Abstract
A total of 120 beef carcasses were analyzed during processing at a slaughterhouse in southern Brazil. The carcasses were sampled by swab at three different steps of the slaughter line and then they were tested for Salmonella and E. coli. The Salmonella isolates were also examined for antimicrobial susceptibility. Salmonella prevalence distribution was modeled and the probability of contamination was simulated using @Risk program and 10,000 interactions. Results demonstrated that 4 beef carcasses (3.3%) were positive for Salmonella only in the first point. The six isolates of Salmonella were classified: S. Newport (n = 3), S. Saintpaul (n = 2) and S. Anatum (n = 1). No Salmonella strains exhibited resistance to any of the antimicrobials tested. As expected, the most contaminated point with E. coli was the first point (hide), presenting counts from 0.31 to 5.07 log cfu/100 cm2. Much smaller E. coli counts were observed in the other points. Results indicated low levels of Salmonella and E. coli on the beef carcasses analyzed and also low probability of contamination of the carcasses by Salmonella, suggesting adequate microbiological quality.
Keywords: Salmonella, E. coli, beef carcasses, slaughter line, antimicrobial susceptibility
Introduction
According to the United States Department of Agriculture (USDA), Brazil is the leading exporter of beef in the world. In 2011, Brazil exported 1.8 million tons of beef and the main markets were Russia, Iran, Hong Kong, Egypt and Venezuela (USDA, 2011). Due to its importance in the international market, the quality and food safety of Brazilian beef must meet well-established international requirements. In order to evaluate microbiological quality of meat, the quantification of E. coli and the presence of Salmonella have frequently been used because these microorganisms are considered good indicators of quality and food safety worldwide. The Food Safety and Inspection Service (FSIS) in the United States of America has established the criteria of a maximum 2.7% of Salmonella for cow and bull carcasses tested per set and a maximum of 102 cfu/cm2 of E. coli biotype I. The results of the quantitative analysis of carcasses at a specified frequency for Escherichia coli biotype I have been used to guide the maintenance of sanitary conditions during slaughter (USDA, 1996).
Salmonella spp. has been identified as the most important contaminant of food and the leading bacterial agent responsible for foodborne outbreaks in several countries (Majowicz et al., 2010). In the last years, global surveillance data indicated that the number of salmonellosis has increased mainly associated with the consumption of raw or undercooked eggs, poultry, meat or dairy products, demonstrating the importance of controlling this pathogen in food production (Braden, 2006; Kimura et al., 2004; Zhao et al., 2001). In the European Union, meat products were the second most common food group contributing to human salmonellosis in 2005 (Norrung and Buncic, 2008).
In Brazil, a remarkable increase in the incidence of foodborne salmonellosis has been reported (Brasil, 2009). According to the Surveillance Service of Brazil, among the 6,349 foodborne outbreaks registered in this country from 1999 to 2009, Salmonella was the most implicated bacterial agent, and was responsible for almost 20.7% of the reported outbreaks (Brasil, 2009). Corroborating these data, in Rio Grande do Sul (RS), southern Brazil, Salmonella was identified as the main causative agent of foodborne diseases, responsible for 35.7% (116 outbreaks) of the total of 323 outbreaks investigated during 1997 to 1999, and the meat products were the third most common food involved (Costalunga and Tondo, 2002).
The increased use of antimicrobial agents in animal production and human medicine as a mean of preventing and treating diseases is a significant factor in the emergence of antibiotic-resistant Salmonella. Therefore, resistant Salmonella which develops as a result of antibiotic use in animal production can be transferred to humans through the food chain, and it is a problem to be considered. Contamination of food with antibiotic-resistant bacteria can be a major threat to public health, causing more difficulties in the treatment of infectious diseases (Arslan and Eyi, 2010).
It is well know that the microbiological quality of meat depends on the control measures implemented in the slaughter lines, and in slaughterhouses working 24 h a day the contamination events can occur very often. In order to correct identify contamination procedures and establish control measures, beef carcasses may be evaluated during different steps of the slaughter.
The aim of this study was to determine the occurrence and antimicrobial susceptibility of Salmonella spp. and the contamination with E. coli on beef carcasses during different steps of slaughter in a slaughterhouse in southern Brazil.
Materail and Methods
Slaughterhouse and animals
Samples were taken at a slaughterhouse located in Rio Grande do Sul, regulated by the Brazilian Federal Inspection Service. The abattoir slaughtered approximately 200 animals daily and the slaughter was done mainly in the morning. The collections were previously scheduled and did not change the routine of the slaughterhouse. During the period from September 2009 to June 2010, thirteen samplings were carried out, analyzing a total of 120 carcasses. The carcasses chosen to be sampled were the first processed at the slaughterhouse on each collection day.
In-plant sampling locations
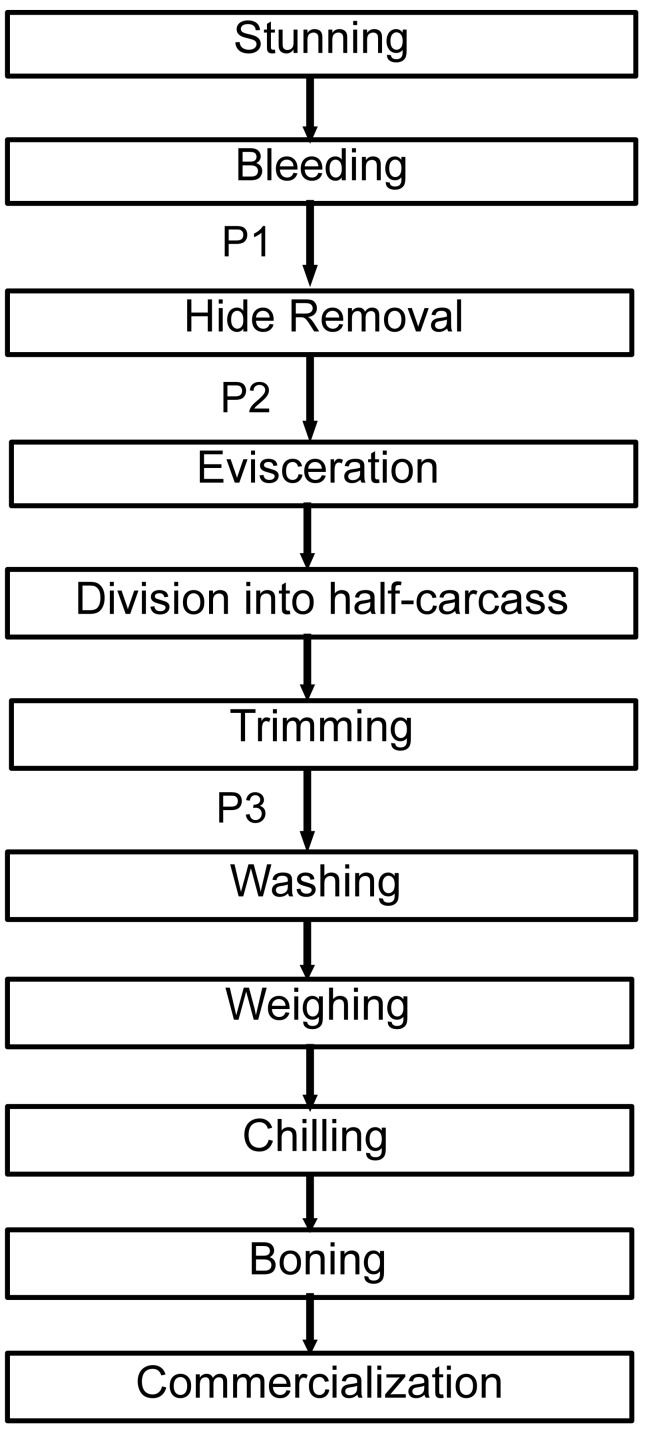
Carcass sampling was carried out at three steps in the processing line: first collection point (P1) was on the hide after bleeding but before hide opening and subsequent hide removal; second collection point (P2) was on the carcass after hide removal but before evisceration; and third collection point (P3) was the half-carcass but before the final washing (Figure 1). The same carcass was sampled at P1, P2 and P3.
Figure 1.
Flowchart of the slaughter line at the Brazilian slaughterhouse: P1, first collection point; P2, second collection point; P3, third collection point.
Carcass sampling areas
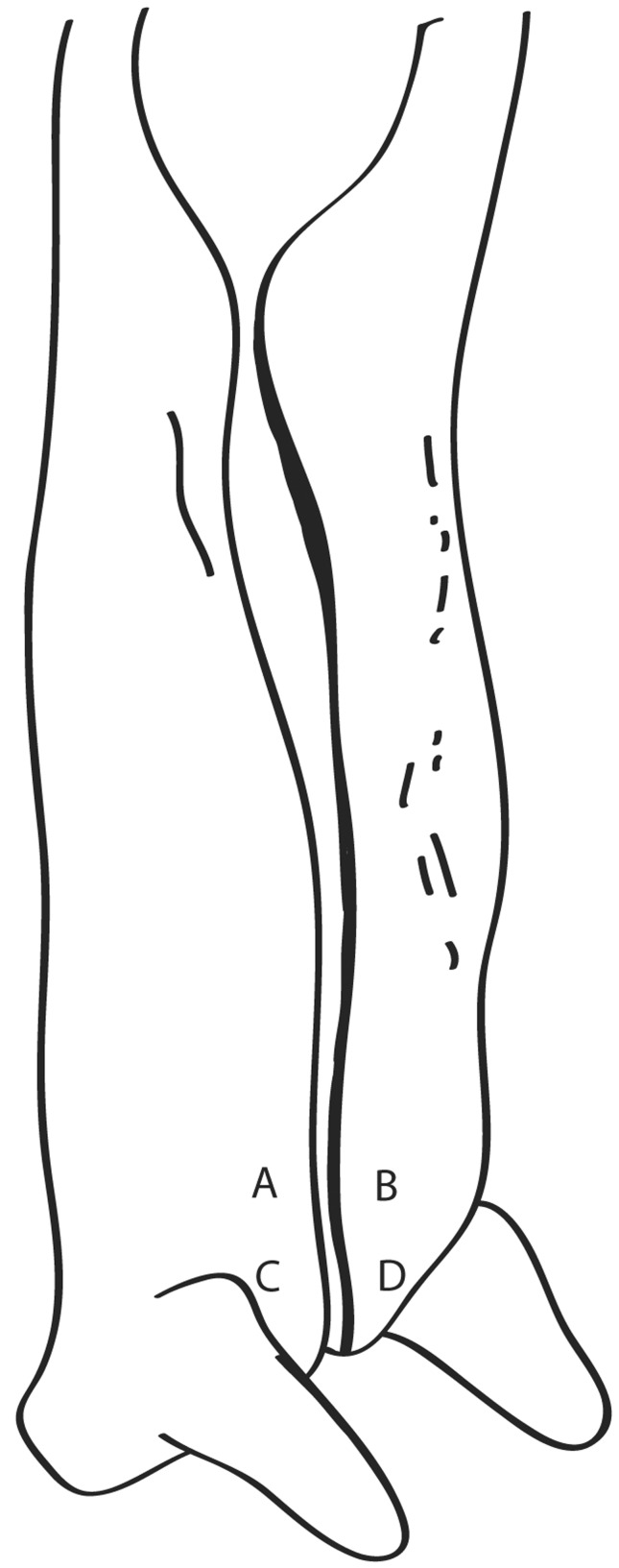
Each carcass was sampled on four regions, i.e. two regions on the right side and other two regions on the left side of the brisket (Figure 2). The area sampled in each region was 100 cm2, resulting in a total area of 400 cm2 sampled. This sampling method was carried out at each one of the three different collection points (P1, P2 and P3). Plain cellulose washing-up sponges (5 × 7 cm; 0.2 cm thickness) containing no antimicrobial additives were sterilized by autoclaving for 15 min at 1 atm and used for sampling. Just before sampling, each cellulose sponge was moistened with 10 mL of sterile saline peptone solution (0.1% bacteriological peptone; 0.85% NaCl) and then placed in a sterile plastic bag. The carcasses were sponge-swabbed by ten consecutive passes at each of the four collecting areas using one cellulose sponge for each sampling region. After sampling, the four cellulose sponges from each point were placed in the same plastic bag and transported under refrigeration (< 7 °C), within 2 h, to the Food Microbiology and Control Laboratory of Food Science and Technology Institute (ICTA/UFRGS) to be analyzed.
Figure 2.
Carcass sampling areas. Each letter indicates a region of 100 cm2 sampled on the carcass: A and C - regions of 100 cm2 sampled on the left side, B and D - regions of 100 cm2 sampled on the right side.
Homogenization of the samples
After the arrival of the samples at the Laboratory, 200 mL of saline peptone solution was added to each plastic bag. The plastic bag was repeatedly squeezed manually for 1 min and further decimal dilutions were made with saline solution to carry out E. coli analysis.
Enumeration of Escherichia coli
The 3M Petrifilm E. coli/Coliform Count Plates (3M, Sumaré, Brazil) were used to enumerate E. coli. From each appropriate decimal dilution, 1 mL was removed, inoculated on a Petrifilm plate and incubated for 48 h at 37 °C. Blue colonies with gas production were counted.
Isolation and enumeration of Salmonella spp
An aliquot of 40 mL of the saline peptone solution was centrifuged at 1000 g for 15 min and the sediment was used to investigate the presence of Salmonella spp.
According to the ISO 6579:2002 method (ISO, 2002), each sediment was incubated in 100 mL of buffered peptone water (Oxoid, São Paulo, Brazil) for 18–24 h at 37 °C. Subsequently, 1 mL was transferred to 10 mL of Muller Kauffmann tetrathionate - novobiocin broth (MKTTn, Oxoid) and 0.1 mL was transferred to 10 mL of Rappaport-Vassiliadis Broth with soya (RVS, Oxoid) and incubated at 37 °C and 42.5 °C, respectively, for 24 h. Then, both Xylose Lysine Deoxycholate agar (XLD, Oxoid) and Mannitol Lysine Crystal Violet Brilliant Green agar (MLCB, Oxoid) plates were inoculated with aliquots of the cultures from RVS and MKTTn. All the plates were incubated at 37 °C for 24 h. Suspect colonies (red with or without a black center from XLD; mauve colored colonies with a black center from MLCB) were purified on a Nutrient agar. Then, biochemical recommended tests and serology by agglutination with Poly O antisera (Probac, São Paulo, Brazil) were carried out to confirm the result (MacFaddin, 2000). After these tests, Salmonella spp. isolates were forwarded to the Laboratório de Enterobactérias of the Instituto Oswaldo Cruz (Rio de Janeiro, RJ, Brazil) for serotyping.
To enumerate Salmonella spp., 0.1 mL of the first dilution was spread-plated on XLD agar (Oxoid, São Paulo, Brazil). The plates were incubated for 24 h at 37 °C. Red colonies with or without black centers were counted and later confirmed by biochemical tests.
Antimicrobial susceptibility test
Susceptibility to antimicrobial agents was tested using the disk diffusion method on Muller-Hinton agar (Oxoid, São Paulo, Brazil) plates according to the National Committee for Clinical Laboratory Standards (National Committee for Clinical Laboratory Standards, 2010). The following 15 antimicrobial agents were examined against Salmonella isolates: ampicillin (10 μg), cefoxitin (30 μg), cephalothin (30 μg), cefotaxime (30 μg), imipenem (10 μg), chloramphenicol (30 μg), amikacin (30 μg), gentamicin (10 μg), kanamycin (30 μg), streptomycin (10 μg), nalidixic acid (30 μg), ciprofloxacin (5 μg), tetracycline (30 μg), trimethoprim-sulphamethoxazole (25 μg), and sulfonamides (300 μg). The diameters of the zones of inhibition were recorded to the nearest millimeter and classified as susceptible, intermediate and resistant.
Measurement of the levels of free residual chlorine in the washing animal water
The levels of free residual chlorine in the animal washing water were provided by the Inspection Service of the slaughterhouse. The values were measured at the beginning of each slaughter process. The measurement was done using a C401 colorimeter (Eutech Instruments, Vernon Hills, U.S.A.).
Statistical analysis
E. coli and Salmonella spp. counts were calculated per 100 cm2 and converted to log counts before statistical analysis. The mean values were calculated and the analysis of variance (ANOVA) and a Tukey test were carried out to compare the differences between the mean values. The differences were considered significant with p values less than 0.05. For the Salmonella spp. analysis, data were reported as the percentage of samples positive for the pathogen. Salmonella prevalence distribution was modeled and used as an output to simulate the probability of contamination (P). The simulations were carried out using @Risk program student version number 5.7, with 10,000 interactions. Prevalence distribution was simulated using binomial distribution considering positive samples (x) and the sample number (n). Beta distribution was used to calculate the probability of contamination by Salmonella, considering the uncertainties of the sampling (β and α factors). For this purpose we followed the formula below:
where p is the probability, x is the number of positive samples for Salmonella spp., n is the number of carcasses sampled.
Results and Discussion
Enumeration of Escherichia coli
The general mean values of E. coli were 2.57, 0.46 and 0.40 log cfu/100 cm2 at P1, P2 and P3, respectively. There were significant differences in counts observed in P1 and P2 and P1 and P3, but no significant difference was observed between the counts of P2 and P3. The low counts in P2 and P3 could possibly be explained due to proper hide removal and the fact that carcass tissues are considered sterile. The significant reduction in counts when P1 was compared with the other two points and the low counts observed in P2 and P3 could indicate that manufacturing practices were adequate during processing in the slaughterhouse.
The counts of E. coli on animal hides (P1) ranged from 0.31 to 5.07 log cfu/100 cm2 (Table 1). The high variability in E. coli counts at P1 could be explained by the different geographic origin of the animals, the age of animals, cleanliness of hides and breeds of animals (Antic et al., 2010; Davies et al., 2000; Hancock et al., 1997; Mcevoy et al., 2000). Other studies reported higher E. coli counts on hides than the results obtained in this study. For example, Bacon et al. (2000) had higher mean values of E. coli ranging from 5.5 to 7.5 log cfu/100 cm2. Arthur et al. (2004) have demonstrated higher mean values of E. coli on animal hides in two commercial fed-beef processing plants. The counts ranged from 6.6 to 8.0 log cfu/100 cm2 in one of the plants and 4.9 to 5.8 log cfu/100 cm2 in the other plant. The microbiological loads of incoming cattle are important because the external hide is a primary source of fecal contamination, which can be eventually transferred to the underlying sterile carcass tissue (Bacon et al., 2000).
Table 1.
Mean values of E. coli at three points of the slaughter line and levels of free chlorine in animal washing water in one slaughterhouse in Southern Brazil.
| Collection day | Mean values (log cfu/100 cm2) | Level of free chlorine in animal washing water (ppm) | ||
|---|---|---|---|---|
|
| ||||
| Collection pointsd | ||||
|
| ||||
| P1 | P2 | P3 | ||
| 1 | 1.93a | 0.97a | N.Db | 5 |
| 2 | 2.98a | N.Db | 0.25b | 0.54 |
| 3 | 4.11a | 0.93b | 0.93b | 0.42 |
| 4 | 3.34a | 0.42b | 0.32b | 4.3 |
| 5 | 5.07a | 2.42b | 2.10b | 2.01 |
| 6 | 2.18a | 0.40b | 0.18b | 0.62 |
| 7 | 3.35a | 0.36b | 0.26b | 2.65 |
| 8 | 2.27a | 0.10b | N.Db | 1.85 |
| 9 | 2.20a | 0.11b | 0.94c | _e |
| 10 | 0.31a | N.Da | N.Da | _e |
| 11 | 1.39a | N.Db | N.Db | 5 |
| 12 | 2.16a | 0.23b | 0.11b | 0.65 |
| 13 | 2.11a | 0.04b | 0.18b | 3.80 |
| General mean value | 2.57a | 0.46b | 0.40b | |
Values in a row with the same capital letter are not significantly different (p > 0.05; Tukey Test).
P1 = first collection point, P2 = second collection point, P3 = third collection point.
The level of chlorine was not provided by the slaughterhouse.
Not detected (To calculate the mean values were considered 1 CFU, so log 1 = 0).
The results obtained for E. coli at P2 showed a maximum value of 2.42 log cfu/100 cm2. Arthur et al. (2004) reported similar results examining 288 beef carcasses in the USA. On the other hand, Bacon et al. (2000) found higher mean values ranging from 2.6 to 5.3 cfu/100 cm2 analyzing beef carcasses in the USA. In the present study, the microbiological loads at P2 probably reflected the extent of the microbiological contamination originating from the hide, since beef carcass surfaces are generally sterile (Bacon et al., 2000).
Analyzing counts obtained on different sampling days, it could be observed that the fifth collection day showed significantly higher counts (P1 = 5.07, P2 = 2.42 and P3 = 2.10 log cfu/100 cm2) for all three points, compared to the other days, while the tenth collection day demonstrated the lowest mean values. The reasons for these differences were not explored in this study; however they corroborate the idea that the higher the initial contamination, the greater the contamination of the final product. Another important result observed was the significant increase between P2 and P3 on the ninth collection day. It could possibly be explained as a recontamination after evisceration or another failure in processing.
The results indicated that there was no direct correlation between the level of free chlorine in the water used to wash the animals before slaughter and the mean values of E. coli observed in P1. As an example, on the first sampling day, the mean value of 1.93 log cfu/100cm2 was verified with a free chlorine level of 5 ppm in water, however on the twelfth collection day, a mean value of 2.16 log cfu/100cm2 was observed, whilst the level of free chlorine was 0.65 ppm. Even though the chlorine level varied almost 9-fold, the bacterial counts were not significantly different. These results may demonstrate that only washing the animal hide with chlorinated water does not necessarily improve the microbial status of the hide. The risk of contamination of the beef carcasses may still exist, if only this practice is used. In order to improve microbiological quality, the use of multiple-sequential interventions, like pre- and post-evisceration water washing, organic acid solution rinsing and hot water washing could be possible measures to decontaminate beef carcasses during the slaughtering and dressing process (Bacon et al., 2000).
Isolation, enumeration and antimicrobial susceptibility of Salmonella spp
The presence of Salmonella spp. was observed in only four carcasses (3.3%), three sampled on the fifth collection day and one carcass sampled on the eighth collection day. Salmonella spp. was only isolated in P1 from animal hides. Reid et al. (2002) obtained similar results showing a prevalence of 3.3% of Salmonella in 90 beef cattle hides in the south-west of England. A much higher Salmonella prevalence (94.8%) was found by Arthur et al. (2007) analyzing 288 beef cattle hides in the USA. Conversely, Antic et al. (2010) were not able to isolate Salmonella spp. in any of the 40 animal hides sampled in one abattoir in Serbia.
The low level of Salmonella contamination found in this study could be explained by multiple factors. First, the slaughtered cattle had different origins, coming from different farms that could easily have had differences in the prevalence of this pathogen. Secondly, in Brazil, extensive farming is the most prevalent kind of animal exploitation. With this practice, direct contact among animals is very low, which may help to avoid the transmission of Salmonella. There are other factors that could explain the Salmonella prevalence on beef carcasses, including those related to the feed, animals, transport and the environment. All those factors vary largely among countries and geographical regions.
The modeling of probability of contamination by Salmonella on P1 indicated values varying from 0.016% to 0.075% with mean numbers of 0.041%. These numbers were calculated considering the uncertainty of the sampling. Such values mean that in 90% of the sampling on beef carcasses in this slaughter line probably at least 1.6% and in the maximum of 7.5% of the carcasses may be positive for Salmonella. These prevalence values were considered low mainly considering that Salmonella was isolated on animal hides.
Unlike of the hides, there was no Salmonella spp. found at any of the other two points (P2 and P3) analyzed in the present study, and these results were similar with those found by Meyer et al. (2010) who examined 841 beef carcasses in Germany. The absence of Salmonella at P2 and P3 and the low E. coli counts at these points demonstrated that the hygienic conditions and manufacturing practices of the slaughterhouses were adequate.
Six strains of Salmonella spp. were isolated in the present study, and they were classified as three different serovars. The most prevalent serovar was S. Newport, which was found on three animals. The serovar S. Saintpaul was found on two animals, while S. Anatum was identified on only one animal.
It was not possible to enumerate Salmonella spp. in the majority of the samples. This may have occurred because Salmonella was not present on the sampled surfaces or because the amount of Salmonella present was too low to be detectable. Other possibilities to explain this result are the influence of background flora that could inhibit Salmonella multiplication, giving a false negative result or the low sensitivity of the method used to detect stressed cells (Meyer et al., 2010).
All the Salmonella spp. stains were susceptible to the 15 antimicrobials tested. This result is different to those observed by Bacon et al. (2002) who have reported 69.4% of 49 Salmonella strains were resistant at least one antimicrobial tested. Stevens et al. (2006) obtained 99 isolated of Salmonella spp. and the percentages of strains resistant to nitrofurans, sulfamethoxazole, streptomycin, chloramphenicol and nalidixic acid were 36.7, 21.1, 14.1, 2 and 1%, respectively.
The difference in the resistance profile observed in this study and the others reported in the literature could be explained by several factors, especially the type of farming. Extensive farming of cattle has been the most prevalent way of farming in Brazil. In this kind of farming, the use of antimicrobials has been very low; therefore, there is no high selective pressure on bacterial strains. On the other hand, in some animal species, like swine, where animal farming has been done of intensive way, the number of resistant strains has been much higher. In this way of farming, due to the intensification of production methods, many diseases related to the introduced technologies have emerged. The control of these diseases has been done by the use of antimicrobials (Castagna et al., 2001).
A correlation between the presence of Salmonella and the quantitative analysis of E. coli was not observed in this study. Salmonella was detected on the fifth and in the eighth collection days. However, the mean value of E. coli on the fifth collection day was significantly higher than the mean value of E. coli on the eighth collection day (p < 0.05). In this study, the percentage of carcasses contaminated with Salmonella and more than 2 log cfu/100 cm2 E. coli was 3.37%. However the percentage of carcasses with Salmonella and less than 2 log cfu/100 cm2 E. coli was 3.23%. These values did not present a statistically significant difference and these results indicated that there was no correlation between the presence of Salmonella and the quantitative analysis of E. coli. Ruby and Ingham (2009) have suggested that the correlation would be better if the absence of Salmonella spp. were linked to negative Enterobacteriaceae results.
In the present study, low levels of Salmonella and E. coli were found on beef carcasses and also low probability of contamination of the carcasses by Salmonella, suggesting that adequate slaughter procedures were carried out in the slaughterhouse analyzed. However, the isolation of these microorganisms on the animal hides indicated that the risk of the contamination still exists.
References
- Antic A, Blagojevic B, Ducic M, Nastasijevic I, Mitrovic R, et al. Distribution of microflora on cattle hides and its transmission to meat via direct contact. Food Control. 2010;21:1025–1029. [Google Scholar]
- Arslan S, Eyi A. Occurrence and antimicrobial resistance profiles of Salmonella species in retail meat products. J Food Prot. 2010;73:1613–1617. doi: 10.4315/0362-028x-73.9.1613. [DOI] [PubMed] [Google Scholar]
- Arthur TM, Bosilevac JM, Brichta-Harhay DM, Kalchayanand N, Shackelford SD, Wheeler TL, et al. Effects of a minimal hide wash cabinet on the levels and prevalence of Escherichia coli O157:H7 and Salmonella on the hides of beef cattle at slaughter. J Food Prot. 2007;70:1076–1079. doi: 10.4315/0362-028x-70.5.1076. [DOI] [PubMed] [Google Scholar]
- Arthur TM, Bosilevac JM, Nou X, Shackelford SD, Wheeler TL, Kent MP, et al. Escherichia coli O157 prevalence and enumeration of aerobic bacteria, Enterobacteriaceae, and Escherichia coli O157 at various steps in commercial beef processing plants. J Food Prot. 2004;67:658–665. doi: 10.4315/0362-028x-67.4.658. [DOI] [PubMed] [Google Scholar]
- Bacon RT, Belk KE, Sofos JN, Clayton RP, Reagan JO, et al. Microbial populations on animal hides and beef carcasses at different stages of slaughter in plants employing multiple-sequential interventions for decontamination. J Food Prot. 2000;63:1080–1086. doi: 10.4315/0362-028x-63.8.1080. [DOI] [PubMed] [Google Scholar]
- Bacon RT, Sofos JN, Belk KE, Hyatt DR, et al. Prevalence and antibiotic susceptibility of Salmonella isolated from beef animal hides and carcasses. J Food Prot. 2002;65:284–290. doi: 10.4315/0362-028x-65.2.284. [DOI] [PubMed] [Google Scholar]
- Braden CR. Salmonella enterica serotypes Enteritidis and eggs: a national epidemic in the United States. Clin Infect Dis. 2006;43:512–517. doi: 10.1086/505973. [DOI] [PubMed] [Google Scholar]
- Brasil. Análise epidemiológica de surtos de doenças transmitidas por alimentos no Brasil, 1999–2009, Brasília, DF. 2009. Ministério da Saúde. Agência Nacional de Vigilância Sanitária. Secretaria de Vigilância em Saúde. Coordenação de Vigilância das Doenças de Transmissão Hídrica e Alimentar. [Google Scholar]
- Castagna SMF, Bessa MC, Carvalho DA, Cardoso M, et al. Resistência a antimicrobianos de amostras de Salmonella sp. Isoladas de suínos abatidos no estado do Rio Grande do Sul. Arquivos da Faculdade de Veteterinária da UFRGS. 2001;29:44–49. [Google Scholar]
- Costalunga S, Tondo EC. Salmonellosis in Rio Grande do Sul, Brazil, 1997 to 1999. Braz J Microbiol. 2002;33:342–346. [Google Scholar]
- Davies MH, Webster SD, Hadley PJ, et al. Production factors that influence the hygienic condition of finished beef cattle. Vet Rec. 2000;146:179–183. doi: 10.1136/vr.146.7.179. [DOI] [PubMed] [Google Scholar]
- Hancock DD, Besser TE, Rice DH, Herriott DE, et al. A longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiol Infect. 1997;118:193–195. doi: 10.1017/s0950268896007212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISO. ISO 6579:2002 - Microbiology of food and animal feeding stuffs - Horizontal method for the detection and enumeration of Salmonella spp 2002 [Google Scholar]
- Kimura AC, Reddy V, Marcus R, Cieslak PR, Mohle-Boetani JC, Kassenborg HD, et al. Chicken consumption is a newly identified risk factor for sporadic Salmonella enterica serotype Enteritidis infections in the United: a case-control study in FoodNet sites. Clin Infect Dis. 2004;38(Suppl 3):244–252. doi: 10.1086/381576. [DOI] [PubMed] [Google Scholar]
- MacFaddin JF. Biochemical tests for identification of medical bacteria. Lippincott Williams & Wilkins; Baltimore: 2000. [Google Scholar]
- Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- Mcevoy JM, Doherty AM, Finnerty M, Sheridan JJ, Mcguire L, Blair IS, et al. The relationship between hide cleanliness and bacterial numbers on beef carcasses at a commercial abattoir. Lett Appl Microbiol. 2000;30:390–395. doi: 10.1046/j.1472-765x.2000.00739.x. [DOI] [PubMed] [Google Scholar]
- Meyer C, Thiel S, Ullrich U, et al. Salmonella in raw meat and by-products from pork and beef. J Food Prot. 2010;73:1780–1784. doi: 10.4315/0362-028x-73.10.1780. [DOI] [PubMed] [Google Scholar]
- National Committee for Clinical Laboratory Standards (NCCLS) Twentieth informational supplement (M100 - S20) CLSI; Wayne: 2010. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- Norrung B, Buncic S. Microbial safety of meat in the European Union. Meat Sci. 2008;78:14–24. doi: 10.1016/j.meatsci.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Reid C-A, Small A, Avery SM, et al. Presence of food-borne pathogens on cattle hides. Food Control. 2002;13:411–415. [Google Scholar]
- Ruby JR, Ingham SC. Use of Enterobacteriaceae analysis results for predicting absence of Salmonella serovars on beef carcasses. J Food Prot. 2009;72:260–266. doi: 10.4315/0362-028x-72.2.260. [DOI] [PubMed] [Google Scholar]
- Stevens A, Kaboré Y, Perrier-Gros-Claude J-D, Millemann Y, Brisabois A, Catteau M, et al. Prevalence and antibiotic-resistance of Salmonella isolated from beef sampled from the slaughterhouse and from retailers in Dakar (Senegal) Int J Food Microbiol. 2006;110:178–186. doi: 10.1016/j.ijfoodmicro.2006.04.018. [DOI] [PubMed] [Google Scholar]
- USDA, Food Safety and Inspection Service. Pathogen reduction; hazard analysis and critical control point (HACCP) systems; final rule. U.S. Fed. Regist. 1996;61:38806–38989. [Google Scholar]
- USDA. [Accessed 21 Feb 2012];Cattle and beef data and statistics. 2011 Available at: http://www.usda.gov.
- Zhao C, Ge B, De Villena R, Sudler R, Yeh E, et al. Prevalence of Campylobacter spp., Escherichia coli, and Salmonella serovars in retail chicken, turkey, pork, and beef from the greater Washington, D. C., area. Appl and Environ Microbiol. 2001;67:5431–5436. doi: 10.1128/AEM.67.12.5431-5436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]