Abstract
The chromatin remodeling complex ACF helps establish the appropriate nucleosome spacing for generating repressed chromatin states. ACF activity is stimulated by two defining features of the nucleosomal substrate: a basic patch on the histone H4 N-terminal tail and the specific length of flanking DNA. Yet the mechanisms by which these two substrate cues function in the ACF remodeling reaction is not well understood. Using electron paramagnetic resonance spectroscopy with spin-labeled ATP analogs to probe the structure of the ATP active site under physiological solution conditions, we identify a closed state of the ATP-binding pocket that correlates with ATPase activity. We find that the H4 tail promotes pocket closure. We further show that ATPase stimulation by the H4 tail does not require a specific structure connecting the H4 tail and the globular domain. In the case of many DNA helicases, closure of the ATP- binding pocket is regulated by specific DNA substrates. Pocket closure by the H4 tail may analogously provide a mechanism to directly couple substrate recognition to activity. Surprisingly, the flanking DNA, which also stimulates ATP hydrolysis, does not promote pocket closure, suggesting that the H4 tail and flanking DNA may be recognized in different reaction steps.
Keywords: human hACF, spin-labeled nucleotide, EPR spectroscopy, nucleosome, basic patch
Introduction
Chromatin remodeling enzymes alter histone-DNA contacts to enable both activation and repression of transcription. The ATP-dependent chromatin assembly factor (ACF) is implicated in gene silencing 1. The gene-silencing function of ACF is postulated to rest in its ability to generate evenly spaced nucleosomes, which are thought to enable the formation of heterochromatic structures 2,3. At least two classes of mechanisms can explain how the activity of ACF is regulated such that it specifically acts at loci that need to be silenced 1. One class of mechanisms entails the recruitment of ACF via sequence-specific DNA binding factors. The other class of mechanisms relies on the intrinsic ability of ACF to distinguish between different types of nucleosomes based on specific nucleosomal cues. Here, we address the latter class of mechanisms in the context of human ACF (hACF).
hACF is part of the ISWI family of remodeling complexes, which contain one ATPase subunit with homology to the Drosophila ISWI ATPase and up to five accessory subunits. Human ACF contains the ATPase subunit SNF2h and the accessory subunit Acf1. Previous work has shown that ISWI family complexes use two distinct features of a nucleosome in their reaction cycle: the length of DNA flanking the nucleosome, and a basic patch of amino acids on the N-terminal histone H4 tail 4–7,8–11. The reliance on these two features is thought to allow down-regulation of ISWI activity in two different in vivo contexts. In actively transcribed genes, the H4 tail is acetylated at lysine 16, a residue in the critical epitope of the H4 tail, the basic patch 1. This post-translational mark reduces the activity of ISWI family enzymes in vitro and may analogously inhibit ISWI complexes at active loci in vivo 6,12–14. In folded chromatin the H4 tail is postulated to interact with an acidic patch on a neighboring nucleosome, and the flanking DNA is also thought to become sterically less accessible 15–17. Such physical occlusion may prevent the ISWI enzyme from mobilizing nucleosomes that are already compacted. Since the H4 tail and flanking DNA act as signals that the enzyme must recognize and integrate in order to act appropriately, we call them ‘substrate cues.’
Interestingly, both substrate cues contribute more to the rate of remodeling by ISWI family enzymes than to their binding affinity 5,10,11,18. Lengthening the DNA flanking the nucleosome up to 60 base pairs stimulates hACF remodeling rates, and beyond 60 bp hACF remodeling rates remain largely constant 10,11. Previous studies further imply that the HAND-SANT-SLIDE domain of ISWI ATPases interacts with flanking DNA 19–21. Within the H4 tail four amino acids, 16–19 (KRHR) are critical for maximal ISWI activity, with the two arginines being the most important 4,6. Further, the specific location of the H4 tail on the nucleosome seems to be important, as swapping the locations of the H4 and H3 tails showed a significant decrease in activity4,6. This result may be explained by the proposed binding site of the ATPase domain of ISWI ATPases, which is two helical DNA turns from the nucleosomal dyad at Superhelical Location 2 [SHL(±2)] 8,20,22–24,25. This is the region where the N-terminal H4 tail emerges from the nucleosome core particle. Complicating the mechanistic interpretation of the roles of flanking DNA and the H4 tail is the observation that hACF functions as a dimer in which the two protomers appear to bind the nucleosome on opposite sides 25. Thus, each protomer has access to flanking DNA and one of the two H4 tails. Current models suggest that such a dimeric architecture allows hACF to achieve bi-directional movement of nucleosomes in a processive manner because the two protomers can take turns moving the nucleosome in one direction 25,26.
Given that both the flanking DNA and the H4 tail have catalytic effects, both cues must either directly or indirectly regulate the ATPase active site. However, the nature of the communication between the substrate cues and the active site is not known. More fundamentally, there is a lack of understanding about the types of conformational changes that occur in the ATP- binding pocket of chromatin-remodeling enzymes and how these conformational changes correspond to changes in nucleosome structure. Because they participate in catalysis, the H4 tail and flanking DNA offer the opportunity to address: (1) the linkage between conformational changes in the ATP active site and physical changes in the nucleosome substrate and, (2) the energetic contributions to catalysis of critical nucleosome epitopes that are known to lead to specificity. A better understanding of how hACF activity is regulated by variable features of a nucleosome has broader significance for understanding how chromatin-remodeling enzymes as a class may be regulated by mechanisms other than simple recruitment.
In this study we use ATP analogs containing an environmentally sensitive spin probe to detect a conformational change in the ATP-binding pocket under physiological solution conditions. Our results suggest that two structural features of the nucleosome that hACF is known to use, the H4 tail and flanking DNA, make qualitatively different contributions to catalysis raising the possibility that the two substrate cues act in different reaction steps.
Results
The effect of the H4 tail and flanking DNA on nucleosome remodeling
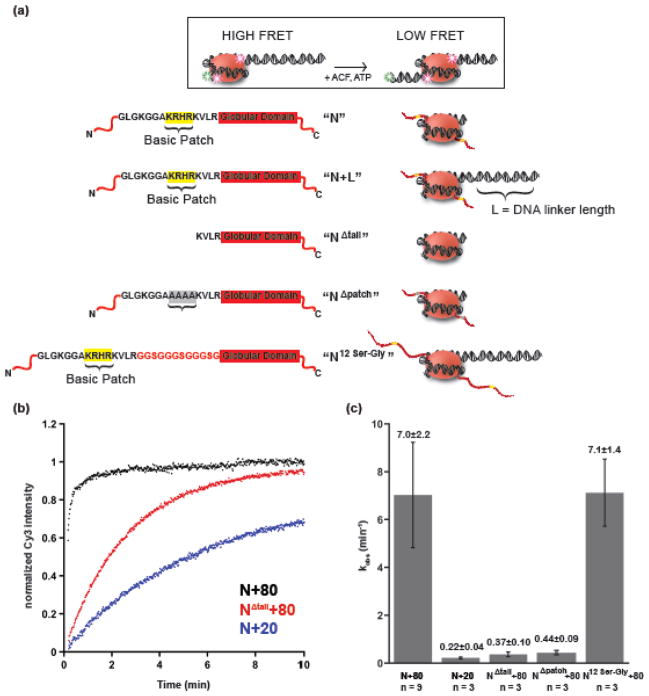
To better understand how these two classes of substrate cues affect activity, we first compared their catalytic effects on nucleosome remodeling. We measured remodeling rate constants for hACF with nucleosome substrates lacking each cue using a fluorescence resonance energy transfer (FRET)-based assay developed previously 11. In this assay, we use nucleosomes containing a Cy3 label on one DNA end and a Cy5 label at amino acid 120 of H2A of the histone octamer by engineering in a cysteine substitution at this position. When an end-positioned nucleosome moves towards the center, this results in a loss of FRET (Fig. 1a). We monitor the change in FRET by following unquenching of the Cy3 dye with time (Fig. 1a, b). Removing the H4 tail in the context of nucleosomes containing 80 base pairs of flanking DNA reduces the maximal rate constant of remodeling by 19-fold under single turnover conditions (saturating and excess ATP and hACF over nucleosomes, Fig. 1b). Mutating the basic patch of the H4 tail, K16R17H18R19, to alanines gives a similar rate defect as removing the H4 tail (16- vs. 19-fold respectively; Fig. 1a,c,). Shortening the flanking DNA from 80 to 20 base pairs reduces the maximal rate of remodeling by 32-fold (Fig. 1a,c). Thus, overall, individually mutating each cue gives reductions in rate of similar magnitude. For technical reasons, we were not able to reliably measure rates of nucleosome remodeling under conditions where both cues are simultaneously removed. The larger reduction in rate demanded substantially longer time-courses and made it difficult to ensure that the enzyme maintained the same specific activity for the duration of the experiment.
Figure 1. H4 tail and flanking DNA length affect hACF nucleosome remodeling.
a. Nucleosome constructs used for FRET experiments. Histone octamers with a cysteine engineered at residue 120 of H2A were labeled with Cy5 and assembled onto DNA templates containing the 601 nucleosome positioning sequence and a proximal Cy3 dye, and either 20 or 80 base pairs of DNA flanking the 601 positioning sequence on one side. The ‘NΔtail’ mutant lacks the first 19 amino acids of the H4 tail, including the basic patch of residues K16R17H18R19. In the ‘NΔpatch’ mutant, the basic patch residues are replaced with alanines. In the ’ N12 Ser-Gly ’ construct, a 12 amino-acid linker (Gly-Ser-Gly-Gly)3 is inserted between residues 24 and 25 of histone H4. b. Representative time courses from the FRET based assay monitoring un-quenching of the Cy3 donor dye are shown with saturating hACF and ATP-Mg. c. Rate constants for remodeling (kobs) with hACF for the nucleosome constructs under saturating hACF and ATP. The average values of the individual rate constants in min−1 with standard deviation are shown on top of each bar.
We next investigated how the H4 tail might stimulate ATP hydrolysis. It has been suggested that interaction of the H4 tail with histone-bound DNA at SHL±2 may drive a region of the H4 tail (residues 16–24) containing the basic patch (16–19) into an alpha-helical conformation 27,28. We therefore asked whether a particular secondary structure adopted by the H4 tail at SHL±2 might be required for the reaction. To test this possibility we inserted a 12 amino acid unstructured serine-glycine linker between the H4 tail and the globular domain (Fig. 1a,c; N12 ser-gly+80). Such a linker should disrupt any continuous secondary structure and also increase the total distance of the basic patch from the globular region. Previous calculations for the length of unstructured peptides suggest that the 12 amino-acid linker should increase the length between the histone octamer core and the basic patch by at least 24 angstroms29–31. Interestingly, the 12 amino-acid Ser-Gly insertion does not alter the maximal hACF remodeling rates (Fig. 1c). The comparison of the basic-patch mutant with the 12 Ser-Gly insertion mutant (Fig. 1c, NΔpatch+80 vs. N12 Ser-Gly+80) along with previous tail-swapping experiments 4,6, suggest that the location of the basic-patch epitope near the SHL±2 position of the nucleosome is more important for function than the precise structure of its connection to the nucleosome.
Conformational changes at the ATP-binding site measured by EPR spectroscopy
The stimulatory effects of the H4 tail basic patch and flanking DNA on hACF activity suggest that these substrate cues allosterically or directly regulate the ATP-binding site. Our goal was to determine how the two substrate cues affect the conformation of the ATP-binding pocket. One of the challenges to understanding how substrate cues contribute to catalysis in motor proteins is that these enzymes tend to adopt multiple conformations that depend both on the nucleotide state and interactions with the second substrate32. Even in cases where there are multiple high resolution crystal structures defining distinct conformations, such as with myosin and kinesin, the functional significance of these structures in a reaction cycle can be difficult to resolve 33,34. The problem is greater for chromatin remodeling enzymes because crystallography of chromatin remodeling enzymes bound to the nucleosome substrate has been technically challenging. Electron paramagnetic resonance (EPR) spectroscopy provides a powerful means to assign functional states to static structures 33–35. EPR spectroscopy can be used to resolve multiple protein conformations in equilibrium, under a wide variety of conditions including physiological reaction conditions. The traditional approach for EPR measurements is site-directed spin labeling where a single cysteine is incorporated into the protein at the location of interest, and then is covalently modified by a reactive spin probe. We have previously used this technique to study the conformational changes of the H4 tail of the nucleosome in the presence of SNF2h, the ATPase subunit of hACF25. Here our goal was to monitor changes in the ATP-binding site, so we instead used spin-labeled ATP-analogs. The analog specifically places the reporter group at the ATP-binding site without modifying the protein. The EPR spectrum of the spin probe provides information on the conformational restriction felt by the probe, thus monitoring changes in the nucleotide-binding pocket as we have shown previously for kinesin and myosin motors33,34. Here we used spin-labeled ATP (2′3′SLATP), where the reporter spin moiety is attached to the ribose ring of ATP (Fig. 2a, inset box).
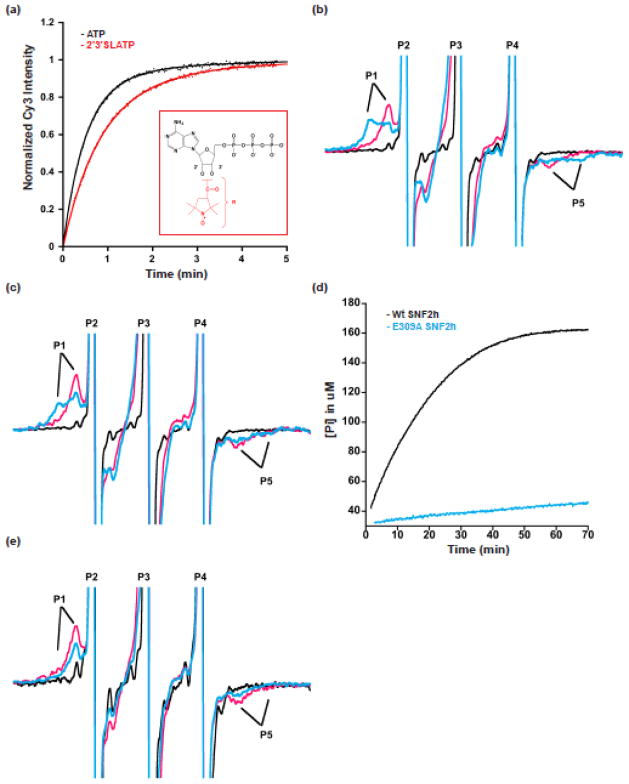
Figure 2. Spin-labeled ATP analogues reveal conformational changes in the ATP-binding pocket of SNF2h.
a. 2′3′SLATP supports SNF2h nucleosome remodeling. Remodeling kinetics of N+80 nucleosomes (5 nM) with 150 nM of SNF2h, 1mM MgCl2 and 0.5mM of ATP or 2′3′SLATP (kobs for n=3 is 2.0±0.06 min−1 and 1.2±0.22 min−1, respectively for ATP and 2′3′SLATP). Representative remodeling traces are shown. Inset: 2′3′SLATP analog (nitroxyl-radical spin probe shown in red). b. Representative EPR spectra of 2′3′SLADP and 2′3′SLADP-BeFx with SNF2h and N+60 nucleosomes. The spectrum of 2′3′SLADP alone (without SNF2h and nucleosomes) is shown in black. The presence of nucleosomes and SNF2h (pink spectrum) produces a bound peak (P1) in the 2′3′SLADP spectrum with a high-field to low-field (P1–P5) splitting of 4.8mT (118.8° cone angle). When the analog 2′3′SLADP•BeFx is used with nucleosomes and SNF2h (blue spectrum) this produces a second, more immobilized peak (P1, blue) with a P1–P5 splitting of 6.2mT (68° cone angle). Spectral peaks are truncated to enhance resolution of bound probe. c. Representative spectra of 2′3′SLADP (pink) and 2′3′SLADP•BeFx (blue) bound to SNF2h in the presence saturating concentrations of a 60 base pair DNA fragment. The black spectrum (2′3′SLADP alone) is from panel b for comparison. d. Comparison of the ATPase rates of wildtype enzyme (black) and the E309A mutant (blue) in the presence of N+60 nucleosomes. The curves level off due to depletion of MESG, the Enzchek assay detection reagent (See Methods). e. Representative spectra of 2′3′SLADP (pink) and 2′3′SLADP•BeFx (blue) bound to the E309A SNF2h mutant in the presence of N+60 nucleosomes.
In all the EPR experiments, we used SNF2h, the ATPase subunit of the hACF complex, on its own because of the technical challenge of obtaining the whole complex at the concentrations required for taking EPR spectra. SNF2h recapitulates all the core functions of the hACF complex (Fig. 1b) 11,36–38. In addition, we confirmed that 2′3′SLATP supports remodeling by SNF2h at similar rates as ATP (Fig. 2a, time course, kobs = 2.0±0.06 min−1 and 1.2±0.22 min−1, for ATP and 2′3′SLATP respectively). The spin probe in solution yields a spectrum consisting of three sharp lines (Fig. 2b, black). The spectrum of SNF2h-2′3′SLADP bound to nucleosomes with 60 base pairs of flanking DNA (SNF2h-N+60 complex) shows more restricted mobility compared to when free in solution (Fig. 2b, pink). This is indicated by a broadening of the spectrum and the outward movement of the low-field peak (P1) and, high-field dip (P5). The observed P1–P5 splitting is 4.8mT (Table 1). A similar spectrum and splitting is obtained with SNF2h-2′3′SLADP alone (Table 1). The mobility of the probe can be modeled as a cone within which the probe diffuses due to thermal motion, where the vertex angle of the cone correlates with the magnitude of the P1–P5 splitting 39. For the SNF2h-N+60-2′3′SLADP complex, the vertex angle is 117.5° (Table 1). The P2–P4 sharp peaks corresponding to unbound nucleotide can be resolved from the broader peaks of the bound nucleotide at low and high field. Competition with unlabeled ATP eliminated the P1 and P5 components and restored the spectrum to that of freely tumbling 2′3′SLADP (data not shown), indicating that the probe is bound at the active site of SNF2h.
TABLE 1.
| SNF2h | Nucleosome | Analogue | Fraction Mobile | Keq (6.2mT/4.8 mT) | Splitting (mT) | Cone Angle |
|---|---|---|---|---|---|---|
| Wt | N+60 | ADP | 0.84 (0.83,0.85) | 0.19 | 4.77 | 117.5° |
| Wt | N+60 | ADP* BeFx | 0.25 (0.21,0.28,0.25) | 3.1±0.6 | 6.22, 4.79 | 67.4°, 117.0° |
| Wt | N+60 | ADP* AlFx | 0.19 (0.19,0.19) | 4.3 | 6.38, 4.73 | 61.2°, 119.1° |
| Wt | None | ADP | 0.87 (0.89,0.84) | 0.16 | 4.72 | 119.1° |
| Wt | None | ADP* BeFx | 0.94 (0.91,0.97) | 0.065 | 4.74 | 118.8° |
| Wt | N | ADP* BeFx | 0.12 (0.11,0.17,0.1) | 7.4±2.2 | 6.29, 4.88 | 64.4°, 116.0° |
| Wt | N | ADP* AlFx | 0.056 (0.04,0.07) | 19 | 6.46, 4.81 | 57.4°, 116.4° |
| Wt | gH4+60 | ADP | 0.81 (0.78,0.85) | 0.24 | 4.76 | 118.1° |
| Wt | gH4+60 | ADP* BeFx | 0.56 (0.50,0.56,0.63) | 0.8±0.2 | 6.31, 4.72 | 63.9°, 119.1° |
| Wt | gH4+60 | ADP* AlFx | 0.74 (0.78,0.71) | 0.35 | 5.90, 4.73 | 79.6°, 120.3° |
| Wt | bp+60 | ADP* BeFx | 0.61 (0.55,0.67) | 0.66 | 6.27, 4.80 | 65.6°, 116.8° |
| Wt | 60mer DNA | ADP | 0.83, 0.82 (0.83) | 0.2 | 4.74 | 117.3° |
| Wt | 60mer DNA | ADP* BeFx | 0.37 (0.32,0.41) | 1.8 | 6.28, 4.78 | 65.0°, 117.3° |
| E309A | N+60 | ADP | 0.77 (0.8, 0.74) | 0.3 | 4.82 | 116.3° |
| E309A | N+60 | ADP* BeFx | 0.79 (0.76, 0.81) | 0.27 | 4.78 | 117.7° |
| E309A | N+60 | ADP* AlFx | 0.64 (0.8, 0.49) | 0.65 | 4.81 | 116.1° |
We next determined if we could identify changes in the conformation of the ATP-binding pocket by following changes in the mobility of the probe upon changing the ATP state. ADP•BeFx is a stable, bipartite analogue that can mimic different nucleotide states depending on the structural context. These states include ground state ATP-like structures, transition state-like structures and ADP-Pi product like structures 40–42. When we used 2′3′SLADP•BeFx in complex with SNF2h and N+60, we observed large changes in the EPR spectrum. The spectrum revealed a new subpopulation of spin probes with a greater P1–P5 splitting of 6.2mT, in addition to the previously observed population with a splitting of 4.8mT (Fig. 2b, cyan, Table 1). This new component is indicated by the additional P1 and P5 components in the spectrum. A greater splitting corresponds to a more restricted or more immobile probe, with a corresponding cone angle of mobility of 67.4°. Another bi-partite nucleotide analogue that mimics activated states of ATP, 2′3′SLADP•AlFx, gives similar results as 2′3′SLADP•BeFx (Fig. 2b, Table 1 and Supplementary Fig. 1). When BeFx is added to the SNF2h-2′3′SLADP complex in the absence of the nucleosome, the immobile component does not increase (Table 1). The multi-component spectra were deconvolved to determine the fraction of mobile vs. immobile probes (Fig. 2b, Table 1; Calculated Keq = fraction immobile/fraction mobile, see Methods).
The differences in probe mobility suggest two conformations of the ATP-binding pocket: a more open (mobile probes) state and a more closed (immobile probes) state. The differences between these two states could reflect local changes in the ATP-binding pocket or large-scale changes involving domain rearrangements. By either interpretation, in the absence of the nucleosome, the closed state is largely absent. SNF2h is also largely inactive in ATP hydrolysis in the absence of nucleosomes (Fig. 3b) 11. The presence of the nucleosome shifts the equilibrium 48-fold toward the closed state, and ATP hydrolysis is correspondingly stimulated (Fig. 2b, Table 1: Keq of 3.1 with the nucleosome, 6.5×10−2 without the nucleosome, Fig. 3b). This comparison raised the possibility that formation of the closed state correlates with ATPase activity.
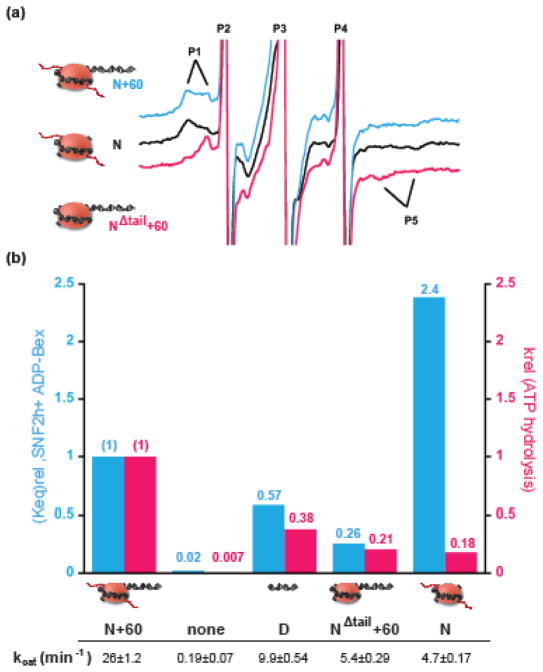
Figure 3. Effects of substrate cues on the immobile/mobile probe equilibrium and on ATP hydrolysis.
a. Representative spectra of 2′3′SLADP•BeFx bound to SNF2h in the presence of different types of nucleosomes. The equilibrium between the immobile and mobile populations of bound probe shifts in favor of the more immobile conformation when flanking DNA is removed (N+60, blue spectrum vs. N, black spectrum). In contrast, the equilibrium shifts towards the more mobile conformation when the H4 tail is removed (N+60, blue spectrum vs. NΔtail+60, pink spectrum). b. Top panel: comparison of effects on ATP hydrolysis and Keq. Blue bars: Keq values relative to the N+60 substrate. Keq is obtained as defined in Table 1 in the presence of 2′3′SLADP•BeFx. Pink bars: kcat values for ATP hydrolysis relative to the N+60 substrate. Bottom panel: The values of kcat for ATP hydrolysis under multiple-turnover conditions.
Effect of naked DNA on conformational changes in the ATP-binding site
Naked DNA is another substrate known to stimulate ATP hydrolysis by SNF2h10,36 (Fig. 3b). We therefore tested if this substrate can also promote formation of the closed state. We examined the spectrum of 2′3′SLADP•BeFx in the context of a SNF2h-DNA complex, using a 60 base-pair long duplex DNA (no nucleosome). We observed an increase in the immobile component for the SNF2h-DNA complex relative to SNF2h alone (Fig. 2c). The presence of DNA shifts the equilibrium 28-fold toward the more immobile component (Table 1, Keq of 1.8 of SNF2h with DNA, 6.5×10−2 without DNA or nucleosome). Thus, naked DNA both stimulates ATP hydrolysis by SNF2h and drives the conformation of the nucleotide-binding pocket of SNF2h to the closed state.
Effects of active site mutation on conformational changes in the ATP-binding site
To further test if the inferred conformational change in the ATP-binding pocket of SNF2h correlates with activity, we characterized the behavior of a SNF2h mutant that is deficient in ATP hydrolysis. The ATP- binding site of SNF2h contains the characteristic Walker A and Walker B motifs present in many ATPases, including the Super-Family 2 (SF2) of helicases to which ATP-dependent chromatin remodeling enzymes belong43. The conserved aspartate and glutamate (DE) residues in the Walker B motif help coordinate the catalytic Mg2+ ion and water molecules for ATP hydrolysis43,46. Mutation of the D or E residue inhibits ATP hydrolysis in other SF2 ATPases43. We mutated glutamate 309 in the Walker B motif of SNF2h to alanine (E309A) to generate a SNF2h mutant that is defective in ATP hydrolysis (Fig. 2d). This mutant is also deficient in formation of the closed state in the presence of 2′3′SLADP-BeFx and N+60 nucleosomes (Fig. 2e). The observation that perturbation of a catalytic residue on the ATP-binding pocket also reduces formation of the closed state further suggests that formation of the closed state is important for catalysis.
The roles of the H4 tail and flanking DNA in nucleotide pocket closure
The apparent correlation between ATP hydrolysis and a closed conformation of the ATP-binding pocket in the presence of ADP•BeFx and ADP•AlFx provided a meaningful context to test the energetic contributions of the substrate cues, the H4 tail and flanking DNA. We first looked at the effect of the H4 tail. The spectrum of 2′3′ SLADP bound to SNF2h complexed with nucleosomes lacking the H4 tail (NΔtail+60) shows as the dominant component, a P1–P5 splitting of 4.8mT corresponding to a cone angle of mobility of 118.1° (Table 1). When BeFx is added, an additional subpopulation of spin probes with a greater P1–P5 splitting of 6.3mT is observed. However, the immobile/mobile equilibrium for the complex between ADP•BeFx, SNF2h and NΔtail+60 is reduced ~4-fold relative to the corresponding complex with N+60 (Fig. 3a, pink vs. cyan spectra respectively). We observed a similar reduction of probes in the immobile conformations upon mutating the basic-patch of the H4 tail (Table 1: NΔpatch+60). These data suggest that the basic patch of the H4 tail promotes a closed conformation of the ATP-binding pocket of SNF2h.
The effect of the H4 tail on the equilibrium between closed and open states of the ATP-binding pocket in the presence of ADP•BeFx (Fig. 3a, 1/Keqrel ~ 4-fold) is of similar magnitude to the effect on ATP hydrolysis (Fig. 3b, 1/krel ~5-fold). This quantitative correlation raises the possibility that the H4 tail promotes ATP hydrolysis by stabilizing a closed conformation of the ATP-binding pocket.
Like the H4 tail, the presence of DNA flanking the nucleosome promotes ATP hydrolysis by SNF2h10,11 (Fig. 3b). We therefore next examined the effects of flanking DNA on the conformation of the nucleotide-binding pocket. The EPR spectrum of 2′3′SLADP•BeFx bound to SNF2h with nucleosomes lacking flanking DNA but containing the H4 tail (N nucleosomes) shows primarily an immobile component, with a P1–P5 splitting of 6.3mT corresponding to a 64.4° cone angle of mobility (Fig. 3a, black spectrum, Table 1). Deconvolution of the spectrum gives a Keq value of ~7 for the closed/open equilibrium, which is slightly greater than the Keq value for nucleosomes with flanking DNA (Table 1, N vs. N+60 with ADP-BeFx). Thus even though the H4 tail and flanking DNA both stimulate ATP hydrolysis, the H4 tail stabilizes the closed conformation of the ATP-binding pocket while the flanking DNA appears to slightly destabilize the closed conformation.
Discussion
A central question in understanding the mechanism of chromatin remodeling enzymes is how they are able to recognize which nucleosomes to move. Previous studies have indicated that recognition of the H4 tail and flanking DNA by ISWI enzymes is critical for catalysis but not for ground state binding 5,10,11,18. The mechanistic basis for how these cues act in the reaction cycle is not understood. To understand how hACF uses the H4 tail and flanking DNA cues during catalysis, we adapted an EPR based method to assay for conformational changes in the ATP-binding pocket of SNF2h, the ATPase subunit of hACF. Using the spin-labeled nucleotide analogues ADP•BeFx and ADP•AlFx, we observe a conformational change in the nucleotide-binding pocket of SNF2h under physiological, solution conditions. In one conformation, the spin probe on the nucleotide is more restricted (6.2mT, more immobile) than in the other conformation (4.8mT, more mobile). A 60bp double-stranded DNA fragment and the nucleosome, both of which stimulate ATP hydrolysis, shift the equilibrium towards the restricted conformation of the probe (Fig. 2b and c). This shift in equilibrium requires ADP•BeFx and is not detected with ADP alone. A Walker B mutant (E309A), which shows defects in ATP hydrolysis, shifts the equilibrium towards the more mobile conformation of the probe. While ADP•BeFx has been shown to mimic the ATP ground state in many ATPases, it has also been shown to mimic transition state like structures and the ADP-Pi product in some systems 40–42. Given the absence of high-resolution structures of chromatin remodeling ATPases bound to nucleotide analogs we do not know which state is mimicked by ADP•BeFx in the context of SNF2h. Nevertheless the state mimicked by this analog stabilizes a conformation of the SNF2h active site that correlates with ATP hydrolysis. This correlation suggests that ADP-BeFx mimics either the ground state ATP structure or a transition state like structure in the context of SNF2h.
At a physical level what could the conformational change in the active site represent? The spin probe reports on environmental changes near the 2′3′hydroxyl groups of the nucleotide. These changes could reflect local effects around the 2′3′ hydroxyl groups or larger conformational changes in the active site. The ATPase domain of chromatin remodeling enzymes consists of two RecA-like lobes separated by a flexible linker. ATP binds in a cleft between these lobes. In many distantly related SF1 and SF2 family helicases, binding of DNA or RNA promotes closure of the two RecA-like lobes of the ATPase domain onto ATP, thus promoting catalysis 44–46. Work on Sulfolobus solfataricus SWI2/SNF2, which is part of the chromatin remodeling superfamily, suggests that binding of the enzyme to DNA can alter the orientation of the two RecA-like lobes relative to one another 47,48.
We find that the substrate cue provided by the basic patch on the H4 tail increases ATPase rates and correspondingly shifts the equilibrium towards a more closed active site. Rate enhancement by the H4 tail is maintained upon inserting a flexible linker between the H4 tail and the globular domain. This result is consistent with a model in which the H4 tail stabilizes a particular conformation of the enzyme through specific binding interactions rather than acting as a mechanical element for the transduction of force. In the case of the related chromatin remodeling enzyme, Chd1, the two RecA-like lobes of the enzyme are physically propped open by a third domain, and thus in an autoinhibited state where they cannot close and hydrolyze ATP until the nucleosome is bound49. It is possible that binding the H4 tail helps relieve an analogous auto-inhibited conformation of SNF2h, and allows for closure of the ATP-binding pocket. Indeed, a recent study of ISWI, the Drosophila homolog of SNF2h, suggests that the H4 tail plays a role in relieving auto-inhibition by displacing a basic-patch mimic sequence on an auto-inhibitory domain of ISWI 50. Further, work with Drosophila ISWI has shown that DNA binding induces a conformational change in the ATPase domain51. Given these previous observations and the results presented here, we speculate that the H4 tail stabilizes a closed conformation of the two RecA-like lobes of SNF2h in which the catalytic residues are properly aligned for ATP hydrolysis, thereby directly coupling substrate recognition to activity.
In contrast to the H4 tail, the substrate cue provided by the flanking DNA increases ATP hydrolysis but does not shift the equilibrium towards a closed active site. The simplest interpretation of the difference between the H4 tail and the flanking DNA is that these substrate cues act in different reaction steps. It is possible that the flanking DNA acts in a different rate-limiting step prior to ATP hydrolysis. For example, such a step could entail a conformational change in SNF2h that allows the proper engagement of the HAND-SANT-SLIDE domain with flanking DNA. However substantial future work will be needed to determine the exact nature of the reaction step that is stimulated by flanking DNA.
Methods
Protein purification
Both human SNF2h and human hACF complex were expressed in SF9 cells and purified as described previously 11. For EPR experiments, SNF2h was concentrated using the centrifugal filter unit Microcon YM-100 (Millipore).
Nucleosome Assembly
Nucleosomes were assembled using the 601 positioning sequence, containing a Pst1 site 18bp in from one end, and 0, 20, 60, and 80bp of DNA flanking the nucleosome on the other end as described previously 11. To measure kinetics of nucleosome remodeling by FRET, H2A containing a unique engineered cysteine at residue 120 was labeled under denaturing conditions with Cy5-maleimide and then assembled by salt dialysis into octamers. H2A-120C-Cy5 octamers were then assembled into nucleosomes using Cy3 end labeled DNA as described previously11.
FRET-based Nucleosome Remodeling Assay
Kinetics of nucleosome remodeling were measured as previously described on an ISS K2 fluorometer, and fit as previously described to two exponential phases using Kaleidagraph 11. Samples were excited at 520nm, using a 400nm longpass filter, and emission of the donor Cy3 dye was measured at 565nm (Fig. 1b), using a 495 longpass filter. All reactions were performed at 30°C, with 5nM nucleosomes, and varying concentrations of hACF enzyme in excess of nucleosomes, in the following reaction buffer: 60mM KCl, 2mM MgCl2, 0.32mM EDTA, 12% (v/v) glycerol, 12mM HEPES (pH 7.9), 4mM Tris (pH7.9), 0.02% (v/v) Nonidet P-40, 0.5mg/ml Flag peptide, 0.4mM DTT, 0.4mM BenzHCl. After a 10 minute incubation at 30°C, reactions were initiated by the addition of 2mM ATP-Mg. Saturation of nucleosomes by hACF was confirmed by varying the concentration of hACF.
ATPase Assay
ATP hydrolysis rates were measured using the Enzchek assay kit (Invitrogen) under multiple turnover conditions with saturating ATP and nucleic acid ligand over SNF2h (3 mM ATP-Mg, either 400 nM nucleosomes or 8uM gel-purified 60bp duplex DNA and 200 nM of SNF2h). Absorbance measurements were taken on a Flexstation 3 platereader (Molecular Devices). Reactions were performed at 30°C in the following reaction buffer: 60mM KCl, 3mM MgCl2, 20mM Tris (pH7.5), 0.02% Nonidet P-40, 200mM 2-amino-6-mercapto-7-methyl-purine riboside (MESG), 1U/mL purine nucleoside phosphorylase. After a 10 minute incubation at 30°C, reactions were initiated by the addition of 3mM ATP-Mg. Using a serial dilution of KH2PO4 as substrate for the Enzchek reaction, a standard curve of A360/Pi (uM) was used to convert absorbance values into the concentration of Pi in micromolar. The value of kcat was obtained by taking the initial slope of each trace (in uM/min) and dividing it by the concentration of SNF2h (0.2 uM).
EPR Spectroscopy
EPR measurements were performed with a Bruker Instruments EMX EPR spectrometer (Billerica, MA) as desc ribed previously 25. First derivative, X-band spectra were recorded in a high-sensitivity microwave cavity using 50-s, 10.0mT wide magnetic field sweeps. For clarity only 8.0mT wide fields are shown in Figures. The instrument settings were as follows: microwave power, 25 mW; time constant, 164 ms; frequency, 9.83 GHz; modulation, 0.1mT at a frequency of 100 kHz. Each spectrum used in the data analysis is an average of 50–150 sweeps from an individual experimental preparation. 2′3′SLATP was prepared as described previously52. 2′3′SLADP was prepared by the same method, but replacing ATP with ADP as a starting material52. Spectra of 10uM of the nucleotide analogue 2′3′ SLADP were measured with 16uM SNF2h (or 32uM SNF2h for the Walker B mutant, E309A) with or without 8uM nucleosomes or 16 uM 60 bp DNA in the following buffer conditions: (12mM HEPES pH 7.9, 4mM Tris pH 7.5, 60mM KCl, 0.2–1mM MgCl2, 0.32mM EDTA, 12% glycerol, 0.4mM DTT, 0.4mM BenzHCl, 0.4 mg/ml Flag Peptide). For experiments with beryllium fluoride, 0.5mM BeFx (made immediately before use by combining BeCl2 and NaF to final concentrations of 0.5mM and 2.5mM respectively) was added, and for experiments with AlFx, 0.5mM AlFx (made immediately before use by combining AlCl3 and NaF to final concentrations of 0.5mM and 2.5mM respectively) was added. Samples were incubated at room temperature for 10 minutes before taking the first scan in the EPR machine. Nucleosome and DNA concentrations were varied to ensure that the SNF2h was saturated with either ligand.
Deconvolution of spectra
Experimental spectra were deconvolved into mobile, immobile and unbound probe components using a least-squares minimization FORTRAN program employing the Levenberg-Marquardt algorithm. Basis spectra for the immobile and mobile components in the least-squares minimization were determined by a subtraction procedure using experimentally determined spectra 53. The unbound basis was determined from solution measurement of the spectrum of spin probe. Double integration of the first-derivative spectra was used to determine the fractions of probes involved in the individual components. As we are interested in the equilibrium between immobile and mobile components, they were weighted 100 times relative to the free probe basis in the minimization.
Supplementary Material
Highlights.
The nucleosomal H4 tail and flanking DNA promote catalysis of chromatin remodeler ACF
ACF requires the H4 basic patch but tolerates changes in rigidity and length of H4 tail linkage
A Spin-labeled ATP analogue reveals two distinct conformations of the SNF2h ATP-pocket
The H4 tail but not flanking DNA, promotes the more restricted ATP-pocket state
The H4 tail and flanking DNA may play qualitatively different roles during remodeling
Acknowledgments
This work was supported by NIH grant GM073767 to G.J.N. and AR053720 to N.N., and E.P. We thank Julia Tretyakova for preparing histones, Daniele Canzio and Coral Y. Zhou for helpful comments on the manuscript, and the Narlikar laboratory members for helpful discussions.
Glossary
- hACF
human ATP-dependent Chromatin Assembly and Remodeling Factor
- ISWI
Imitation SWItch enzyme
- SNF2h
Sucrose Nonfermenting 2 Homolog enzyme
- SHL(±2)
Superhelical Location (±2)
- FRET
Fluorescence Resonance Energy Transfer
- EPR
electron paramagnetic resonance spectroscopy
- 2′3′SLATP
2′3′ Spin-Labeled Adenosine Triphosphate
- SF1
Superfamily 1 helicase
- SF2
Superfamily 2 helicase
- SWI2/SNF2
Switch/Sucrose Non-Fermentable enzyme
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 2.Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 2004;18:170–83. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chioda M, Vengadasalam S, Kremmer E, Eberharter A, Becker PB. Developmental role for ACF1-containing nucleosome remodellers in chromatin organisation. Development. 2010;137:3513–22. doi: 10.1242/dev.048405. [DOI] [PubMed] [Google Scholar]
- 4.Hamiche A, Kang JG, Dennis C, Xiao H, Wu C. Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc Natl Acad Sci U S A. 2001;98:14316–21. doi: 10.1073/pnas.251421398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang W, Kagalwala MN, Bartholomew B. Regulation of ISW2 by concerted action of histone H4 tail and extranucleosomal DNA. Mol Cell Biol. 2006;26:7388–96. doi: 10.1128/MCB.01159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clapier CR, Nightingale KP, Becker PB. A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 2002;30:649–55. doi: 10.1093/nar/30.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clapier CR, Langst G, Corona DF, Becker PB, Nightingale KP. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol Cell Biol. 2001;21:875–83. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zofall M, Persinger J, Bartholomew B. Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2. Mol Cell Biol. 2004;24:10047–57. doi: 10.1128/MCB.24.22.10047-10057.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem. 2006;281:16279–88. doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He X, Fan HY, Narlikar GJ, Kingston RE. Human ACF1 alters the remodeling strategy of SNF2h. J Biol Chem. 2006;281:28636–47. doi: 10.1074/jbc.M603008200. [DOI] [PubMed] [Google Scholar]
- 11.Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat Struct Mol Biol. 2006;13:1078–83. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- 12.Corona DF, Clapier CR, Becker PB, Tamkun JW. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 2002;3:242–7. doi: 10.1093/embo-reports/kvf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira H, Flaus A, Owen-Hughes T. Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms. J Mol Biol. 2007;374:563–79. doi: 10.1016/j.jmb.2007.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zlatanova J, Leuba SH, Yang G, Bustamante C, van Holde K. Linker DNA accessibility in chromatin fibers of different conformations: a reevaluation. Proc Natl Acad Sci U S A. 1994;91:5277–80. doi: 10.1073/pnas.91.12.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McBryant SJ, Klonoski J, Sorensen TC, Norskog SS, Williams S, Resch MG, Toombs JA, 3rd, Hobdey SE, Hansen JC. Determinants of histone H4 N-terminal domain function during nucleosomal array oligomerization: roles of amino acid sequence, domain length, and charge density. J Biol Chem. 2009;284:16716–22. doi: 10.1074/jbc.M109.011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–3. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 18.Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. Embo J. 2004;23:2092–104. doi: 10.1038/sj.emboj.7600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grune T, Brzeski J, Eberharter A, Clapier CR, Corona DF, Becker PB, Muller CW. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell. 2003;12:449–60. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 20.Dang W, Bartholomew B. Domain Architecture of the Catalytic Subunit in the ISW2-Nucleosome Complex. Mol Cell Biol. 2007;27:8306–17. doi: 10.1128/MCB.01351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada K, Frouws TD, Angst B, Fitzgerald DJ, DeLuca C, Schimmele K, Sargent DF, Richmond TJ. Structure and mechanism of the chromatin remodelling factor ISW1a. Nature. 2011;472:448–53. doi: 10.1038/nature09947. [DOI] [PubMed] [Google Scholar]
- 22.Schwanbeck R, Xiao H, Wu C. Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J Biol Chem. 2004;279:39933–41. doi: 10.1074/jbc.M406060200. [DOI] [PubMed] [Google Scholar]
- 23.Whitehouse I, Stockdale C, Flaus A, Szczelkun MD, Owen-Hughes T. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol Cell Biol. 2003;23:1935–45. doi: 10.1128/MCB.23.6.1935-1945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–46. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 25.Racki LR, Yang JG, Naber N, Partensky PD, Acevedo A, Purcell TJ, Cooke R, Cheng Y, Narlikar GJ. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature. 2009;462:1016–21. doi: 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blosser TR, Yang JG, Stone MD, Narlikar GJ, Zhuang X. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–7. doi: 10.1038/nature08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baneres JL, Martin A, Parello J. The N tails of histones H3 and H4 adopt a highly structured conformation in the nucleosome. J Mol Biol. 1997;273:503–8. doi: 10.1006/jmbi.1997.1297. [DOI] [PubMed] [Google Scholar]
- 28.Johnson LM, Fisher-Adams G, Grunstein M. Identification of a non-basic domain in the histone H4 N-terminus required for repression of the yeast silent mating loci. EMBO J. 1992;11:2201–9. doi: 10.1002/j.1460-2075.1992.tb05279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan HS, Dill KA. Polymer principles in protein structure and stability. Annu Rev Biophys Biophys Chem. 1991;20:447–90. doi: 10.1146/annurev.bb.20.060191.002311. [DOI] [PubMed] [Google Scholar]
- 30.Goldenberg DP. Computational simulation of the statistical properties of unfolded proteins. J Mol Biol. 2003;326:1615–33. doi: 10.1016/s0022-2836(03)00033-0. [DOI] [PubMed] [Google Scholar]
- 31.Miller WG, Goebel CV. Dimensions of protein random coils. Biochemistry. 1968;7:3925–35. doi: 10.1021/bi00851a021. [DOI] [PubMed] [Google Scholar]
- 32.Spudich JA. Molecular motors: forty years of interdisciplinary research. Mol Biol Cell. 2011;22:3936–9. doi: 10.1091/mbc.E11-05-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naber N, Minehardt TJ, Rice S, Chen X, Grammer J, Matuska M, Vale RD, Kollman PA, Car R, Yount RG, Cooke R, Pate E. Closing of the nucleotide pocket of kinesin-family motors upon binding to microtubules. Science. 2003;300:798–801. doi: 10.1126/science.1082374. [DOI] [PubMed] [Google Scholar]
- 34.Naber N, Purcell TJ, Pate E, Cooke R. Dynamics of the nucleotide pocket of myosin measured by spin-labeled nucleotides. Biophys J. 2007;92:172–84. doi: 10.1529/biophysj.106.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, Pate E, Cooke R, Taylor EW, Milligan RA, Vale RD. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–84. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 36.Aalfs JD, Narlikar GJ, Kingston RE. Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J Biol Chem. 2001;276:34270–8. doi: 10.1074/jbc.M104163200. [DOI] [PubMed] [Google Scholar]
- 37.Fan HY, He X, Kingston RE, Narlikar GJ. Distinct strategies to make nucleosomal DNA accessible. Mol Cell. 2003;11:1311–22. doi: 10.1016/s1097-2765(03)00192-8. [DOI] [PubMed] [Google Scholar]
- 38.Fan HY, Trotter KW, Archer TK, Kingston RE. Swapping function of two chromatin remodeling complexes. Mol Cell. 2005;17:805–15. doi: 10.1016/j.molcel.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 39.Griffith OH, Jost PC. Spin Labeling Theory and Applications. Academic Press; NY: 1976. Lipid spin labels in biological membranes; pp. 454–532. [Google Scholar]
- 40.Del Campo M, Lambowitz AM. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–34. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rayment I, Smith C, Yount RG. The active site of myosin. Annu Rev Physiol. 1996;58:671–702. doi: 10.1146/annurev.ph.58.030196.003323. [DOI] [PubMed] [Google Scholar]
- 43.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–24. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mallam AL, Jarmoskaite I, Tijerina P, Del Campo M, Seifert S, Guo L, Russell R, Lambowitz AM. Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc Natl Acad Sci U S A. 2011;108:12254–9. doi: 10.1073/pnas.1109566108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–60. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 47.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–73. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 48.Lewis R, Durr H, Hopfner KP, Michaelis J. Conformational changes of a Swi2/Snf2 ATPase during its mechano-chemical cycle. Nucleic Acids Res. 2008;36:1881–90. doi: 10.1093/nar/gkn040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hauk G, McKnight JN, Nodelman IM, Bowman GD. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell. 2010;39:711–23. doi: 10.1016/j.molcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clapier CR, Cairns BR. Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature. 2012;492:280–4. doi: 10.1038/nature11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mueller-Planitz F, Klinker H, Ludwigsen J, Becker PB. The ATPase domain of ISWI is an autonomous nucleosome remodeling machine. Nat Struct Mol Biol. 2013;20:82–9. doi: 10.1038/nsmb.2457. [DOI] [PubMed] [Google Scholar]
- 52.Crowder MS, Cooke R. Orientation of spin-labeled nucleotides bound to myosin in glycerinated muscle fibers. Biophys J. 1987;51:323–33. doi: 10.1016/S0006-3495(87)83338-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purcell TJ, Naber N, Franks-Skiba K, Dunn AR, Eldred CC, Berger CL, Malnasi-Csizmadia A, Spudich JA, Swank DM, Pate E, Cooke R. Nucleotide pocket thermodynamics measured by EPR reveal how energy partitioning relates myosin speed to efficiency. J Mol Biol. 2011;407:79–91. doi: 10.1016/j.jmb.2010.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.