–14:00 h), water temperature, dissolved oxygen and conductivity were recorded in situ with an YSI model 85; pH was measured in the laboratory using a Cole-Parmer 5800 pH-meter.
Yeast isolation
Three serial dilutions in peptone water were performed from each sediment sample (10−1, 10−2, and 10−3). Equal volumes (200 μL) from each dilution were spread over the surface of glucose-peptone-yeast extract agar plates (GPY, glucose 2%, peptone 2%, yeast extract 1%, agar 2%) supplemented with 25 mg/L of penicillin and chloramphenicol. A total of 500 μL of each water sample, without dilution, were spread over the same media and all the plates were incubated for three days at room temperature (c. 25 °C).
The yeasts were chosen for isolation based on colony morphology. Yeast CFUs were registered for quantitative analysis of yeast occurrence during the third day of incubation. All the isolates were preserved in GPY broth supplemented with 30% glycerol at −20 °C in the Universidad del Valle’s culture collection.
All yeasts were preliminarily grouped based on their cultural morphology. The groups were confirmed genetically by PCR fingerprinting, using MSP-PCR (Silva-Filho et al., 2005).
DNA Extraction
The DNA extraction was performed according to Osorio-Cadavid et al. (2009). DNA quantization and purity was performed spectophotometrically at a 260-nm wavelength and with the spectophotometric relation 260/280 nm, respectively, using a NanoDrop 2000 (v. 1.0, Termo Scientific, United States).
PCR Fingerprinting
The synthetic oligonucleotide (GTG)5 was used in MSP-PCR experiments. The PCR reactions were performed according to Silva-Filho et al. (2005). Yeast strains with 80% or more similar DNA banding patterns were grouped and initially considered as belonging to the same species as stated by Silva-Filho et al. (2005). At least one representative strain from each MSP-PCR group was subjected to sequence analysis of the D1/D2 domains of the large subunit of the rRNA gene, as described ahead.
Yeast molecular identification
The D1/D2 variable domains of the large subunit of the rRNA gene were amplified as described previously by López et al. (2010) by using the NL-1 (5-GCATATCAATAAGCGGAGGAAAAG-3) and NL-4 (5-GGTCCGTGTTTCAAGACGG-3) primers. Sequencing of the D1/D2 domains was performed directly from PCR products. The sequences obtained were compared to those included in the GenBank database by using the Basic Local Alignment Search Tool (BLAST at http://www.ncbi.nlm.nih.gov). Identification of the strains to species level was carried out according to Kurtzman et al. (2011).
Statistical analysis
Species diversity was measured in terms of the richness, evenness, and dominance given by three indexes: Shannon H, Simpson’s index, and Dominance. All results were obtained with 95% confidence, and bootstrap values were calculated from 1000 iterations. Species richness refers to the number of species in a community, and species dominance refers to the contribution of individuals. The index calculations, Nonmetric Multi-Dimensional Scaling (N-MDS) and one-way similarity analysis (ANOSIM) were performed by using the PAST computer program, version 2.14 (Hammer et al., 2001).
The physicochemical data were processed in an Excel (2004) spreadsheet and a t test was performed to analyze the differences between both lakes, based on the statistical hypothesis that no significant differences exist between them.
Results
Yeast sampling
An average of the physicochemical data obtained during samplings is displayed in Table 1. The statistical analysis performed presented significant differences (p < 0.05) between both lakes in all the parameters. The Central Lake has higher temperature and dissolved oxygen but lower pH and conductivity than Lake Estación.
Table 1.
Average of physicochemical parameters analyzed in the Central Lake and Lake Estación
| Parameter | Lake Estación | Central Lake | p-value | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| Water | Sediment | Water | Sediment | Water | Sediment | |
| D. Ox. (mg/L) | 4.92 | - | 5.3 | - | 4.75E-04 | - |
| Temperature (ºC) | 25.2 | 23.5 | 25.8 | 22.9 | 5.95E-04 | 5.37E-05 |
| Conductivity | 60.8 | - | 52.8 | - | 7.24E-06 | - |
| pH | 6.8 | 6.8 | 5.6 | 5.5 | 3.23E-03 | 4.26E-03 |
Yeast isolation
A total of 138 isolates were obtained: 74 from the Central Lake and 64 from Lake Estación. The average yeast count for the sediment of the Central Lake was 1300 cells/g and 1100 cells/g for Lake Estación. In water, the average yeast counts were 8.5 × 104 cells/l for Central Lake and 7.5 × 104 cells/l for Estación Lake. Based on colony and cell morphology, 16 groups were formed: 14 with several similar isolates and two with unique isolates.
MSP-PCR Fingerprinting
The average DNA concentration obtained from the isolates was 1200 ng/μL, with a purity of 2.14. By using GTG5 as single primer to determine the genetic profiles of the groups made from morphological characteristics, one to 10 DNA bands per isolate were visualized with a molecular weight ranging from 200 to 2200 bp.
Characterization via MSP-PCR fingerprinting allowed the formation of seven groups with similar DNA banding patterns and the discrimination of unique isolates (data not shown).
Sequencing analysis
Some unique isolates and several isolates from each major group were chosen for molecular identification. The isolates identified their accession number from GenBank, the species they matched and the percentage of identity they presented with the type of strain sequence of each species is shown in Tables 2 and 3 for each lake.
Table 2.
Sequence analysis with BLAST algorithm for the 19 isolates sequenced.
| Isolate Accession | Species | % identity | Type strain accession |
|---|---|---|---|
| JQ672604 | Rhodotorula mucilaginosa | 99% | AF070432 |
| JQ672605 | Candida glabrata | 99% | U44808 |
| JQ672606 | Torulaspora delbrueckii | 99% | U72156 |
| JQ672607 | Issatchenkia sp. | 99% | AB247501 |
| JQ672608 | Candida diversa | 99% | U71064 |
| JQ672609 | Saccharomyces cerevisiae | 99% | AY048154 |
| JQ672610 | Torulaspora pretoriensis | 100% | U72157 |
| JQ672615 | Candida pseudolambica | 99% | U71063 |
| JQ672617 | Cryptococcus podzolicus | 99% | AF075481 |
| JQ672619 | Hanseniaspora uvarum | 99% | U84229 |
| JQ672588 | Yarrowia lipolytica | 99% | U40080 |
| JQ672589 | Trichosporon jirovecii | 100% | AF105398 |
| JQ672590 | Williopsis saturnus | 99% | U75958 |
| JQ672591 | Candida albicans | 99% | AY497682 |
| JQ672594 | Hanseniaspora thailandica | 99% | DQ404527 |
| JQ672597 | Trichosporon laibachii | 99% | AF075514 |
| JQ672598 | Bullera sp. | 94% | AF363660 |
| JQ672599 | Cryptococcus laurentii | 99% | AB35042 |
| JQ672600 | Cryptococcus rajasthanensis | 100% | AM262324 |
Table 3.
Species and number of isolates per species found in Central and Estación lakes.
| Species | Lakes | Species | Lakes | ||
|---|---|---|---|---|---|
|
|
|
||||
| Central | Estación | Central | Estación | ||
| Y. lipolytica | 2 | 0 | H. uvarum | 0 | 11 |
| W. saturnus | 8 | 0 | C. rajasthanensis | 6 | 6 |
| T. laibachii | 1 | 0 | C. podzolicus | 0 | 5 |
| T. jirovecii | 3 | 0 | C. laurentii | 7 | 0 |
| T. pretoriensis | 0 | 3 | Candida sp. 7 | 2 | 1 |
| T. delbrueckii | 5 | 1 | Candida sp. 16 | 1 | 0 |
| S. cerevisiae | 3 | 1 | Candida sp. 13 | 4 | 0 |
| R. muscilaginosa | 0 | 3 | C. pseudolambica | 0 | 5 |
| Pichia sp. 9 | 3 | 1 | C. glabrata | 0 | 4 |
| Pichia sp. 11 | 2 | 0 | C. diversa | 16 | 2 |
| Pichia sp. 10 | 3 | 4 | C. albicans | 3 | 0 |
| Issatchenkia sp. | 0 | 17 | Bullera sp. | 3 | 0 |
| H. thailandica | 2 | 0 | Total | 74 | 64 |
A total of nineteen species were identified. The species obtained and their numbers of isolates for each lake are presented in Figure 1. Twenty-two isolates belonging to six groups could not be sequenced and were transiently classified as Pichia sp. or Candida sp.
Statistical analysis
The Shannon, Simpson, and Dominance indexes and their p-values are shown in Table 4. These values (p < 0.05) indicate significant differences between the diversities of the lakes.
Table 4.
Statistical analysis of yeast diversity between Central and Estación lakes.
| Index | Central Lake | Lake Estación | p-value |
|---|---|---|---|
| Taxa | 18 | 14 | - |
| # of Individuals | 74 | 67 | - |
| Shannon H | 2.625 | 2.32 | 0.036 |
| Simpson | 0.91 | 0.87 | 0.038 |
| Dominance | 0.095 | 0.13 | 0.038 |
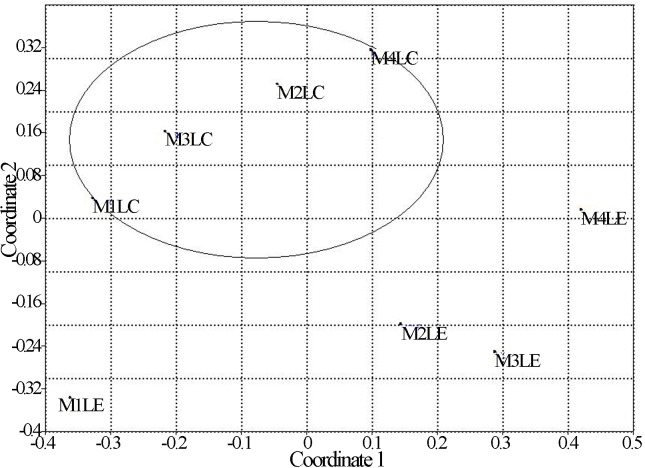
The similarity analysis, ANOSIM, reported R = 0.642 with a p-value < 0.05. The N-MDS analysis shown in Figure 2 also displays significant differences between the communities of the lakes.
Figure 2.
N-MDS analysis. The circled group represents Central Lake (LC). M1, M2 and M3 are the sediment samples from Central Lake and Estación Lake (LE). M4 are the water samples from each lake.
Discussion
The count of yeast cells per liter showed that the abundance of yeast in the water of the lakes at Universidad del Valle is high and consistent with a eutrophic ecosystem, by being above 1000 cells/L (Hagler, 2006). This high count in water indicates that the isolation method used was efficient, considering that only a small fraction of known organisms can be cultivated on artificial media and under laboratory conditions.
The analysis of the D1/D2 region sequences proved a very efficient technique to identify yeast species. It allowed identifying to species level with 99% or more percentage of identity for all isolates tested, when compared with sequences stored in GenBank and subsequently with sequences of the type of strains of each of their species. Several isolates coincided with species that have not yet been officially described by the researchers who discovered them (Issatchenkia sp. and Bullera sp.) and we expect to contribute with some data to the taxonomic description of these species.
The diversity of species in each lake showed significant differences according to the Shannon index (p < 0.05). This index reveals that both lakes have low diversities because they are between 0 and 3, but that they are not equal because of the higher number of species found in the Central Lake (18 species). Possibly, the significant differences found in the physicochemical parameters between these two environments allow each lake to provide the resources and conditions necessary to sustain the same yeast species richness but not the same diversity of species.
Although the ecological flexibility of yeast allows them to occupy all aquatic habitats because of their ability to tolerate wide ranges of salinity, temperature, and acidity in the surrounding medium (Boguslawska-Was and Dabrowski, 2001), some may be more sensitive to these changes. Possible examples of yeast with high ecological flexibility can be the ones isolated from both lakes. Species composition was similar between the lakes by 23%, sharing the C. diversa, C. rajasthanensis, T. delbrueckii species and three that were not possible to identify to species level, belonging to the Pichia and Candida genera. One possible cause of the presence of these species in both lakes could be the fact that there is a flow of water from Central Lake to Lake Estación, which can serve as a means of transport, especially for species found in water.
It is important to note that the vast majority of species shared was in greater abundance in the Central Lake and that not all species found in the Central Lake water were found in Lake Estación (Table 3). This implies that the significant differences obtained in the physicochemical parameters of water, plus the ecological flexibility inherent to certain species, may also be selecting the species that manage to move to Lake Estación. These differences in water conditions can explain the result of N-MDS analysis (Figure 2), where a separate group for each lake was formed and was statistically validated by the ANOSIM, revealing that there are significant differences in the composition of yeast species between the lakes.
The groups formed in the N-MDS analysis also match the results obtained for Simpson and Dominance indexes (Table 4), where different species are predominant in each lake. The most predominant species are Issatchenkia sp. for Lake Estación and C. diversa for the Central Lake. In second place of dominance is H. uvarum for Lake Estación and W. saturnus for the Central Lake, followed by unique species for each lake found in lower abundance.
The majority of yeasts isolated are usually found in nature, mainly on the ground; the species with pathogenic potential are often associated with animal feces and anthropogenic pollution (Hagler, 2006). Among the species identified, several have been reported with biotechnological, industrial, and bioremediation potential. This is the case of W. saturnus found in the Central Lake. This is a species with worldwide distribution, frequently isolated from soil, fresh water, and litter that has shown potential as a biocatalyst to oxidize secondary racemic alcohols, contributing to the development of models to understand the activity of these microorganisms and to compare the substrate specificity requirements of the enzymes involved (Carballeira et al., 2004).
From the Torulaspora genus, T. delbrueckii and T. pretoriensis species were isolated. T. delbrueckii is a globally distributed species that has been isolated from soil, tree bark, fruits, and fermented foods (Kurtzman et al., 2011). This species was also isolated from inflorescences of Mango trees (Mangifera indica) (Gaviria, 2012), which is the second-most abundant tree around the Central Lake. It is possible that the abiotic conditions found in the Central Lake allow this yeast species to remain and reproduce in the water and sediment after falling with some inflorescences or being washed from them with rain. For their ability to ferment, their little loss of viability after long periods stored at freezing temperatures, and the contribution of new flavors and odors to wine, this species is interesting for bread, pastry, and wine production industries (Alves-Araújo et al., 2004; Bely et al., 2008; Holm-Hansen et al., 2001). T. pretoriensis has also been isolated from soil and some cocoa fermentation, but needs further study (Kurtzman et al., 2011).
Hanseniaspora uvarum has been isolated from soil, insects, fruits, marine and freshwater and it is considered a species with global distribution (Kurtzman et al., 2011). It was also specifically isolated from Pomarroso fruits (Syzygium malaccense) (Usman 2012, unpublished data) the most abundant tree species around the Central Lake. However, this species was not present in isolates from this lake, but it was isolated from Lake Estación where this tree is not present, leading to other possible sources of origin different from the fruits of this tree. It is possible that this and other species, previously isolated from soil, can live in the sediment and suspended in the water column. Some strains of this species have enological importance for the production of enzymes responsible for releasing aromatic compounds in wines (Kurtzman et al., 2011).
In addition to species with biotechnological potential, potentially pathogenic species recognized as bioindicators of water quality were also isolated. Candida sp., Rhodotorula sp., Cryptococcus sp., and Trichosporon sp. are yeasts genera that can be used as bio-indicators, but their presence should not only be limited to their analysis as indicators (Hagler et al., 2006). Yeasts belonging to all these genera were found in both lakes and it is feasible that they come from the contaminated waters of the Meléndez River or from the feces of the fauna associated with each lake.
In the Central Lake, more potentially pathogenic species such as C. albicans, C. laurentii, T. jirovecii, and T. laibachii were found. All these species have been isolated from clinical samples but are also common in nature, like C. albicans. This species is known worldwide for producing fungal infections in humans, especially candidiasis in people with compromised immune systems (Cooper, 2011), but it has also been isolated from decaying litter (Kurtzman et al., 2011) and Pomarroso fruits (Syzygium malaccense) (Usman 2012, unpublished data), the most abundant tree around the Central Lake.
In Lake Estación, a pathogenic yeast belonging to the genus Candida was also found. Candida glabrata is considered an emerging pathogen, common in nosocomial infections from immune-compromised patients. It usually affects the oral cavity, urogenital tract, and bloodstream and has an innate resistance to “azole” antifungals (Kurtzman et al., 2011).
Other non-pathogenic species of Candida were also found in the lakes. C. pseudolambica is regarded as a cosmopolitan species that has been isolated from soil and from bivalves feeding from Mangrove trees (Rhizophora mangle) (Kurtzman et al., 2011). C. diversa has been isolated from different grape musts and orange juices and it is known for its inability to assimilate all carbohydrates except standard glucose. These species have been poorly studied and there is no information about their ecology or their potential uses in biotechnology (Kurtzman et al., 2011).
Cryptococcus laurentii is another potentially pathogenic species that has been isolated from soil, seawater, decaying wood (Kurtzman et al., 2011; Mussa et al., 2000), and recently from Mango fruits (Mangifera indica) (Usman 2012, unpublished data), which occurs frequently around the Central Lake. This species is also important in biotechnology because some isolates have presented potentially desirable characteristics for yeast and fungal contaminant control in various food processes and products (Filonow, 2001).
Species from the Trichosporon genus are in second place of pathogenicity after the Candida species (Chagas-Neto et al., 2008). In the Central Lake, the T. jirovecii and T. laibachii species were found and both are considered pathogenic because they are frequently isolated from clinical and animal feces samples. The T. jirovecii species was described from a strain isolated from human nails (Kurtzman et al., 2011), indicating possible anthropogenic contamination of the Melendez River water that feeds the Central Lake. T. laibachii has been isolated from bat guano (Sugita et al., 2005), mammals that are possibly abundant around the Central Lake because of the amount of fruit trees such as Mango and Pomarroso that exist on its banks.
Rhodotorula mucilaginosa is possibly the most ubiquitous species of yeast. It has been found in all possible habitats, the atmosphere (Kurtzman et al., 2011), and in oligotrophic lakes (Brandão et al., 2011). It is also considered an indicator species for causing some human mycoses, along with others from the genre whose main characteristic is the production of carotenoid compounds that give them their distinctive pink color (Hagler, 2006). By being indicator species and easy to distinguish based on their color, pink colony count has been proposed as a rapid method for the analysis of water quality (Hagler, 2006).
Besides being considered pathogenic, studies have shown that species of the Cryptococcus and Trichosporon genera may also play an important role in bioremediation of polycyclic aromatic hydrocarbons and heavy metals, given that they tend to incorporate them in their metabolism; thus, extracting them from the environment (Botha, 2006; MacGillivray and Shiaris, 1993). Isolates of C. podzolicus were found able to withstand up to 500 ppm Copper (Cornelissen et al., 2003) and several isolates have presented production of epoxide hydrolases with promising characteristics to obtain enantiopure epoxides and vicinal diols in enantio-selective hydrolysis of racemic epoxides for the preparation of biologically active compounds (Botes et al., 2005).
Another species considered an emerging pathogen, but with proven applications in bioremediation and biotechnology is Yarrowia lipolytica (Kurtzman et al., 2011). This species is predominantly found in marine environments (Hagler and Mendonça-Hagler, 1979) and it is considered an emerging pathogen because it has recently been found associated with some human mycoses (Kurtzman et al., 2011). Y. lipolytica is known to degrade fats and n-paraffins (Peters and Nelson, 1948; Tsugawa et al., 1969), desirable characteristics for bioremediation processes and their presence is taken as a possible indicator of industrial pollution of water and soil (Hagler, 2006). Some isolates of this species have shown tolerance to high concentrations of chromium (III) (Raspor and Zupan, 2006).
Hanseniaspora thailandica is a new species recently described by Jindamorakot et al. (2009). These researchers isolated this species from various natural substrates like insect feces, flowers of Sonneratia caseolaris, lichens, and decomposed fruits of Psidium guajava. This species was also isolated by Usman (2012, unpublished data) from Mango (Mangifera indica) and Pomarroso fruits (Syzygium malaccense), the two most-abundant tree species around the Central Lake. This is the first report of this species in sediments from an artificial lake.
Conclusions
The Central Lake and Lake Estación have different species composition despite their proximity and water flow from the Central Lake to Lake Estación. Probably, the significant differences in the physicochemical parameters of the lakes may be selecting the yeast species that inhabit them. Similarly, the fauna and flora around the lakes and the Rio Melendez’s water can influence the diversity of yeast in them.
Two yeast strains isolated from the lakes were found to be undescribed new species and the species identified have important potential uses in industry, biotechnology, and bioremediation.
References
- Alves-Araújo C, Almeida MJ, Sousa MJ, Leão C. Freeze tolerance of the yeast Torulaspora delbrueckii: cellular and biochemical basis. FEMS Microbiol Lett. 2004;240:7–14. doi: 10.1016/j.femsle.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Bely M, Stoeckle P, Masneuf-Pomarède I, Dubourdieu D. Impact of mixed Torulaspora delbrueckii-Saccharomyces cerevisiae culture on high-sugar fermentation. Int J Food Microbiol. 2008;122:312–320. doi: 10.1016/j.ijfoodmicro.2007.12.023. [DOI] [PubMed] [Google Scholar]
- Boguslawska-Was E, Dabrowski W. The seasonal variability of yeasts and yeast-like organisms in water and bottom sediment of the Szczecin Lagoon. Int J Hyg Envir Heal. 2001;203:451–458. doi: 10.1078/1438-4639-00056. [DOI] [PubMed] [Google Scholar]
- Botes AL, Lotter JO, Rhode HJ, Botha A. Interspecies differences in the enantioselectivity of epoxide hydrolases in Cryptococcus laurentii (Kufferath) C.E. Skinner and Cryptococcus podzolicus (Bab’jeva & Reshetova) Golubev. Syst Appl Microbiol. 2005;28:27–33. doi: 10.1016/j.syapm.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Botha A. Yeasts in soil. In: Gábor P, Rosa C, editors. Biodiversity and Ecophysiology of Yeasts. Springer; Berlin: 2006. pp. 221–240. 2006. [Google Scholar]
- Brandão LR, Libkind D, Vaz ABM, Espírito Santo LC, Moliné M, de García V, van Broock M, Rosa CA. Yeasts from an oligotrophic lake in Patagonia (Argentina): diversity, distribution and synthesis of photoprotective compounds and extracellular enzymes. FEMS Microbiol Ecol. 2011;76:1–13. doi: 10.1111/j.1574-6941.2010.01030.x. [DOI] [PubMed] [Google Scholar]
- Carballeira JD, Quezada MA, Álvarez E, Sinisterra JV. High Throughput Screening and QSAR-3D/CoMFA: Useful Tools to Design Predictive Models of Substrate Specificity for Biocatalysts. Molecules. 2004;9:673–693. doi: 10.3390/90800673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagas-Neto TC, Chaves GM, Colombo AL. Update on the Genus Trichosporon. Mycopathologia. 2008;166:21–132. doi: 10.1007/s11046-008-9136-x. [DOI] [PubMed] [Google Scholar]
- Cooper CR. Yeast pathogenic to humans. In: Kurtzman CP, Fell JW, Boekhout T, editors. The yeasts: A taxonomic study. Elsevier; The United States: 2011. pp. 9–19. [Google Scholar]
- Cornelissen S, Botha A, Conradie WJ, Wolfaardt GM. Shifts in community composition provide a mechanism for maintenance of soil yeast activity in the presence of elevated copper levels. Can J Microbiol. 2003;49:425–432. doi: 10.1139/w03-057. [DOI] [PubMed] [Google Scholar]
- Filonow AB. Butyl Acetate and yeasts interact in adhesion and germination of Botrytis cinerea conidia in vitro and in fungal decay of Golden Delicious Apple. J Chem Ecol. 2001;27:831–844. doi: 10.1023/a:1010314305461. [DOI] [PubMed] [Google Scholar]
- Gaviria J, Osorio-Cadavid E. Diversidad de levaduras asociadas a inflorescencias de mango y flores de “Lulo Arbóreo”. Rev Bio Agro. 2012;10:160–169. [Google Scholar]
- Hagler AN. Yeasts as Indicators of Environmental Quality. In: Gábor P, Rosa C, editors. Biodiversity and Ecophysiology of Yeasts. Springer; Berlin: 2006. pp. 515–532. 2006. [Google Scholar]
- Hagler AN, Mendonça-Hagler LC. Candida lipolytica isolated from Guanabara Bay and its ability to grow in marine and estuarine conditions. Rev Bras Pesqui Med Biol. 1979;12:273–277. [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica. 2001;4:9. http://palaeo-electronica.org/2001_1/past/issue1_01.htm. [Google Scholar]
- Holm-Hansen E, Nissen P, Sommer P, Nielsen JC, Arneborg N. The effect of oxygen on the survival of non-Saccharomyces yeasts during mixed culture fermentations of grape juice with Saccharomyces cerevisiae. J Appl Microbiol. 2001;91:541–547. doi: 10.1046/j.1365-2672.2001.01426.x. [DOI] [PubMed] [Google Scholar]
- Jindamorakot S, Ninomiya S, Limtong S, Yongmanitchai W, Tuntirungkij M, Potacharoen W, Tanaka K, Kawasaki H, Nakase T. Three news pecies of bipolar budding yeasts of the genus Hanseniaspora and its anamorph Kloeckera isolated in Thailand. FEMS Yeast Res. 2009;9:1327–1337. doi: 10.1111/j.1567-1364.2009.00568.x. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Fell JW, Boekhout T, editors. The yeasts: A taxonomic study. Elsevier; USA: 2011. p. 2100. [Google Scholar]
- López-Arboleda WA, Ramírez-Castrillón M, Mambuscay-Mena LA, Osorio-Cadavid E. Diversidad de levaduras asociadas a chichas tradicionales de Colombia. Rev Colomb Biotecnol. 2010;12:176–186. [Google Scholar]
- MacGillivray AR, Shiaris MP. Biotransformation of Polycyclic Aromatic Hydrocarbons by yeasts isolated from coastal sediments. Appl Environ Microb. 1993;59:1613–1618. doi: 10.1128/aem.59.5.1613-1618.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros AO, Kohler LM, Hamdan JS, Missagia BS, Barbosa FAR, Rosa CA. Diversity and antifungal susceptibility of yeasts from tropical freshwater environments in Southeastern Brazil. Water Res. 2008;42:3921–3929. doi: 10.1016/j.watres.2008.05.026. [DOI] [PubMed] [Google Scholar]
- Meyers SP, Ahearn DG, Cook WL. Mycological studies of lake Champlain. Mycologia. 1970;62:504–515. [Google Scholar]
- Mussa AY, Randhawa HS, Khan ZU. Decaying wood as a natural habitat of melanin-forming (Mel+) variant of Cryptococcus laurentii. Current Sci India. 2000;79:1471–1474. [Google Scholar]
- Osorio-Cadavid E, Ramírez M, López WA, Mambuscay LA. Estandarización de un protocolo sencillo para la extracción de ADN genómico de levadura. Rev Colomb Biotecnol. 2009;11:125–131. [Google Scholar]
- Peters II, Nelson FE. Factors influencing the production of Lipase by Mycotorula lipolytica. J Bacteriol. 1948;55:581–591. doi: 10.1128/jb.55.5.581-591.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspor P, Zupan J. Yeast in extreme environments. In: Gábor P, Rosa C, editors. Biodiversity and Ecophysiology of Yeasts. Springer; Berlin: 2006. pp. 371–417. [Google Scholar]
- Silva-Filho EA, dos Santos SKB, Resende AM, de Morais JOF, de Morais MA, Simões DA. Yeast population dynamics of industrial fuel-ethanol fermentation process assessed by PCR fingerprinting. Antonie van Leeuwenhoek. 2005;88:13–23. doi: 10.1007/s10482-004-7283-8. [DOI] [PubMed] [Google Scholar]
- Sugita T, Kikuchi K, Makimura K, Urata K, Someya T, Kamei K, Niimi M, Uehara Y. Trichosporon species isolated from guano samples obtained from bat-inhabited caves in Japan. Appl Environ Microbiol. 2005;71:7626–7629. doi: 10.1128/AEM.71.11.7626-7629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa R, Nakase T, Koyabashi T, Yamashita K, Okumura S. Fermentation of n-paraffins by yeast. Part III. α-Ketoglutarate productivity of various yeasts. Agr Biol Chem. 1969;33:929–938. [Google Scholar]
- van Uden N, Ahearn DG. Occurrence and population densities of yeast species in a fresh-water lake. Antonie van Leeuwenhoek J Microbiol Serol. 1963;29:308–312. doi: 10.1007/BF02046074. [DOI] [PubMed] [Google Scholar]
- Wurzbacher CM, Bärlocher F, Grossart HP. Fungi in lake ecosystems. Aquat Microb Ecol. 2010;59:125–149. [Google Scholar]