Abstract
Background
Sinus node (SN) dysfunction is observed in some Long QT syndrome (LQTS) patients, but has not been studied as a function of LQTS genotype. LQTS6 involves mutations in the hERG β-subunit MiRP1, which also interacts with hyperpolarization-activated, cyclic nucleotide gated (HCN) channels - the molecular correlate of SN pacemaker current (If). An LQTS registry search identified a 55 year male with M54T MiRP1 mutation, history of sinus bradycardia (39–56 bpm), and prolonged QTc.
Objective
We tested if LQTS6 incorporates sinus bradycardia due to abnormal If.
Methods
We transiently co-transfected neonatal rat ventricular myocytes (to study currents in a myocyte background) with human HCN4 (hHCN4, primary SN isoform) or human HCN2 (hHCN2) and one of the following: empty vector, wildtype hMiRP1 (WT), M54T hMiRP1 (M54T). Current amplitude, voltage dependence and kinetics were measured by whole cell patch clamp.
Results
M54T co-expression decreased HCN4 current density by 80% compared to hHCN4 alone or with WT, and also slowed HCN4 activation at physiologically relevant voltages. Neither WT nor M54T altered HCN4 voltage dependence. A computer simulation predicts that these changes in HCN4 current would decrease rate and be additive with published effects of M54T mutation on hERG kinetics on rate.
Conclusions
We conclude that M54T LQTS6 mutation can cause sinus bradycardia through effects on both hERG and HCN currents. Patients with other LQTS6 mutations should be examined for SN dysfunction, and the effect on HCN current determined.
Keywords: long-QT syndrome 6, sinus node dysfunction, sinus bradycardia, pacemaker current, M54T mutation, MiRP1, KCNE2, HCN4, HCN2
INTRODUCTION
The long-QT syndrome (LQTS) predisposes to cardiac arrhythmias and is characterized by prolongation of the QT interval on ECG. One of the leading causes of sudden death in the young, LQTS is caused by mutations in genes encoding ion channels controlling ventricular repolarization, or mutations in proteins that affect the function of these channels. Patients with LQTS may present with syncope, seizures or sudden death.
Some studies suggest that average resting heart rate is lower in LQTS patient,; especially children compared with normal control subjects1–3. In the study by Schwartz et al in 1985 proposing the first set of diagnostic criteria for LQTS, low heart rate in children is one of the minor criteria4. In the 1993 update to this study, a resting heart rate below the second percentile still remains a diagnostic criteria5. In addition to being a strong clue to diagnosis in the pediatric age group, sinus bradycardia is a risk factor for cardiac events in family members of LQTS patients6.
Molecular genetic studies have identified up to 12 genetic loci associated with LQTS7. Mutations in potassium channel subunits KVLQT1 (LQTS1), hERG (LQTS2), KCNE1 (LQTS5) and KCNE2 (LQTS6) diminish the potassium current and cause delayed repolarization of the myocardium8–10 prolonging the QT interval. Of these, the KCNE2 gene encodes a single transmembrane domain protein, MinK related Peptide 1 (MiRP1), which assembles with pore-forming α subunits to establish the attributes of characteristic channels in vivo. MiRP1 functions as a β subunit of hERG (human ether-a-go-go channel) potassium channel11, and together these proteins form the functional channel that generates IKr rapid delayed rectifier potassium current. The QT prolongation in LQTS6 is ascribed to the effect of MiRP1 mutations on IKr.
MiRP1 combines with and alters the function of not only hERG11, but also other K channels (Kv4.212, KvLQT113,14, Kv1.515) as well as the HCN (hyperpolarization activated cyclic nucleotide gated channel) family of pacemaker channels16–17. MiRP1 has been shown to modulate HCN channel expression and gating properties in myocytes and heterologous expression systems16–18. However, these effects vary with different isoforms and species. High levels of both MiRP1 and HCN subunits (predominantly HCN4, with HCN1 or HCN2 also present, depending on the species) are expressed in the cardiac SN16,19–21, where pacemaker current contributes to impulse initiation. Mutations in the HCN gene are associated with familial sinus bradycardia22. The question of whether human MiRP1 mutations would differentially affect HCN current compared to WT hMiRP1 protein in a manner that could contribute to sinus bradycardia has not been addressed.
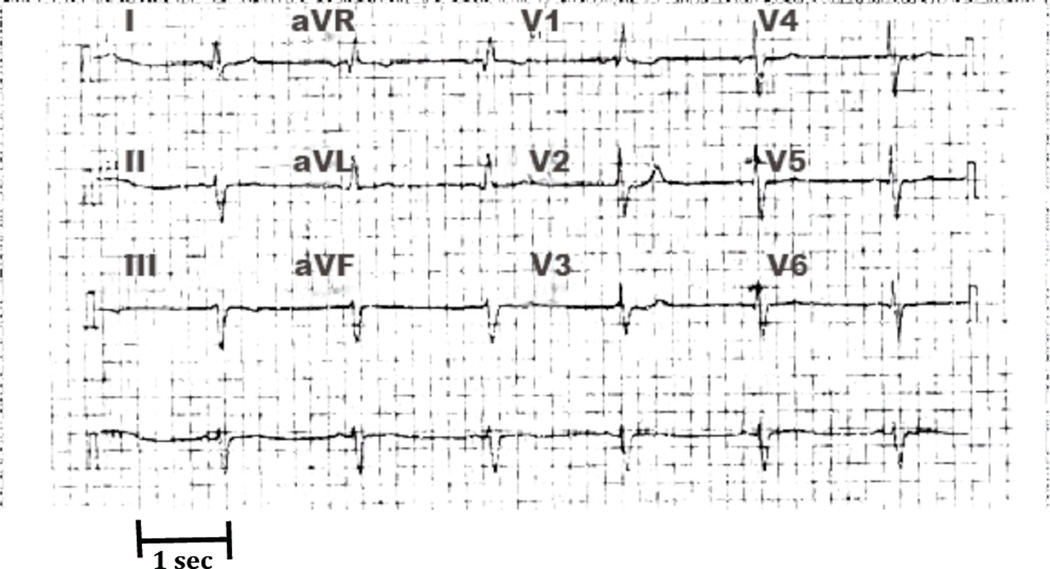
The International Long-QT syndrome Registry, with its expanding number of genotyped families, has provided an opportunity to study the clinical aspects and explore the genotype-phenotype relationships in LQTS23. A search of the Long-QT syndrome Registry identified a 55-year-old patient with the M54T MiRP1 mutation (first reported by Abbott et al11) with sinus bradycardia. This patient’s previously healthy daughter died suddenly at the age of 13. No abnormalities were found at autopsy, but a subsequent blood test confirmed the M54T mutation. Following this result, the index patient tested positive for the M54T mutation. A 12-lead ECG from this patient at age 51 years shows heart rate of 39 bpm (Fig 1) with a corrected QT interval of 476 milliseconds. This patient had telemetry strips from age 7 years that show underlying sinus bradycardia with heart rates between 31–56 bpm.
Figure 1.
ECG of the index patient at 51 years showing a heart rate of 39. The corrected QT interval was 476 ms. Each grid box represents 0.2 sec.
We thus asked if this M54T MiRP1 mutation associated with LQTS6 affects HCN function differently than the wildtype hMiRP1 (WT). This difference if present could possibly explain SN dysfunction in this subgroup of LQTS patients. If there is no difference it could mean that any observed SN dysfunction in LQTS6 patients arises from effects on SN targets other than HCN. Previous studies16–18 have used different species isoforms to study effects of MiRP1 on HCN; for example Qu et al18 tested the effects of rat MiRP1 on mouse HCN2. Abbott et al11 have shown that when different species of MiRP1 interact with hERG, the functional outcome is different. Therefore, in this paper we study the effects of human WT MiRP1 and its M54T mutant on human HCN isoforms.
METHODS
Molecular cloning and mutagenesis
Mammalian expression vectors: The complete human HCN2 and human HCN4 sequences were packaged in pDC515 and pDC516 (AdMax, Microbix Biosystems) respectively to create the pDC515hHCN2 and pDC516hHCN4 vectors driven by CMV promoter. For hMiRP1, the pCINeo vector (a gift from Dr. Geoffrey Abbott, University of California, Irvine) was used as a carrier. To identify transfected cells in all groups, the gene encoding the green fluorescent protein (GFP) was co-transfected using the pmaxGFP vector (Amaxa kit, Lonza technology).
The M54T mutant was created using Quikchange Site-directed mutagenesis kit (Stratagene). In the M54T mutant: threonine replaces methionine in the transmembranous region. The primers (Genelink) designed for mutagenesis are shown in Table 1. Successful mutagenesis and the mutant DNA sequence were verified by sequencing at the Columbia University DNA sequencing facility.
Table 1.
The oligonucleotides used mutagenesis. Mutant nucleotides are in bold and underlined.
| Primers | Sequence |
|---|---|
| M54Tmutants | Forward TGTCATCCTGTACCTCACGGTGATGATTGGAATGT Reverse ACATTCCAATCATCACCGTGAGGTACAGGATGACA |
Preparation of Genomic DNA and Sequencing of HCN4
Informed consent was obtained from the subject. The protocol for performing skin biopsies on patients and families with inherited heart disease has been reviewed and approved by Columbia University Institutional Review Board (IRB-AAAD5685). A 3 mm punch biopsy was obtained from the subject and dermal fibroblasts were derived in tissue culture. Genomic DNA was derived from cultured fibroblasts using QiAmp DNA Mini Kit (Qiagen). HCN4 coding sequences were amplified by PCR using FastStart Taq polymerase (Roche). Primers selected are listed in Table 2. PCR products were analyzed by DNA sequencing (Bio-Fab Research).
Table 2.
Primers used for screening HCN4
| HCN4 | Amplicon | Primer Forward | Primer Reverse | bp |
|---|---|---|---|---|
| Exon 1.1 | GCAAAGGACGCGTCCCC | GCGACTGCAGGCGCTTC | 395 | |
| 1.2 | GCAAGTCCAGCACGAACGG | GCGACCAGATCCTCCCGG | 354 | |
| 1.3 | GGACACCGCTATCAAAGTGG | CCGGAGTTAACTTTCCTGCC | 330 | |
| Exon 2.1 | TCCTCTCTCTCTTCCTGGCGACT | CTGGACCCGCAGCGGATTAA | 260 | |
| 2.2 | GTGGTGGAGGACAACACAGA | TGGTCAGTGCAAACCTCTTG | 280 | |
| Exon 3 | CCAGGGCAGGGAAAGTGGC | GGTGGCGAGATGTGGACTTG | 315 | |
| Exon 4 | TTCCCTCTCATCCACTGTCCC | GGAGCACCCGCACATTGGTC | 295 | |
| Exon 5 | GGAACCAAGTTTAGCCAGGA | ACCACCTCCTTCCCGTGT | 353 | |
| Exon 6.1 | GTGTCCCAATCCACCCTGT | GACTACATCATCCGGGAAGG | 166 | |
| 6.2 | AGCTGCGTTTCGAGGTCTT | GACAGGGCAGCTGCTCCC | 192 | |
| Exon 7.1 | AGGCTGTGCATGCCTCAT | CAATGAGGTGCTGGAGGAGT | 149 | |
| 7.2 | GACACCTACTGCCGCCTCTA | TGAGTGCCTGTTCTTGTTCTG | 249 | |
| Exon 8.1 | CTGCACCTGATCTCCTTCC | TAGCCCTCACCCACCACC | 311 | |
| 8.2 | CGTCATCTGGACCCCGCTGAT | AGCAGCCCGTCCCAGGTG | 236 | |
| 8.3 | GGCTGCAGTCCCTGATCCC | CCCCGCTGCAGCCAGGC | 314 | |
| 8.4 | TCCACAAGGCGCTGGGTGG | CTCCCCGAGGAGGTCTCAG | 349 | |
| 8.5 | GTCCCTTGTGGCAGGGGC | CTCATCTCCGCGTCTCAGCC | 261 | |
| 8.6 | CCAGGACCTCAAGCTCATCT | GTCTTTGTTTGGGGCAAGAG | 275 | |
| 8.7 | TCCTCAGGTTCTTTGCCACC | CTCTCCCTTCCTTCTTCCTT | 198 |
Cell culture and Transfection
Cultures of newborn rat ventricular myocytes (NRVM) were prepared as previously described24. Briefly, 1–2 day-old Wistar rats were euthanized by decapitation in accordance with protocols approved by the Institutional Animal Care and Use Committee of Columbia University. Hearts were quickly removed and ventricles were dissociated by a standard trypsinization procedure. Myocytes were harvested and plated in protamine-sulfate coated dishes. On the day of the experiment, the cell monolayer was resuspended by brief exposure to 0.1% trypsin and the cells then re-plated onto fibronectin-coated coverslips.
Transfection was performed on day 0 by electroporation using the Rat Cardiomyocyte Neo Nucleofector® Kit (Lonza technology). The NRVM (2×106 cells) were transfected with a total of 3.5µg of DNA as follows: either pDC516hHCN4 (1.5µg) or pDC515hHCN2 (1.5µg) with one of the following: 1) pCINeoWThMiRP1 (1.5µg), 2) pCINeoM54ThMiRP1 (1.5µg), 3) pDC516 (“Empty”; 1.5µg empty plasmid) to maintain equal DNA load during electroporation. To identify the transfected cells for electrophysiology, all the cell groups were co-transfected with GFP (green florescent protein; 0.5 µg).
Electrophysiology
The whole-cell HCN current from myocytes was recorded at 35°C. Extracellular solution contained (mM): NaCl, 140; NaOH, 2.3; MgCl2, 1; KCl, 10; CaCl2, 1; HEPES, 5; glucose, 10; MnCl2 (2 mM) and BaCl2 (4 mM) were included to eliminate ICa and IK1, respectively; pH 7.4. The pipette solution included (mM): aspartic acid, 130; KOH, 146; NaCl, 10; CaCl2, 2; EGTA-KOH, 5; Mg-ATP, 2; HEPES-KOH, 10; pH 7.2. The pipette resistance was typically 3–5 MΩ. An Axopatch-200B amplifier and pClamp9.2 software (Molecular Devices) were used for data acquisition and analysis.
The normalized plot of tail current versus test voltages was fitted with a Boltzmann function and then the voltages of half maximal activation and slope factor were determined from the fitting. Time constants of activation were obtained by fitting the early time course of current traces with a monoexponential function; the initial delay and any slow activation phase were not fit. Deactivation time constants were obtained by a single exponential fit of the time course of the current trace at each test voltage after activation by a prepulse to −125 mV. For both activation and deactivation, the duration of the trace being fit was at least three times the measured time constant to ensure accuracy. Data analysis was done using Origin software version 7. Pooled data are expressed as mean±SEM; n denotes number of cells. Statistical significance was determined by the t test or ANOVA, as appropriate; p<0.05 was considered significant.
HCN4 current was defined as the time-dependent component taken at the end of 9-second hyperpolarizing test voltage steps from −35 to −115 mV with increments of 10mV from the holding potential of −35 mV. The hyperpolarizing step was followed by a 9-second step to −110 mV to record the tail current and then a 0.5 sec pulse to −5 mV to ensure full deactivation. Current-voltage relationships were obtained by measuring the current amplitude after application of 9-second hyperpolarizing test pulses and normalization to the fully activated current at −110 mV. The HCN2 protocol varied from the above in that the hyperpolarizing step duration was 6 seconds and the tail current was measured at −100 mV where current was fully activated, reflecting the faster activation kinetics of this isoform.
Computer simulation
We employed the sinoatrial node model of Kurata et al25. The magnitude, midpoint and slope of the pacemaker current activation curve were modified to match the experimentally observed effect of the M54T mutation on HCN4. We also modified IKr to reflect the published report of the effect of M54T on this current26. The Kurata model represents significant improvements over earlier models as it provides well-integrated explanations of the electrophysiological behavior of primary pacemaker cells in the rabbit SN and incorporates If not included in previous models and expression of kinetics of IKr in addition to other currents25,27,28.
RESULTS
Effect of M54T MiRP1 on biophysical properties of HCN4 currents expressed in NRVM
Since we had previously shown that HCN channel biophysics varies with cellular context29,30 we studied biophysical properties of HCN4 and HCN2 current in NRVM to provide a cardiac background. HCN2 or HCN4 was co-expressed with empty plasmid (the HCN group), WT hMiRP1 (the +WT group) and M54T hMiRP1 (the +M54T group) and current density, kinetics and voltage dependence were compared.
M54T decreases HCN4 amplitude
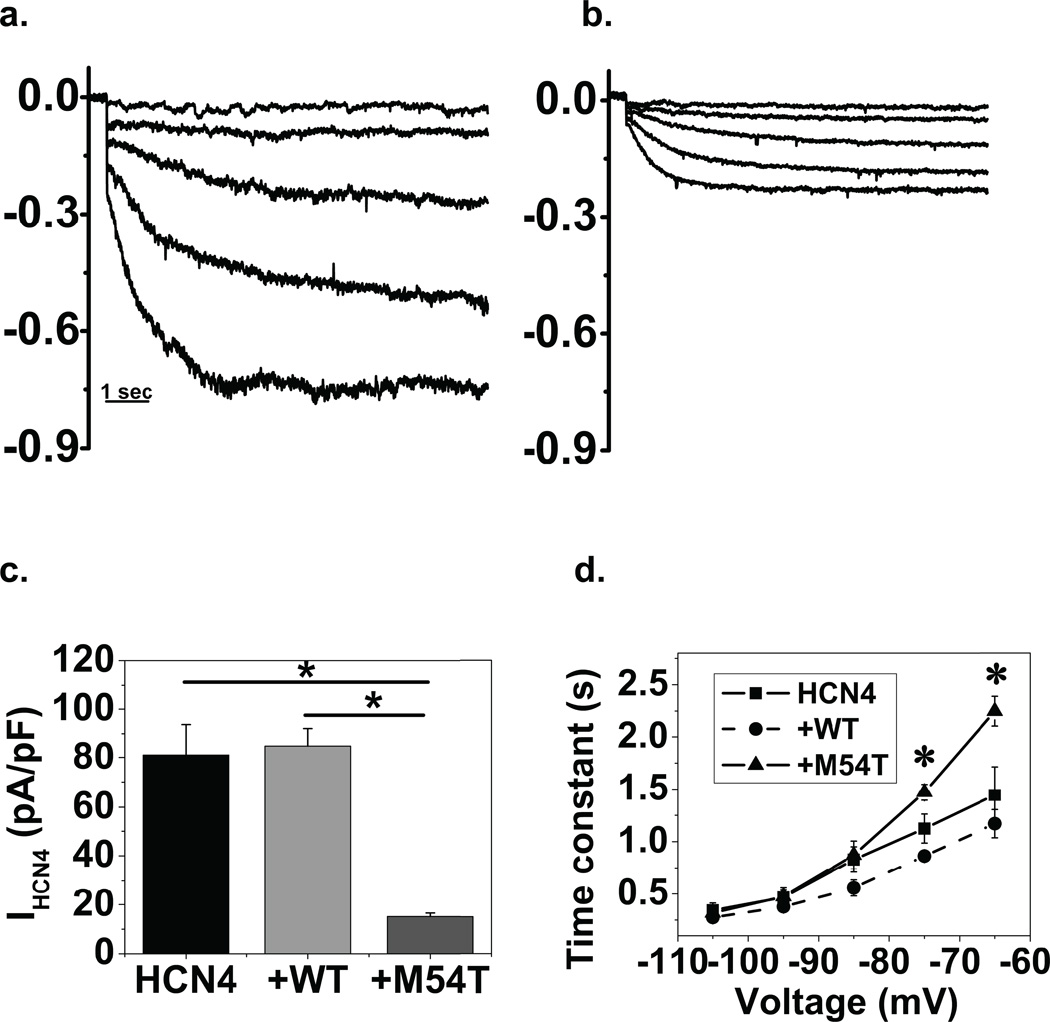
Representative current recordings in NRVM for +WT and +M54T group are shown in Figs 2a and 2b. Tail currents were measured at −110 mV where the current was fully activated and to avoid contaminating currents at less negative voltages. Co-expression of M54T (15±1.4 pA/PF) decreases HCN4 tail current density by 80% compared with HCN4 alone (81±13 pA/pF) as well as with +WT (85±7 pA/pF) as shown in Fig 2c. The magnitudes of the time dependent currents for M54T are significantly different from the HCN4 and +WT groups (n=6–10) by ANOVA, p<0.001. WT co-expression did not have a significant effect on the HCN4 current amplitude.
Figure 2. M54T mutant alters HCN4 current density and kinetics when co-expressed in myocytes.
HCN4+WT (a.) and HCN4+M54T (b.) representative current traces in myocytes. Current traces (in nA) of HCN4 were generated by applying hyperpolarizing steps to −45, −55, −65, −75, −85 mV; tail currents at −110 mV not shown. The representative traces show that co-expression with M54T decreases current amplitude, as illustrated from tail current measurements (panel c); n=6–10, p<0.05 by ANOVA (*). d. M54T activation kinetics curve differs significantly from the +WT and HCN4 group (*) at physiologically relevant voltages. e. The deactivation of the M54T group did not alter significantly from the +WT and HCN4 group.
M54T slows activation kinetics of HCN4 current
Fig 2d plots average time constants of activation as a function of voltage. It is evident that the M54T mutant, when co-expressed with HCN4, slows the kinetics in comparison with HCN4 alone or +WT. These differences are statistically significant for M54T at −65 and −75 mV (by pairwise multiple comparison, Bonferroni t test, p<0.05; n=6–10), voltages relevant to the membrane potential in sinus node. Tau at −65 mV was 2246±142 ms for M54T compared with 1443±271 ms for HCN4 or 1167±135 for +WT; n=6–10, p<0.01. We did not find any statistically significant difference between kinetics of the HCN4 group and +WT group. We separately studied the effect of the M54T mutation on HCN4 deactivation kinetics and found no significant difference between the three groups (Fig 2e). Tau at −65 mV was 2660±204 ms for M54T compared with 2506±202 ms for HCN4 or 2721±288 for +WT; n=8–10, p>0.05. Further, the M54T mutant did not affect voltage dependence of HCN4 current. The midpoints of activation of the three groups: HCN4 alone, +WT, +M54T, were −69±4, −71±3, −68±3 mV (p>0.05 by ANOVA, n=6–10). There also was no statistically significant difference between the groups in the slope factors of the fractional activation curves (data not shown).
Effect of WT and M54T MiRP1 on biophysical properties of HCN2 currents expressed in NRVM
M54T did not alter HCN2 amplitude
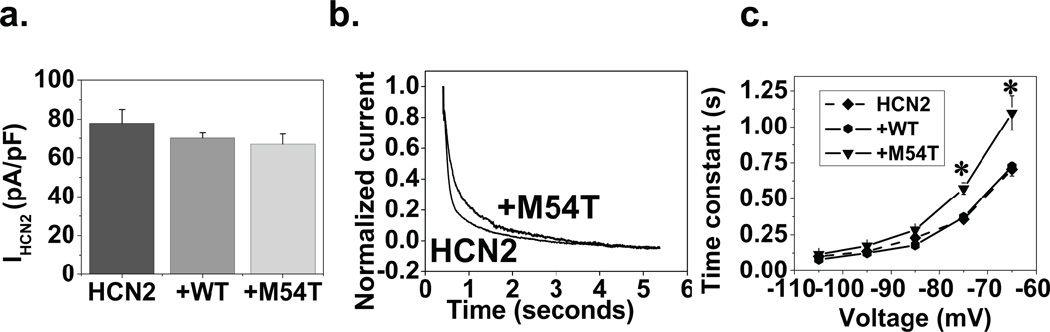
To determine if the effects of M54T were specific to the HCN4 isoform, we repeated these studies with hHCN2. In contrast to HCN4, when measuring HCN2 tail currents (at −100 mV), the M54T mutant (67±5 pA/pF) had no significant effect on HCN2 amplitude in comparison with HCN2 (78±7 pA/pF) or +WT (70±2 pA/pF), p>0.05 by ANOVA, n=6–10 (Fig 3a).
Figure 3. M54T affects kinetics but not current density of HCN2.
a. Current amplitude was determined from tail currents (at −100 mV). M54T did not alter HCN2 current density. There was no difference between the three groups (HCN2, +WT, +M54T), p>0.05 by ANOVA, n=6–10. b. Normalized current traces from the HCN2 and +M54T group at −75mV showing faster activation kinetics of HCN2 than the +M54T cell. This is represented in graph form in panel c. M54T also slows activation kinetics when compared to HCN2 or +WT group. M54T is significantly different from the other two groups at −65 mV and −75 mV(*).
M54T mutant slows activation kinetics of HCN2 current
Normalized current traces of sample cells of the HCN2 and +M54T group at −75 mV are presented in Fig 3b showing slower activation kinetics with the +M54T group. In Fig 3c, the HCN2 alone and +WT curves did not differ significantly; however, there was a statistically significant difference between the M54T curve in comparison with the former two groups (p<0.05 by ANOVA, n=6–10). For M54T, tau at −65 mV was 1190±126 ms compared with 705±46 ms for HCN2 or 725±29 for +WT; n=6–10, p<0.01. Thus, similar to what was observed for HCN4, the activation kinetics for HCN2 were slower when co-expressed with M54T mutant in comparison with HCN2 or +WT.
In addition to amplitude and activation kinetics, the other biophysical property we studied was voltage dependence. The mean V1/2 for the HCN2 was −64±3 mV versus +WT (−66±3 mV) or +M54T (−66±2 mV); no statistical difference was found between the three groups, n=6–10. The slope factor of the fractional activation curves did not differ significantly between the groups (data not shown).
Computer simulation of effect of M54T mutation on SN automaticity
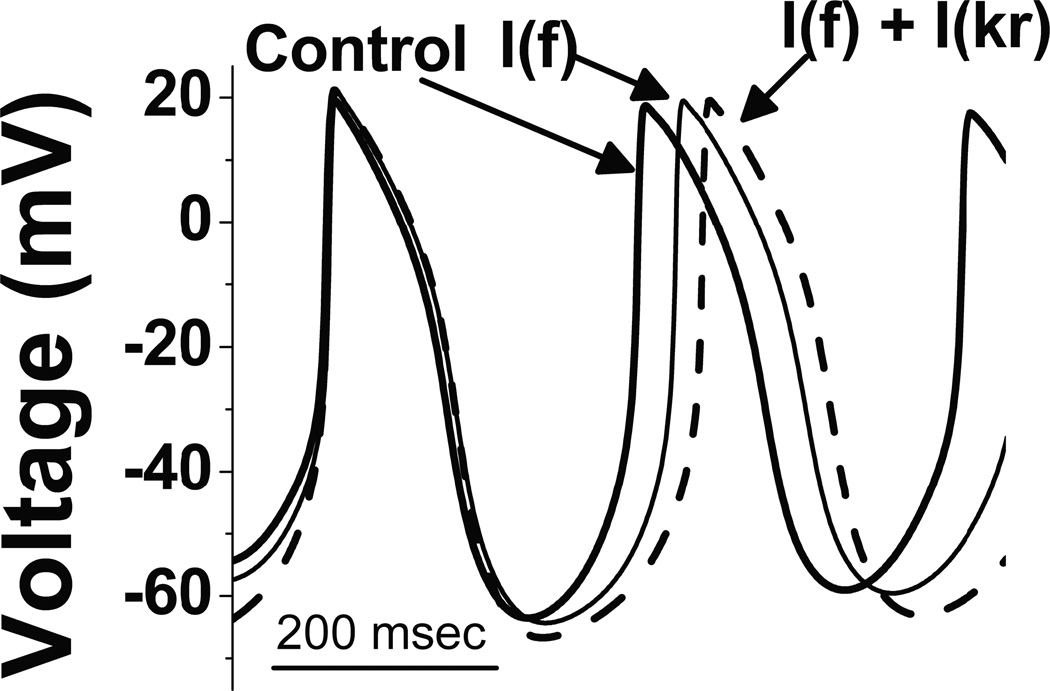
To investigate the possible impact of the change of amplitude and kinetics of HCN by the M54T mutant on spontaneous rate, we employed the Kurata computer model of SN. This model is based on the rabbit sinus node, the tissue with the most complete dataset since the rabbit sinus node is the prototypical experimental model for the study of sinus node channel function. As such, our goal was not to quantitatively replicate what occurs in a human subject, but rather to determine if our experimentally measured changes in pacemaker current could qualitatively slow rate. Simulated action potentials are shown in Fig 4. M54T mutation decreased the HCN4 amplitude by 80% and slowed activation kinetics. Using these HCN experimental data, the cycle length (CL) with M54T mutation was longer at 331 ms compared to control whose CL was 307 ms. The M54T mutant effect on Ikr increased the CL to 324 ms. This is secondary to M54T slowing the deactivation kinetics of Ikr when co-expressed with hERG in comparison with WTMiRP1 co-expressed with hERG26. Combining both the effects of If and Ikr the CL changed to 358 ms reflecting a greater slowing of rate than the individual effects of If or Ikr.
Figure 4. Computer simulations.
In the SN computer simulation model, the control cycle length was 307 ms (dark trace). M54T mutant, due to its effects on HCN4 amplitude (decrease) and activation kinetics (slowing), increases the cycle length (referring to slowing of the heart rate) from 307 ms to 331 ms (light trace; labeled I(f)). Combining the effects of M54T mutant on both If and Ikr the cycle length of the control changed from 307 ms (dark trace) to 358 ms (dashed trace; labeled I(f) + I(kr)). This translates into slowing of heart rate by 14% relative to control.
HCN4 sequence analysis from subject patient
The heterologous expression studies demonstrate that the M54T mutation of MiRP1 can result in altered HCN4 current, and the computer simulation suggests that when this is combined with the known effect of this mutation on hERG the result is a measureable slowing of rate. However, it is possible that there are other genetic factors in our subject that contribute to the marked bradycardia. The most obvious candidate would be a mutation in HCN4, since such mutations have been linked to familial sinus bradycardia22. We therefore sequenced the HCN4 gene in our subject. We identified three single-nucleotide polymorphisms (SNPs; Table 3), one of which leads to a missense (G36E) and two to synonymous mutations (L520L and P1200P), and all previously reported (http://www.ncbi.nlm.nih.gov/projects/SNP). Of the three SNPs found, only 107G>A, generating the missense mutation G36E, has a Minor Allele Frequency (MAF) just below 0.05, the value assumed to separate rare variants from common polymorphisms31. To our knowledge, no pathological effect of any of these SNPs is reported in the literature.
Table 3.
SNPs found in HCN4 sequence. Data extracted from http://www.ncbi.nlm.nih.gov/projects/SNP.
| HCN4 | Location | Nucleotide | a.a. | M.A.F. | dbSNP |
|---|---|---|---|---|---|
| Exon 1 | 107G>A | G36E | 0.0435 | rs143090627 | |
| Exon 4 | 1558 C>T | L520L | 0.0536 | rs12909882 | |
| Exon 8 | 3600A>G | P1200P | 0.1310 | rs529004 |
Clinical Implication
We had identified our index patient, a 55-year-old Caucasian male with LQTS6. The patient’s daughter died of sudden death at age 13. Subsequent postmortem genetic testing revealed the M54T mutation. She did not have any ECG prior to her death. The mother’s ECG was normal and she tested negative for the mutation. Our patient who tested positive for the M54T mutation had resting bradycardia in the 30s from a young age. Holter monitoring showed an average heart rate of 43 bpm, with lowest heart rate of 30 bpm and highest rate of 125 bpm along with 17 pauses. The underlying rhythm was sinus bradycardia. Our results are consistent with the bradycardia arising, at least in part, from the effect of the M54T mutation on If and Ikr currents contributing to SN automaticity.
DISCUSSION
Long-QT syndrome type 6 is a rare type of LQTS. The majority of the patients have LQTS mutations in the first three genes (KVLQT1, hERG and SCN5A). Nonetheless, MiRP1 protein has been found in the SN and interacts with the pacemaker channels, functioning as a β subunit. The mutations in this gene affecting the pacemaker channel function are interesting as these modulate HCN properties different than the WT. Most SN studies in LQTS patients have been done in patients with LQTS 1–332,33. We focused on LQTS6 mutation M54T and its effect on biophysical properties of HCN isoforms, providing an underlying mechanism for sinus bradycardia seen in our index patient. None of the previous studies have explored pacemaker current as a cause of sinus bradycardia in LQTS.
We over-expressed HCN channels in NRVM to investigate the biophysical properties of HCN currents alone, when co-expressed with WT, and in comparison with co-expression of HCN and the M54T mutant. We used this preparation to provide a myocardial context, since we previously demonstrated that heterologous HCN currents differ biophysically when expressed in myocytes versus non-myocytes29. However, we did not use these automatic cells to mimic SN function, since there are clear differences in factors contributing to automaticity in NRVM and SN, such as the contribution of a Na current and absence of Ca-stimulated adenylyl cyclase isoforms in NRVM34,35. These rat cells do, however, express endogenous HCN and MiRP118 that could interact with the exogenous human species we are expressing. However, we previously demonstrated that the exogenous HCN2 and MiRP1 proteins are expressed in these cells at levels far above those of endogenous proteins18. Thus, the endogenous rMiRP1 cannot saturate the high levels of expressed hHCN4 in these experiments. Our results show that: (1) M54T slows activation kinetics of HCN2 and HCN4 at physiologically relevant diastolic potentials, but does not alter HCN4 deactivation kinetics (2) M54T markedly decreases HCN4 but not HCN2 current amplitude (3) Voltage dependence of neither HCN2 nor HCN4 was altered by M54T.
These findings were used in generating a SN computer simulation25,36. The simulation predicts that the observed changes in HCN4 current slow rate 7%, that the published effects of the M54T mutation on hERG kinetics26 separately slows rate 5%, and the combined effects of M54T mutant on If and Ikr current together slow rate by 14%. It should be remembered that the contribution of If to SN pacemaking, and therefore the magnitude of effect caused by M54T, varies with the specific computer simulation employed25,37. Therefore, the quantitative result is less important than the qualitative one, namely that the experimental effects of the M54T mutation on HCN and hERG are additive in terms of impact on SN automaticity. It also should be noted that MiRP1 is reported to affect additional K currents10–14 that could further impact automaticity, but it is not known if these actions are affected by the M54T mutation.
This study investigated if SN dysfunction in LQTS6 could be explained based on an effect of the LQTS6 M54T mutation on the biophysical properties of the predominant HCN isoform in the SN namely HCN4, with HCN2 studied for comparison. Previous studies17,18,38 found effects of MiRP1 on HCN1, HCN2 and HCN4. However, our study does not show an effect of WT on HCN2 or HCN4 in any of the studied biophysical properties. The differences in our results of hMiRP1 not accelerating kinetics and increasing amplitude of hHCN2 and hHCN4 could be explained by the difference in the species specificity11. Rat and human MiRP1 show 82% identity and 97% homology11. Human HCN2 sequence has 90% identity to mouse HCN2. The cell line employed also may affect outcome. Brandt et al38 used hMiRP1 with mouse HCN2 and hHCN4 with hMiRP1 in CHO cells. We conducted all our studies with hHCN and hMiRP1 in NRVM. However, it also is possible that the NRVM cellular environment differs from that of SN myocytes in important ways that impact HCN-MiRP interaction and function. However, we did observe clear differences between the effect of M54T and WT hMiRP1 on HCN current, and interestingly these differences were HCN-isoform specific.
A limitation of this study is that the bradycardia in our patient may only partly relate to the M54T mutation and pacemaker current. It is certainly possible that this particular patient has other background characteristics that combine with M54T to result in the marked bradycardia, although we eliminated the most obvious candidate by sequencing the HCN4 gene. Importantly, M54T does result in a measureable change in HCN4 function and thus is a plausible explanation for the bradycardia. The Kurata computer simulation predicts the altered current will slow SN rate, and that this effect is additive with that of MiRP1-M54T on hERG current. However, the bradycardia in the computer simulation is modest, suggesting there may be effects of MiRP1-M54T on other currents in SN and/or limitations of the existing rabbit based SN computer model. Additional studies will be required to establish a causal relation between the effects of M54T on SN currents and the marked bradycardia observed in this patient.
Bradycardia remains an important risk factor for sudden cardiac events in patients with LQTS. A recent Scientific Statement from the American Heart Association (AHA)39 recommends that hospital patients should receive QT interval monitoring if certain conditions are present: sinus bradycardia is one of those conditions. This study is the first to explore the cause of this risk factor in LQTS6 at a cellular level as an effect on pacemaker current. The functional effects of these mutants on pacemaker channel could translate into subtle clinical differences in SN function that may be otherwise missed in the LQTS6 group. However, we believe there are broader implications for other mutations related to LQT or other arrhythmias where the mutation lies not in a channel but in an interacting subunit, because such subunits often interact with multiple ion channels and therefore can have secondary effects. Comparable secondary effects could occur in other LQT subtypes, namely type 4 and 9 where ankyrin and caveolin 3 subunits also interact with HCN channels. The data suggest that as arrhythmia predisposition genotyping becomes more common, asymptomatic individuals with potentially pathological MiRP1 variants in such pedigrees should be aware of important associated abnormalities like sinus bradycardia. Further, there are other LQTS types that affect non-channel proteins and which may impact the HCN or other SN currents; these also may merit exploration in terms of SN function.
Acknowledgments
Supported in part by the American Heart Association (Clancy), research grants HL-33843/HL-51618 (Moss) and HL-28958 (Robinson) from the National Institutes of Health, and Columbia University Department of Pediatrics.
List of abbreviations
- SN
Sinus node
- LQTS
Long QT syndrome
- HCN channel
Hyperpolarization-activated, cyclic nucleotide gated channel
- hERG channel
Human ether-go-go channel
- MiRP1
MinK-related peptide 1
- QTc
Corrected QT Interval
Footnotes
No disclosures.
References
- 1.Beinder E, Grancay T, Menendez T, Singer H, Hofbeck M. Fetal sinus bradycardia and the long QT syndrome. Am J Obstet Gynecol. 2001;185:743–747. doi: 10.1067/mob.2001.117973. [DOI] [PubMed] [Google Scholar]
- 2.Hofbeck M, Ulmer H, Beinder E, Sieber E, Singer H. Prenatal findings in patients with prolonged QT interval in the neonatal period. Heart. 1997;77:198–204. doi: 10.1136/hrt.77.3.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz PJ, Periti M, Malliani A. The long Q-T syndrome. Am Heart J. 1975;89:378–390. doi: 10.1016/0002-8703(75)90089-7. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz PJ. Idiopathic long QT syndrome: progress and questions. Am Heart J. 1985;109:399–411. doi: 10.1016/0002-8703(85)90626-x. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz PJ, Moss AJ, Vincent GM, Crampton RS. Diagnostic criteria for the long QT syndrome. An update. Circulation. 1993;88:782–784. doi: 10.1161/01.cir.88.2.782. [DOI] [PubMed] [Google Scholar]
- 6.Zareba W, Moss AJ, le Cessie S, Locati EH, Robinson JL, Hall WJ, Andrews ML. Risk of cardiac events in family members of patients with long QT syndrome. J Am Coll Cardiol. 1995;26:1685–1691. doi: 10.1016/0735-1097(95)60383-2. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz PJ, Stramba-Badiale M, Crotti L, Pedrazzini M, Besana A, Bosi G, Gabbarini F, Goulene K, Insolia R, Mannarino S, Mosca F, Nespoli L, Rimini A, Rosati E, Salice P, Spazzolini C. Prevalence of the congenital long-QT syndrome. Circulation. 2009;120:1761–1767. doi: 10.1161/CIRCULATIONAHA.109.863209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Splawski I, Tristani-Firouzi M, Lehmann MH, Sanguinetti MC, Keating MT. Mutations in the hminK gene cause long QT syndrome and suppress IKs function. Nat Genet. 1997;17:338–340. doi: 10.1038/ng1197-338. [DOI] [PubMed] [Google Scholar]
- 9.Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, Shen J, Timothy KW, Vincent GM, de Jager T, Schwartz PJ, Toubin JA, Moss AJ, Atkinson DL, Landes GM, Connors TD, Keating MT. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- 11.Abbott GW, Sesti F, Splawski I, Buck ME, Lehmann MH, Timothy KW, Keating MT, Goldstein SA. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell. 1999;97:175–187. doi: 10.1016/s0092-8674(00)80728-x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang M, Jiang M, Tseng GN. minK-related peptide 1 associates with Kv4.2 and modulates its gating function: potential role as beta subunit of cardiac transient outward channel? Circ Res. 2001;88:1012–1019. doi: 10.1161/hh1001.090839. [DOI] [PubMed] [Google Scholar]
- 13.Tinel N, Diochot S, Borsotto M, Lazdunski M, Barhanin J. KCNE2 confers background current characteristics to the cardiac KCNQ1 potassium channel. Embo J. 2000;19:6326–6330. doi: 10.1093/emboj/19.23.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang M, Xu X, Wang Y, Toyoda F, Liu XS, Zhang M, Robinson RB, Tseng GN. Dynamic partnership between KCNQ1 and KCNE1 and influence on cardiac IKs current amplitude by KCNE2. J Biol Chem. 2009;284:16452–16462. doi: 10.1074/jbc.M808262200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roepke TK, Kontogeorgis A, Ovanez C, Xu X, Young JB, Purtell K, Goldstein PA, Christini DJ, Peters NS, Akar FG, Gutstein DE, Lerner DJ, Abbott GW. Targeted deletion of kcne2 impairs ventricular repolarization via disruption of I(K,slow1) and I(to,f) Faseb J. 2008;22:3648–3660. doi: 10.1096/fj.08-110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H, Wu J, Potapova I, Wymore RT, Holmes B, Zuckerman J, Pan Z, Wang H, Shi W, Robinson RB, El-Maghrabi MR, Benjamin W, Dixon J, McKinnon D, Cohen IS, Wymore R. MinK-related peptide 1: A beta subunit for the HCN ion channel subunit family enhances expression and speeds activation. Circ Res. 2001;88:E84–E87. doi: 10.1161/hh1201.093511. [DOI] [PubMed] [Google Scholar]
- 17.Decher N, Bundis F, Vajna R, Steinmeyer K. KCNE2 modulates current amplitudes and activation kinetics of HCN4: influence of KCNE family members on HCN4 currents. Pflugers Arch. 2003;446:633–640. doi: 10.1007/s00424-003-1127-7. [DOI] [PubMed] [Google Scholar]
- 18.Qu J, Kryukova Y, Potapova IA, Doronin SV, Larsen M, Krishnamurthy G, Cohen IS, Robinson RB. MiRP1 modulates HCN2 channel expression and gating in cardiac myocytes. J Biol Chem. 2004;279:43497–43502. doi: 10.1074/jbc.M405018200. [DOI] [PubMed] [Google Scholar]
- 19.Shi W, Wymore R, Yu H, Wu J, Wymore RT, Pan Z, Robinson RB, Dixon JE, McKinnon D, Cohen IS. Distribution and prevalence of hyperpolarization-activated cation channel (HCN) mRNA expression in cardiac tissues. Circ Res. 1999;85:e1–e6. doi: 10.1161/01.res.85.1.e1. [DOI] [PubMed] [Google Scholar]
- 20.DiFrancesco D. Pacemaker mechanisms in cardiac tissue. Annu Rev Physiol. 1993;55:455–472. doi: 10.1146/annurev.ph.55.030193.002323. [DOI] [PubMed] [Google Scholar]
- 21.Accili EA, Proenza C, Baruscotti M, DiFrancesco D. From funny current to HCN channels: 20 years of excitation. News Physiol Sci. 2002;17:32–37. doi: 10.1152/physiologyonline.2002.17.1.32. [DOI] [PubMed] [Google Scholar]
- 22.Milanesi R, Baruscotti M, Gnecchi-Ruscone T, DiFrancesco D. Familial sinus bradycardia associated with a mutation in the cardiac pacemaker channel. N Engl J Med. 2006;354:151–157. doi: 10.1056/NEJMoa052475. [DOI] [PubMed] [Google Scholar]
- 23.Moss AJ, Schwartz PJ. 25th anniversary of the International Long-QT Syndrome Registry: an ongoing quest to uncover the secrets of long-QT syndrome. Circulation. 2005;111:1199–1201. doi: 10.1161/01.CIR.0000157069.91834.DA. [DOI] [PubMed] [Google Scholar]
- 24.Protas L, Robinson RB. Neuropeptide Y contributes to innervation-dependent increase in I(Ca, L) via ventricular Y2 receptors. Am J Physiol. 1999;277:H940–H946. doi: 10.1152/ajpheart.1999.277.3.H940. [DOI] [PubMed] [Google Scholar]
- 25.Kurata Y, Hisatome I, Imanishi S, Shibamoto T. Dynamical description of sinoatrial node pacemaking: improved mathematical model for primary pacemaker cell. Am J Physiol Heart Circ Physiol. 2002;283:H2074–H2101. doi: 10.1152/ajpheart.00900.2001. [DOI] [PubMed] [Google Scholar]
- 26.Lu Y, Mahaut-Smith MP, Huang CL, Vandenberg JI. Mutant MiRP1 subunits modulate HERG K+ channel gating: a mechanism for pro-arrhythmia in long QT syndrome type 6. J Physiol. 2003;551:253–262. doi: 10.1113/jphysiol.2003.046045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demir SS, Clark JW, Murphey CR, Giles WR. A mathematical model of a rabbit sinoatrial node cell. The American journal of physiology. 1994;266:C832–C852. doi: 10.1152/ajpcell.1994.266.3.C832. [DOI] [PubMed] [Google Scholar]
- 28.Dokos S, Celler B, Lovell N. Ion currents underlying sinoatrial node pacemaker activity: a new single cell mathematical model. J Theor Biol. 1996;181:245–272. doi: 10.1006/jtbi.1996.0129. [DOI] [PubMed] [Google Scholar]
- 29.Qu J, Altomare C, Bucchi A, DiFrancesco D, Robinson RB. Functional comparison of HCN isoforms expressed in ventricular and HEK 293 cells. Pflugers Arch. 2002;444:597–601. doi: 10.1007/s00424-002-0860-7. [DOI] [PubMed] [Google Scholar]
- 30.Qu J, Barbuti A, Protas L, Santoro B, Cohen IS, Robinson RB. HCN2 overexpression in newborn and adult ventricular myocytes: distinct effects on gating and excitability. Circ Res. 2001;89:E8–E14. doi: 10.1161/hh1301.094395. [DOI] [PubMed] [Google Scholar]
- 31.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swan H, Viitasalo M, Piippo K, Laitinen P, Kontula K, Toivonen L. Sinus node function and ventricular repolarization during exercise stress test in long QT syndrome patients with KvLQT1 and HERG potassium channel defects. J Am Coll Cardiol. 1999;34:823–829. doi: 10.1016/s0735-1097(99)00255-7. [DOI] [PubMed] [Google Scholar]
- 33.Lei M, Huang CL, Zhang Y. Genetic Na+ channelopathies and sinus node dysfunction. Prog Biophys Mol Biol. 2008;98:171–178. doi: 10.1016/j.pbiomolbio.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Robinson RB, Legato MJ. Maintained differentiation in rat cardiac monolayer cultures: tetrodotoxin sensitivity and ultrastructure. Journal of molecular and cellular cardiology. 1980;12:493–498. doi: 10.1016/0022-2828(80)90005-x. [DOI] [PubMed] [Google Scholar]
- 35.Kryukova YN, Protas L, Robinson RB. Ca2+-activated adenylyl cyclase 1 introduces Ca2+-dependence to beta-adrenergic stimulation of HCN2 current. Journal of molecular and cellular cardiology. 2012;52:1233–1239. doi: 10.1016/j.yjmcc.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao X, Bucchi A, Oren RV, Kryukova Y, Dun W, Clancy CE, Robinson RB. In vitro characterization of HCN channel kinetics and frequency dependence in myocytes predicts biological pacemaker functionality. J Physiol. 2009;587:1513–1525. doi: 10.1113/jphysiol.2008.163444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Severi S, Fantini M, Charawi LA, DiFrancesco D. An updated computational model of rabbit sinoatrial action potential to investigate the mechanisms of heart rate modulation. J Physiol. 2012;590:4483–4499. doi: 10.1113/jphysiol.2012.229435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandt MC, Endres-Becker J, Zagidullin N, Motloch LJ, Er F, Rottlaender D, Michels G, Herzig S, Hoppe UC. Effects of KCNE2 on HCN isoforms: distinct modulation of membrane expression and single channel properties. Am J Physiol Heart Circ Physiol. 2009;297:H355–H363. doi: 10.1152/ajpheart.00154.2009. [DOI] [PubMed] [Google Scholar]
- 39.Drew BJ, Ackerman MJ, Funk M, Gibler WB, Kligfield P, Menon V, Philippides GJ, Roden DM, Zareba W. Prevention of torsade de pointes in hospital settings: a scientific statement from the American Heart Association and the American College of Cardiology Foundation. Circulation. 2010;121:1047–1060. doi: 10.1161/CIRCULATIONAHA.109.192704. [DOI] [PMC free article] [PubMed] [Google Scholar]