Abstract

We investigated the influence of the protruding domain of Norwalk virus-like particles (NVLP) on its overall structural and mechanical stability. Deletion of the protruding domain yields smooth mutant particles and our AFM nanoindentation measurements show a surprisingly altered indentation response of these particles. Notably, the brittle behavior of the NVLP as compared to the plastic behavior of the mutant reveals that the protruding domain drastically changes the capsid’s material properties. We conclude that the protruding domain introduces prestress, thereby increasing the stiffness of the NVLP and effectively stabilizing the viral nanoparticles. Our results exemplify the variety of methods that nature has explored to improve the mechanical properties of viral capsids, which in turn provides new insights for developing rationally designed, self-assembled nanodevices.
Keywords: AFM, nanoindentation, native mass spectrometry, viral nanoparticles, Noro virus, mechanical properties
Viruses are increasingly being used as functional nanoparticles in medicine and nanotechnology. Applications include the use of viruses as nanoreactors for material synthesis,1 as building blocks in self-assembly and patterning approaches,2,3 as nano-containers for drug delivery, and as platforms for imaging and tumor targeting.4 However, acquiring a thorough knowledge of their physical properties is still in development, and in order to fully exploit the vast potential that viral nanoparticles offer it is essential to elucidate these properties.5 Recently developed atomic force microscopy (AFM) nanoindentation experiments are starting to shed light on the material properties of single viral particles, including their Young’s modulus, breaking force, and resistance to material fatigue.6,7 These characteristics differ significantly between viruses. For instance, their elastic properties range from being as flexible as rubber to as hard as plexiglass. Until now, only viruses with a relatively smooth outer surface have been probed, which generally follow the predictions of thin shell elastic continuum theory.7 However, it is unclear how a heterogeneous surface morphology of the viral particles affects their overall material properties. To study these effects, we have looked at the mechanical properties of the human pathogenic Norwalk virus, which exhibits uneven surface features with distinct hollows and protrusions. Knowledge on its mechanical properties will help us better understand general design principles of icosahedral viruses and will shed light on the binding mechanism of the virus to its host cells. The former will support the rational design of self-assembled nanoparticles whereas the latter will help in developing drugs targeted at disabling or blocking the receptor binding step of the infection cycle.
Norwalk virus is a nonenveloped, icosahedral virus. Its 7654-nucleotide single-stranded RNA genome encodes for various proteins including the major capsid protein VP1 and the minor protein VP2.8,9 Expressing the capsid proteins in insect cells with a recombinant baculovirus yields self-assembled Norwalk virus-like particles (NVLP) with similar morphology and antigenicity as the native Norwalk virus.10–12 VP1 folds into two distinct domains of the capsid: the S (shell) domain forms the smooth icosahedral shell while the P (protruding) domain forms protrusions that radially project outward from the shell, creating cup-like depressions at the 3- and 5-fold symmetry axes. The NVLPs are composed of 180 copies of VP1 with a diameter of 38 nm, exhibiting T = 3 icosahedral symmetry.13 VP2 is present in very small amounts: probably only a few molecules per capsid. Although it is suspected to be internal in the capsid layer, its precise location is still uncertain and it is not seen as part of the capsid in the X-ray crystallographic structure.13,14 The structural requirements for assembly of the Norwalk virus capsid were investigated by mutational analyses on VP1. Complete deletion of the protruding domain leads to the assembly of mutated particles (CT303) which have smooth T = 3 icosahedral shells with a diameter of ~27 nm.12 The structure of the CT303 particles resembles the NVLP without protrusion (see Supporting Information Figure S1) indicating that the shell domain alone contains the determinants required for assembly of the icosahedral capsid. Yet, the icosahedral shell in the CT303 particles is slightly smaller than the contiguous shell made by the shell domain in the native NVLPs. The protruding domain, which contributes to the overall diameter of the particles, is implicated in mediating host cell attachment. However, the effects of the P domain on particle stability have remained unclear.
We investigated the role of the P domain on the structural stability of the capsid by comparing the mechanical properties of the particles formed from the wild type protein and the CT303 truncation mutants. This was done by nanoindentation experiments using AFM.7 Our results suggest that the protruding domain forces the Norwalk virus shell in a state of prestress. Prestress is a common feature in biological structures which can govern the mechanical structure including their stability and integrity. For instance, cells and in vitro actin networks that are under prestress exhibit an increased stiffness.15,16 Furthermore, modeling of spherelike viruses shows that prestress is an important component in the structure of self-assembled icosahedral protein shells.17,18 In addition, it was recently observed that elongated bacteriophages are under an anisotropic prestress.19 The current study indicates that the icosahedral NVLPs are under an isotropic prestress, increasing the radius of the shell domain and resulting in an overall strengthening of the viral particle.
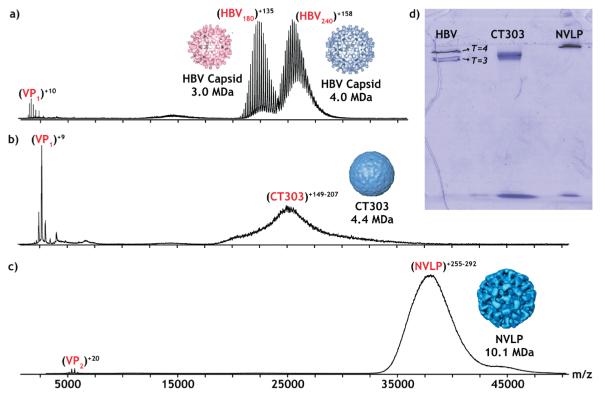
The integrity of NVLP and CT303 particles was first characterized by native mass spectrometry, blue native polyacrylamide gel electrophoresis (BN-PAGE), and AFM imaging (see Supporting Information for Materials and Methods).20–22 Figure 1 shows native mass spectra of Hepatitis B virus (HBV),23 CT303, and NVLP.24 Because the charge state of HBV is well characterized we use its spectra to support our mass estimations of NVLP and CT303. The mass of the Norwalk VP1 (MW = 56077 Da) and CT303 monomer proteins (MW = 24286 Da) was established using this mass spectrometry data. In addition, we estimate that the molecular mass of the intact NVLP and CT303 capsids is 10.1 and 4.4 MDa, respectively, assuming 180 capsid protein subunits. Consistent with this data, analysis of HBV, CT303, and NVLP by native gel electrophoresis revealed that the intact NVLP particles migrated less far than the intact CT303 particles. The CT303 particles moved slightly further than expected when compared to the similarly sized HBV T = 4 capsids (4 MDa),23 which is probably related to its very smooth outer surface. Furthermore, it was shown that the CT303 particles were only stable under a very narrow pH range (5–7) and not above pH 7, whereas in the NVLP sample, there were still particles present at higher pH (8–9).24 VP2 was neither detected by mass spectrometry nor in SDS-PAGE gels.24 Therefore it remains unclear what its stoichiometry and function in capsid assembly is. Next, the immobilized particles were imaged in buffer solution using AFM.22 Figure 2 shows AFM images of NVLP and CT303 particles together with their respective height profiles. The height distribution in Supporting Information Figure S2 shows that the NVLPs have an average diameter of 38.3 ± 0.1 nm, comparable to the size of the particle in the atomic model.13 The size distribution of the CT303 particles shows a uniform distribution with an average diameter of 27.9 ± 0.2 nm. This value is comparable to the size determined from cryoelectron microscopic reconstructions.12
Figure 1.
Representative native nanoelectrospray mass spectra for (a) HBV (0.04 μM capsid concentration) in 200 mM ammonium acetate buffer (pH 6.8), serving as comparison for the other spectra, (b) CT303 (0.1 μM capsid concentration) in 250 mM ammonium acetate buffer (pH 7), and (c) NVLPs (0.2 μM capsid concentration) in 250 mM ammonium acetate buffer (pH 7). (d) Native gel: for each capsid, 25 μg was loaded onto a 4% (w/v) gel where they were analyzed by BN-PAGE to further confirm their integrity. The bands at the bottom of the gel represent monomers/dimers.
Figure 2.
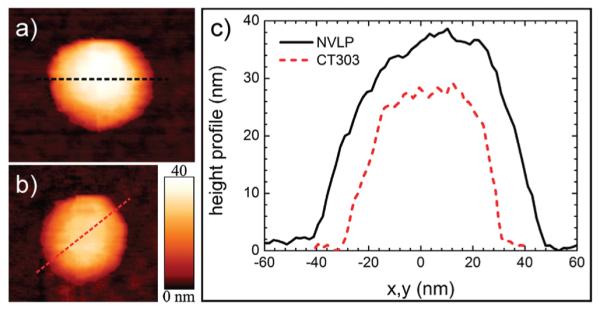
AFM images and height profiles of (a) NVLP and (b) CT303 particles before nanoindentation. The height profiles in (c) are used to estimate the diameter of the particles, yielding 39 and 29 nm for the NVLP and CT303 mutant particles, respectively, in this example.
After imaging, nanoindentation experiments were performed.7 Five successive indentation and retraction curves were recorded for each particle and Figure 3 shows typical force indentation curves for NVLP and CT303 particles. The spring—constant is determined from the linear part of the slope during the first indentation. Its distribution is depicted in Supporting Information Figure S3, showing that NVLPs have a much higher spring constant than the CT303 particles: 0.30 ± 0.01 N/m versus 0.111 ± 0.004 N/m (84 NVLP and 92 CT303 particles were tested). The distribution of spring constants for the NVLPs is broader, likely reflecting the heterogeneity of its surface as compared to the smooth CT303 particles. Linear elasticity theory describes the 3D Young’s modulus E for a thin shell by E = αRk/t2, where k is the spring constant, R is the radius, and t is the thickness of the shell.25 For the proportionality factor α, a value of 1 can be assumed.6,26,27 Whereas technically speaking the CT303 particles cannot be called thin shelled particles (t << R), it has previously been shown that this equation still holds for viral particles with thick shells.23,27,28 Therefore, we apply this theory to get an estimate of E for the homogeneous CT303 particles. These particles are modeled as shells with a thickness of 2.7 nm and effective radius of 12.2 nm, (taken from the middle between the inner and outer radii). Inserting these numbers into the thin shell equation gives us an estimated 3D Young’s modulus of ~0.2 GPa for the CT303 particles, which is comparable to HBV and CCMV,7 two viruses that self-assemble around their genome as Norwalk virus does.
Figure 3.
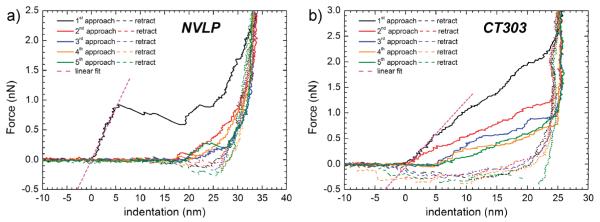
Typical force—indentation curves of (a) NVLP and (b) CT303 particles for five successive nanoindentations. Initially, the force sensed by the AFM tip is zero until it touches the particle’s top surface (indicated by 0 nm in the indentation axis). As the tip pushes further onto the particle, the force gradually rises in a linear fashion from which the particle’s spring constant can be derived. The structural strengths of the particles were tested by indenting further beyond the linear regime leading to nonlinear deformation including plasticity, buckling, or breakage.
Both sets of indentation curves in Figure 3 show an almost vertical rise of the force on the right end of the plots, indicating that the AFM tip has reached a point where the particle cannot be squeezed any further. The CT303 particle has a diameter of about 28 nm thus, the vertical rise of the force at around 24 nm indicates an incompressible protein layer of ~4 nm (Figure 3b). Since the wall thickness of the particle is ~2.7 nm, this indicates that the incompressible layer refers to the two opposite surfaces of the particle being squeezed together and partially deformed. On the contrary, the measured incompressible layer in Figure 3a of about 5 nm leads to the conclusion that it is most likely not a double but only a single surface layer of the NVLP particle (which has a wall thickness of ~7.5 nm), indicating that the top surface has been torn or broken apart. Cuellar et al.29 studied the material properties of NVLPs as a function of pH, showing an increase in compliance at alkaline conditions. Whereas this study is valuable to compare the influence of pH on the relative mechanical stability of NVLPs, it seems difficult to compare the absolute values with our results. In their study the particles are heavily deformed during imaging (up to ~20% decrease in height).29 This could lead to premature damage, before the start of the nanoindentation experiments, explaining the higher spring constant we find.
Figure 3a also shows that NVLPs typically buckle and break during the first indentation as demonstrated by a sharp drop in the measured force after reaching a critical point. The succeeding indentation curves (second—fifth) show no deformation for the first ~18 nm, indicating that structural failure already occurred during the first indentation. This is a common behavior also observed for other icosahedral capsid particles like CCMV,27 Phage λ,30,31 and HSV-1.32 Breakage of NVLPs occurs around a relative deformation of ~22% of the radius, which is comparable to other studied capsids.7 The critical force at which nonlinear deformation occurs during nanoindentation of the NVLPs is 1.1 ± 0.1 nN, whereas for the CT303 particle, it is 0.53 ± 0.03 nN. The indentation at the critical force is similar for both particles at 4.1 ± 0.4 nm and 4.9 ± 0.3 nm, respectively. In contrast to the NVLP, the CT303 particle does not exhibit buckling behavior but instead shows a nonlinear continuous deformation. The particle bounces back to a spherelike shape after being fully compressed, as can be seen in Figure 3b by observing the second approach curve which rises at the same position as the first. However, the slope of the second curve is significantly lower, indicating that despite the full recovery of the particle’s height it did suffer material damage, affecting its spring constant. The successive force—indentation curves show that the particle’s height did decrease after several indentations, as the curves start to rise later. Furthermore, the change in the slope of the linear part of the third to fifth approach curves also shows that the decrease in spring constant continued. Comparing the indentation behavior of all CT303 particles indicates that they directly show a decrease in spring constant after the first indentation, while at the same time only ~75% of the particles show a decrease in height. The other 25% show a decrease in height only after the second indentation. The decrease in diameter and elasticity after several indentations implies that although the particles do not clearly break, they do undergo plastic deformation, as previously observed for HBV.23,28,33 However, HBV deforms plastically above an indentation of ~60% of its radius, whereas the CT303 particles already start to deform nonlinearly above ~36%. Furthermore, the nonlinear deformation of the CT303 particle occurs above an indenting force of ~0.5 nN, while for the HBV T = 3 and T = 4 particles this is ~1.0 nN.23 Moreover, the CT303 particles have a 30% lower height after five indentations. HBV capsids decrease only 10% in height under similar conditions.33 These observations indicate that the CT303 particles are quite prone to plastic deformation in comparison to the HBV capsids.
To further investigate the observed plastic deformation and structural failure of the Norwalk viral shells we use an analysis involving the Föppl-von-Karman number (FvK) γ.28,34 This dimensionless number emerges from linear continuum elasticity theory and is defined by
| (1) |
that is, the ratio of the two-dimensional stretching elasticity Y and bending stiffness κ of homogeneous, isotropic shells of radius R. Even though the underlying FvK theory assumes completely reversible behavior, it has been shown that there is a remarkable similarity between simulated indentation curves using finite element methods and experimental curves that show plastic deformation or failure. Both the experimental and the simulated curves depend on the icosahedral orientation along which the particle is deformed.28,35 However, the global characteristics of the curves are similar for indentation along the 2-, 3- and 5-fold axis. For small γ (<~100), the simulated indentation curves28,34 show linear deformation. For intermediate γ (~100–1000), a nonlinear but continuous indentation behavior is observed. Finally, for large γ (>~1000), buckling events are predicted. Comparing the indentation curves of the NVLP and CT303 particles to simulated curves indicates that the CT303 particle is expected to have a γ of roughly 200 and the NVLP a γ of >1000. Using an approximation of the FvK number γ = 12(1 – ν2)*(R/t)2 (ref 28) with Poisson’s ratio ν and assuming that both particles are made up of material with the same mechanical properties, this implies that the NVLP should be a particle with a thinner shell than the mutant particle. As can be seen in Supporting Information Figure S1 this is the opposite of what is known about these particles, that is, the NVLP has a thicker shell than the CT303. Hence, the addition of the protruding domain affects the material properties of the whole particle in an unexpected way and thus the overall material properties of the shells of NVLP and CT303 are not the same. The measured spring constant of the NVLP is roughly 3 times larger than the spring constant of the CT303 mutant. Linear response theory predicts that this spring constant is proportional to (κY)1/2/R.34 Taking into account the difference in shell thickness, this yields that the product of the bending modulus κ and the 2D Young’s modulus Y should increase with a factor of roughly 10 when the protruding domain is present. Assuming that only Y is changed by the presence of the protruding domain, it indicates that the NVLP would have an FvK number of roughly 2000, which fits the shape of the corresponding experimental and simulated force—indentation curves. However, this raises the question why Y changed this much. In order to dissect the origin of this unexpected behavior, we examine the molecular details of the Norwalk virus capsids.
X-ray crystallography has shown that the dimeric contacts between protruding domains form distinct archlike structures that surround the hollows at the 5- and quasi 6-fold axis of the T = 3 capsid.13 The C-terminal residues of the P domain are coupled via hydrogen bond interactions to the S domain residues. These bonds, observed in the crystal structure of the NVLP, seem to control the size of the viral particle as removing these bonds induces a change in size.12 For instance, it was shown that the shell of the CT303 particles has a smaller radius than the shell domain of the NVLP particles. This surprising finding shows that the equilibrium shape of the shell without the protruding domain is different from its shape when the protruding domain is present. Apparently the bridgelike arches lock the whole shell in a specific shape, which is not the ground state of the shell domain. Therefore the shell domain in the NVLP is being pulled in an outward direction by the protruding domain (Figure 4). Conversely, the shell domain pulls the protruding domain in an inward direction, as the shell domain would prefer its smaller ground state. This tightens the bonds between the protruding domains of the capsid proteins, like wedges being pressed together. It yields a whole shell under continuous prestress and the ensuing balance of forces (Figure 4) seems to result in the observed drastically increased rigidity of the NVLP as compared to the CT303 mutant. This prestress puts the whole particle under tension making it more difficult to pull apart and therefore effectively increases its 2D Young’s modulus Y.
Figure 4.
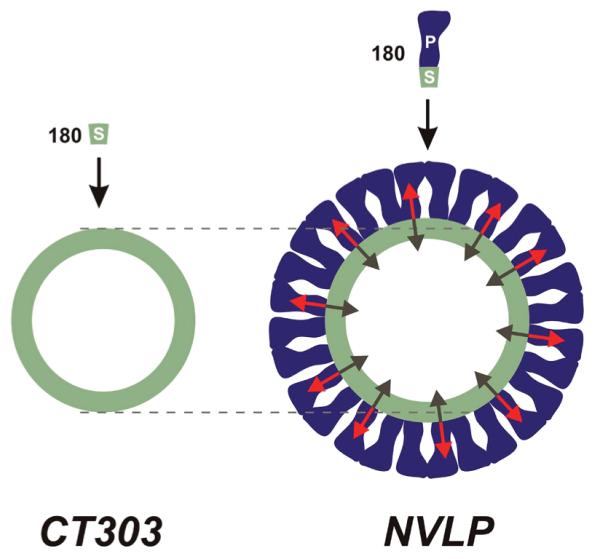
Balance of forces: Mechanical model of the CT303 mutant and NVLP. The red arrows in the NVLP model indicate the prestress acting on the shell domains (S) due to the presence of the protruding domains (P). The dark green arrows indicate the inward force of the shell domain acting on the protruding domain, thereby creating prestress in the latter. The gray dashed lines show the difference in size between the CT303 particles and the shell domain of the NVLP.
In summary, the heterogeneous, rough protrusions in Norwalk virus capsids have various functions. They are essential for successful interfacing of the viral nanoparticles with their host cells as they contain the appropriate binding sites, making them indispensable for infection.36 In this study, we demonstrate that they also have a stabilizing role. We conclude that the protruding domain acts through the generation of prestress in the capsid shell, thereby increasing the particle’s rigidity. The CT303 particles are stable at neutral pH, however, unlike the NVLPs, they completely fall apart by a slight increase in pH. This pH sensitivity would have disastrous effects for the wild-type Norwalk virus particles as the small intestine, in which cells it reproduces, is alkaline. The CT303 particle is characterized by tight and close packing of β-barrels and our results indicate that such a packing leads to a flexible and ductile material. Nanoindentation experiments on viruses with a comparable packing of β-sheets, like for instance, southern bean mosaic virus, will allow for testing of this hypothesis. The presented results on the mechanics of NVLP and CT303 particles seem to indicate that the protruding domain stabilizes the viral particles in an unexpected way by prestress. These novel insights provide handles for improving the material properties of engineered functional nanoparticles by introducing a uniform prestress, effectively strengthening these particles.
Supplementary Material
ACKNOWLEDGMENT
This work is supported by an STW-administered NanoSci-E+ grant and by the “Physics of the genome” program of the Fundamenteel Onderzoek der Materie (FOM) foundation, both to G.J.L.W. G.K.S. was supported by the Natural Sciences and Engineering Research Council through a postdoctoral fellowship grant. A.J.R.H. received support from The Netherlands Organization for Scientific Research ALW-ECHO (819.02.010). G.K.S., C.U., and A.J.R.H. were additionally supported by The Netherlands Proteomics Centre. S.E.C., M.K.E, and B.V.V.P. acknowledge support from NIH grant P01 AI057788, M.K.E. from P30 DK56338, and B.V.V.P. from Robert Welch foundation (Q1279).
Footnotes
Supporting Information. Materials and Methods and three figures: reconstructions, AFM particle size distribution and spring constant distribution of NVLP and CT303. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Douglas T, Strable E, Willits D, Aitouchen A, Libera M, Young M. Adv. Mater. 2002;14(6):415–418. [Google Scholar]
- (2).Lee SW, Mao CB, Flynn CE, Belcher AM. Science. 2002;296(5569):892–895. doi: 10.1126/science.1068054. [DOI] [PubMed] [Google Scholar]
- (3).Steinmetz NF, Bock E, Richter RP, Spatz JP, Lomonossoff GP, Evans DJ. Biomacromolecules. 2008;9(2):456–462. doi: 10.1021/bm700797b. [DOI] [PubMed] [Google Scholar]
- (4).Brunel FM, Lewis JD, Destito G, Steinmetz NF, Manchester M, Stuhlmann H, Dawson PE. Nano Lett. 2010;10(3):1093–1097. doi: 10.1021/nl1002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Mitragotri S, Lahann J. Nat. Mater. 2009;8(1):15–23. doi: 10.1038/nmat2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ivanovska IL, de Pablo PJ, Ibarra B, Sgalari G, MacKintosh FC, Carrascosa JL, Schmidt CF, Wuite GJL. Proc. Natl. Acad. Sci. U. S.A. 2004;101(20):7600–7605. doi: 10.1073/pnas.0308198101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Roos WH, Bruinsma R, Wuite GJL. Nat. Phys. 2010;6(10):733–743. [Google Scholar]
- (8).Hardy ME, Estes MK. Virus Genes. 1996;12(3):287–290. doi: 10.1007/BF00284649. [DOI] [PubMed] [Google Scholar]
- (9).Jiang X, Wang M, Wang KN, Estes MK. Virology. 1993;195(1):51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- (10).Green KY, Lew JF, Xi J, Kapikian AZ, Estes MK. J. Clinic. Microbiol. 1993;31(8):2185–2191. doi: 10.1128/jcm.31.8.2185-2191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Jiang X, Wang M, Graham DY, Estes MK. J. Virol. 1992;66(11):6527–32. doi: 10.1128/jvi.66.11.6527-6532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Bertolotti-Ciarlet A, White LJ, Chen R, Prasad BVV, Estes MK. J. Virol. 2002;76(8):4044–4055. doi: 10.1128/JVI.76.8.4044-4055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Prasad BVV, Hardy ME, Dokland T, Bella J, Rossmann MG, Estes MK. Science. 1999;286(5438):287–290. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- (14).Bertolotti-Ciarlet A, Crawford SE, Hutson AM, Estes MK. J. Virol. 2003;77(21):11603–11615. doi: 10.1128/JVI.77.21.11603-11615.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Wang N, Tolic-Norrelykke IM, Chen JX, Mijailovich SM, Butler JP, Fredberg JJ, Stamenovic D. Am. J. Physiol. 2002;282(3):C606–C616. doi: 10.1152/ajpcell.00269.2001. [DOI] [PubMed] [Google Scholar]
- (16).Gardel ML, Nakamura F, Hartwig JH, Crocker JC, Stossel TP, Weitz DA. Proc. Natl. Acad. Sci. U.S.A. 2006;103(6):1762–1767. doi: 10.1073/pnas.0504777103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Lidmar J, Mirny L, Nelson DR. Phys. Rev. E. 2003;68(5):051910. doi: 10.1103/PhysRevE.68.051910. [DOI] [PubMed] [Google Scholar]
- (18).Zandi R, Reguera D. Phys. Rev. E. 2005;72(2):021917. doi: 10.1103/PhysRevE.72.021917. [DOI] [PubMed] [Google Scholar]
- (19).Carrasco C, Luque A, Hernando-Perez M, Miranda R, Carrascosa JL, Serena PA, de Ridder M, Raman A, Gomez-Herrero J, Schaap IAT, Reguera D, de Pablo PJ. Biophys. J. 2011;100(4):1100–1108. doi: 10.1016/j.bpj.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Heck AJR. Nat. Methods. 2008;5(11):927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- (21).Uetrecht C, Heck AJR. Angew. Chem., Int. Ed. 2011;50(36):8248–8262. doi: 10.1002/anie.201008120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Baclayon M, Wuite GJL, Roos WH. Soft Matter. 2010;6(21):5273–5285. [Google Scholar]
- (23).Uetrecht C, Versluis C, Watts NR, Roos WH, Wuite GJL, Wingfield PT, Steven AC, Heck AJR. Proc. Natl. Acad. Sci. U.S.A. 2008;105(27):9216–9220. doi: 10.1073/pnas.0800406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Shoemaker GK, van Duijn E, Crawford SE, Uetrecht C, Baclayon M, Roos WH, Wuite GJL, Estes MK, Prasad BVV, Heck AJR. Mol. Cell. Proteom. 2010;9(8):1742–1751. doi: 10.1074/mcp.M900620-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Landau LD, Lifshitz EM. Theory of Elasticity. 3rd ed. Elsevier Butterworth-Heinemann; Oxford: 1986. [Google Scholar]
- (26).Gibbons MM, Klug WS. Phys. Rev. E. 2007;75(3):031901. doi: 10.1103/PhysRevE.75.031901. [DOI] [PubMed] [Google Scholar]
- (27).Michel JP, Ivanovska IL, Gibbons MM, Klug WS, Knobler CM, Wuite GJL, Schmidt CF. Proc. Natl. Acad. Sci. U.S.A. 2006;103(16):6184–6189. doi: 10.1073/pnas.0601744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Roos WH, Gibbons MM, Arkhipov A, Uetrecht C, Watts NR, Wingfield PT, Steven AC, Heck AJR, Schulten K, Klug WS, Wuite GJL. Biophys. J. 2010;99(4):1175–1181. doi: 10.1016/j.bpj.2010.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Cuellar JL, Meinhoevel F, Hoehne M, Donath E. J. Gen. Virol. 2010;91:2449–2456. doi: 10.1099/vir.0.021212-0. [DOI] [PubMed] [Google Scholar]
- (30).Evilevitch A, Roos WH, Ivanovska IL, Jeembaeva M, Jonsson B, Wuite GJL. J. Mol. Biol. 2011;405(1):18–23. doi: 10.1016/j.jmb.2010.10.039. [DOI] [PubMed] [Google Scholar]
- (31).Ivanovska I, Wuite G, Jonsson B, Evilevitch A. Proc. Natl. Acad. Sci. U.S.A. 2007;104(23):9603–9608. doi: 10.1073/pnas.0703166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Roos WH, Radtke K, Kniesmeijer E, Geertsema H, Sodeik B, Wuite GJL. Proc. Natl. Acad. Sci. U.S.A. 2009;106(24):9673–9678. doi: 10.1073/pnas.0901514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Arkhipov A, Roos WH, Wuite GJL, Schulten K. Biophys. J. 2009;97(7):2061–2069. doi: 10.1016/j.bpj.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Klug WS, Bruinsma RF, Michel JP, Knobler CM, Ivanovska IL, Schmidt CF, Wuite GJL. Phys. Rev. Lett. 2006;97(22):228101. doi: 10.1103/PhysRevLett.97.228101. [DOI] [PubMed] [Google Scholar]
- (35).Carrasco C, Carreira A, Schaap IAT, Serena PA, Gomez-Herrero J, Mateu MG, Pablo PJ. Proc. Natl. Acad. Sci. U.S.A. 2006;103(37):13706–13711. doi: 10.1073/pnas.0601881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Tan M, Hegde RS, Jiang X. J. Virol. 2004;78(12):6233–6242. doi: 10.1128/JVI.78.12.6233-6242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.