Abstract
Ribonucleotide reductases (RRs) catalyze a crucial step of de novo DNA synthesis by converting ribonucleoside diphosphates to deoxyribonucleoside diphosphates. Tight control of the dNTP pool is essential for cellular homeostasis. The activity of the enzyme is tightly regulated at the S-phase by allosteric regulation. Recent structural studies by our group and others provided the molecular basis for understanding how RR recognizes substrates, how it interacts with chemotherapeutic agents, and how it is regulated by its allosteric regulators ATP and dATP. This review discusses the molecular basis of allosteric regulation and substrate recognition of RR, and particularly the discovery that subunit oligomerization is an important prerequisite step in enzyme inhibition.
1. INTRODUCTION
The process by which biological macromolecules such as proteins transmit regulatory signals upon ligand binding to a distinct site distant from the functional site is known as allostery.1 Allostery is important in governing vital functions of cell metabolism and gene regulation.2,3 Considering that allostery is such an important process, a general understanding of its structural basis has been elusive. In the past four decades, allostery has usually been understood via two classical models: the concerted or symmetrical model also known as the MWC model and the sequential model also known as the KNF model.4,5 The concerted model proposes that regulatory proteins have a quaternary structure consisting of subunits organized into finite assemblies, or oligomers, and that all subunits in an oligomer switch states at the same time.3 This model proposes that the conformational coupling occurs between different parts of the molecule as a complete entity.1 The second model postulates that subunits in the same oligomer can be in different states, but the conversion of one subunit makes the conversion of other subunits in the oligomer more likely. In this model, ligand binding to the allosteric site(s) causes structural changes intrinsic to the protein molecule that affect substrate binding.
Allosteric regulation can occur upon binding of a small molecule at a site distant from the active site. So it is pertinent to ask how the signal is transmitted over such a distance. Ligands that activate and inhibit an enzyme through allostery are called allosteric modulators. A significant amount of information on allosteric mechanisms has come from high-resolution structures determined by X-ray crystallography or NMR spectroscopy. High-resolution structure analysis by these methods has shown how ligand binding triggers the allosteric mechanism in those proteins. Comparison of various structures of apoproteins and the same proteins with bound ligand showed bonds formed or lost on ligand binding that correlated with the functional (T) or relaxed (R) states of the molecule.6,7 However, in this review, we discuss ribonucleotide reductase (RR); an allosteric enzyme follows neither the concerted nor the sequential model.
2. RIBONUCLEOTIDE REDUCTASE
RR is an allosteric enzyme that provides the balanced pool of deoxyribonucleoside triphosphate (dNTPs) to maintain genomic integrity in all cells.8,9 RR catalyzes the rate-determining step of dNTP synthesis and hence provides the essential precursors for DNA replication and repair.10–12
Historically, RR was first characterized by Peter Reichard and group at the Karolinska Institute in Sweden in the 1960s.12–16 Four decades after conducting the first enzymatic studies of RR, we still discover novel mechanisms pertaining to its mode of action.
In 1969, Brown and Reichard showed that RR has the amazing ability to catalyze transformation of all four nucleoside diphosphate substrates to their deoxy forms. They also showed that there were two distinct allosteric sites. The first site bound effectors and determined substrate preferences while the second site bound ATP to activate the enzyme or dATP to inhibit it. Mechanistically, reduction of ribonucleoside to deoxyribonucleoside by RR requires the generation of thiyl radical by cysteine residues near the active site of the enzyme. Metal cofactors are involved in the generation of free radicals that ultimately lead to the formation of the important thiyl radical in the active site.17–19 So far, three classes of RR have been reported based on the metal cofactors that generate their thiyl radicals.9,17,18,20 In this review, we discuss the Class I enzymes that use a tyrosyl free radical for catalysis.
Class I enzymes are expressed in nearly all eukaryotes, in some prokaryotes, and in certain viruses.9,20–23 However, in this review, we focus mostly on human, yeast (eukaryotic), and E. coli (prokaryotic) RR. The active form of the eukaryotic enzyme consists of two proteins, the larger being RR1 (α) and the smaller RR2 (β). These proteins associate in dimeric or multimeric forms such as α2β2 and (α)n(β2)m (where n = 4 or 6 and m = 1, 2, or 3). 24–30 Substrate catalysis occurs in RR1 (α) which contains two allosteric sites, the specificity site (S-site), and the activity site (A-site) in addition to the catalytic site (C-site)29,31–34 (Fig. 14.1). The smaller RR2 (β) subunit is associated with the diferric-oxygen center which generates the stable tyrosyl free radical.35 Class I enzymes are further divided into three subclasses, Class Ia, Ib, and Ic, based on the organization of RR genes, protein topology, and metalo-cluster assembly of the RR2 (β) protein.9,36,37 Class Ia and Class Ib require a diferric-tyrosyl radical cofactor (FeIII FeIII–Y•) associated with their RR2 (β) protein. The Class Ic enzymes do not use Y•, but the synthesis of an active metal cluster in the β subunit requires oxygen. Catalysis by Class Ia enzymes involves reduction of cysteine residues near the C-site brought about by transfer of the radical originating at the diferric-tyrosyl 30–35 Å from the C-site.30,31,38 After catalysis, the cysteines are re-oxidized to regenerate the enzyme.
Figure 14.1.
Structure of large subunit of ScRR1 in dimeric form. ScRR1 monomers are yellow and green; effector dGTP (violet) and substrate ADP (blue) from the effector substrate pair are shown in the specificity (S-site) and catalytic site (C-site). Reproduced with permission from Ref. 32. Copyright (2006) National Academy of Sciences, USA.
The early biochemical studies raised a remarkable question concerning RR: by what molecular mechanism did this enzyme catalyze specific reduction of all four nucleoside diphosphate substrates? In other words, what causes this enzyme to switch its specificity when it binds different effectors? Among the two allosteric sites, one, the A-site, regulates the overall activity and the other, the S-site, regulates the substrate specificity. Binding of ATP or dATP at the A-site increases or reduces, respectively, the overall activity of the enzyme.10,12,33 The S-site has the ability to bind dGTP and TTP in addition to ATP and dATP. RR1 uses allosteric communication to determine C-site substrate preference based on the nucleotide effector bound at the S-site.9,10,21,32,34 Either ATP or dATP binding at the S-site facilitates both CDP and UDP binding at the C-site; TTP binding at the S-site selects for GDP binding at the C-site, and dGTP binding at the S-site promotes ADP binding at the C-site. The determination of the first three-dimensional structure of RR1 from E. coli provided the molecular structure of the two allosteric sites and the C-site.31 Another important study from Hans Eklund’s group in 1997 showed the structure of E. coli RR1 in complex with TTP/GDP for the first time.39 This study illustrated the importance of a polypeptide chain called Loop 2 which was involved in substrate recognition and another called Loop 1 which was important in effector binding. It also laid the groundwork for the definition of the S-site and the C-site. In the same study, they showed how substrate binding at the C-site is structurally regulated by the binding of allosteric effectors.
RR is highly regulated transcriptionally, by subcellular compartmentalization, by small protein inhibitors such as the yeast proteins Sml1 and Spd1 and, during the S-phase of the cell cycle, by allosteric regulation.9 In this review, we discuss the allosteric regulation of RR by which a single protein provides a balanced supply of all four dNTPs. Understanding RR regulation is important in cancer therapy since many of the clinically approved anticancer drugs such as gemcitabine, clofarabine, cladribine, and fludarabine mimic the substrates and allosteric effectors of this enzyme.40,41 RR has a highly sophisticated mechanism for synthesis and regulation of dNTPs, and even after more than four decades of RR discovery, research on this enzyme continues to reveal fascinating new aspects of its action. Recent studies from our group and others advance our understanding of the intriguing mechanisms of allosteric regulation in this enzyme.29,42–46
2.1. Allosteric regulation
Interestingly, the allosteric regulation occurring between the S- and C-sites is shared by both Class I and Class II enzymes. In contrast, Class III enzymes use a slightly different set of substrate selection rules. The A-site is known as the pyrimidine site and can bind either ATP or dATP. Binding of ATP to the pyrimidine site stimulates reduction of pyrimidine ribonucleotides. The purine site resembles the S-site in Class I and Class II enzymes. dGTP and TTP bind at the purine site and select for ATP or GTP at the C-site, respectively. However, the binding of dATP to either the purine or pyrimidine site is always inhibitory. The molecular basis for substrate selections is described in two studies based on the Class II and Class I structures.32,47
2.2. The C- and S-sites of Saccharomyces cerevisiae RR and Homo sapiens RR (hRRM1)
Our lab has produced hRRM1•TTP•GDP and ScRR1•TTP•GDP ternary complexes by soaking cocrystals of hRRM1•TTP and ScRR1•TTP with the substrate GDP. The structures of these ternary complexes showed that the ribose ring of the substrate adopts a puckered 3′-endo conformation similar to that previously observed in the TmNrd1 substrate–effector complex structures.29,32,48 Although in these complexes the C-site interactions with the ribose of the GDP are fairly well conserved, there are differences with those of the Class II TmNrd1 complex. For instance, the sulfur of the catalytic cysteine in the hRRM1 and ScRR1 complexes is within hydrogen bonding distance of the 2′ and 3′ OH groups of the ribose, while the corresponding groups are more than 4 Å apart in the TmNrd1 complexes. Comparison of the S-site of the hRRM1•TTP•GDP complex with those of the TTP complexes formed by eukaryotic ScRR1 and prokaryotic EcRR1 and TmNrd1 shows that TTP binds very similarly in all the structures, with the exception of the EcRR1•TTP•GDP complex in which the base is substantially shifted by about 5 Å.
2.3. The role of Loop 1 and Loop 2 in allosteric regulation of RR1
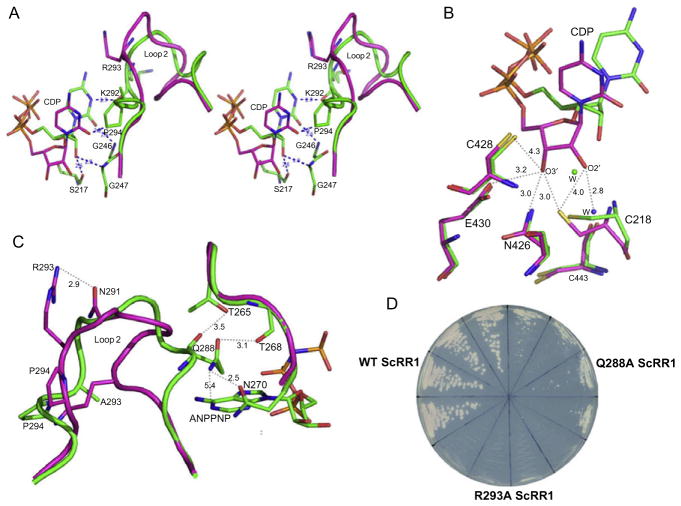
Two very important loops called Loop 1 (residues 245–260) and Loop 2 (residues 286–295) (Fig. 14.1) were identified in the E. coli X-ray structure.39 Nucleoside or dNTP effector binding at the S-site is a prerequisite for the dimerization of the large subunit, which is essential for substrate selection. This is because allosteric communication occurs between subunits by means of Loop 2 (Fig. 14.1) (which connects the S-site on one subunit with the C-site of the adjacent subunit). The mechanism of substrate selection mediated by Loop 2 has been the subject of two crystallographic studies conducted on T. maritima Class II RR and S. cerevisiae Class I RR. These enzymes share a common allosteric mechanism involving key residues in Loop 247 (Fig. 14.2). In the apo form of S. cerevisiae RR, this loop sits in a partially closed conformation, leaving little or no room for substrate binding. Upon nucleoside triphosphate binding to the S-site, Loop 2 shifts toward the effector site, concomitantly creating room for substrate entry and creating a face of the binding site that helps select a particular substrate (Figs. 14.1 and 14.2). Once the substrate is bound, Loop 2 yet again changes conformation and moves back toward the substrate.32 We called this elegant communication between the S-site and C-site “specificity cross talk.” Two residues in the Loop 2, R293 and Q288, have been proposed to be critical for substrate recognition.42,49 A recent study has shown that a number of single point mutations in the ScRR1 Loop 2 have a significant impact on the cellular dNTP pool and the cell growth phenotype.49,50 Structural studies have shown that Q288 and R293 in ScRR1 form hydrogen bonds with the adenine ring, and the R293 makes an additional stacking interaction with the adenine.32 Mutation of R293 in ScRR1 to alanine severely compromised ADP binding at the C-site as evident in the crystal structure of the R293A ScRR1-dGTP complex. Furthermore, isothermal titration calorimetry analysis indicated significant loss of affinity for ADP substrate in the case of R293A ScRR1.42 This is not surprising, as ADP makes a greater number of interactions with residue 293 when bound to wild-type ScRR1 than to the mutant. The Q288A mutation destroys two hydrogen bonds that the glutamine side chain forms with ADP and R293. The latter interaction is important for positioning the guanidinium side chain of R293, to form its hydrogen bond and stacking interaction with ADP.
Figure 14.2.

Substrate selection mediated via Loop 2: effector AMPPNP and substrate ADP binding at the S and C-site, respectively. The key residues on Loop 2 required for substrate selection are Q288 and R293 as shown. Reproduced with permission from Ref. 32. Copyright (2006) National Academy of Sciences, USA.
Mutation at R293 in ScRR1 leads to altered CDP substrate binding, such that CDP binds farther from the C-site when compared to wild-type ScRR1. Moreover, this mutation results in a conformational change in Loop 2 accompanied by a shift toward the C-site. The newly acquired loop conformation would cause a steric clash between P294 and the CDP substrate if the CDP bound in the original position as observed in the wild-type ScRR1. This study shows that residues R293 and Q288 are important not only for facilitating substrate selection but also for stabilizing the conformation of Loop 2. It is evident in these structural studies that Loop 2 conformation is crucial for the communication between the S-site and the C-site (Fig. 14.3A–C). The Chabes group has previously shown that expressing R293A and Q288A ScRR1 mutants in yeast skewed the dNTP pool toward higher dATP/dCTP and dGTP/dCTP ratios. Substitution at multiple positions in Loop 2 also gave rise to mutants with imbalanced dNTP pools; however, only R293A and Q288A mutations exhibited synthetic lethality based on meiotic progeny analysis when the alternative RR1 subunit ScRR3 was deleted,49 indicating the significant role played by Q288 and R293 in ScRR1 (Fig. 14.3D).
Figure 14.3.
Effect of R293A and Q288A mutations on wild-type ScRR1. (A) Structural comparison of AMPPNP- and CDP-bound wild type and R293A ScRR1; stereo view of the substrate CDP binding and its proximity to Loop 2. (B) Comparison of catalytic site of wild type and R293A ScRR1 with CDP bound. The catalytic important residues C218, C428, N426, C428, and E430 are also shown binding to the ribose moiety of wild-type ScRR1 (shown as dashed lines). (C) Effect of R293A and Q288A mutations in the Loop 2 conformation. In these figures, wild-type ScRR1 is colored in magenta and R293A ScRR1 is colored in green. (D) Growth defects in wild-type ScRR1 because of R293A and Q288A mutations; R293A ScRR1 causes lethality whereas Q288A ScRR1 showed slower growth compared to wild-type ScRR1. Reproduced with permission from Ref. 42. Copyright (2012).
2.4. ATP and dATP binding at the A-site (ATP cone)
The A-site in hRRM1 consists of a four-helix bundle. These helices are numbered H1–H4, with H1 comprising residues 15–26, H2 comprising residues 36–46, H3 comprising residues 53–70, and H4 comprising residues 74–90 (Fig. 14.4A and B). This ATP-binding cone is covered at one end by the previously termed “β-cap,” a β-hairpin formed by the first 14 N-terminal residues and residues 48–51.31 Binding of either ATP or dATP requires helix H1 and the β-cap to shift from their positions in the native (TTP-only) form (Fig. 14.4C), involved in some induced fit. However, the two binding modes are markedly different. dATP binds more deeply inside the four-helix bundle and its ribose adopts a half-chair conformation with the C2′ out of plane. In contrast, ATP binds less deeply inside the four-helix bundle and its ribose adopts a 2′-endo conformation (Fig. 14.4). Interestingly, we have observed in our ITC binding experiment that hRRM1 has severalfolds higher affinity for dATP compared to ATP (Ahmad MF, unpublished data). The binding energy of protein–ligand interactions can be expressed as the change in solvent-accessible surface area between the bound and unbound states51; the deeper binding of dATP and its larger buried surface area compared to ATP must contribute to its higher affinity. The chemical difference at the 2′ position on the ribose moiety between ATP and dATP causes the difference in their binding at the A-site. The 2′-OH of ATP sterically precludes the ribose from binding more deeply inside the four-helix bundle, whereas dATP, lacking the 2′-OH, can penetrate the pocket more deeply. In addition, our structural studies on dATP-bound hRRM1 illustrated that residue I18 acts as a steric gate, as it would clash with the 2′-OH of ATP binding at the dATP position. In the similar study, we proposed the molecular basis for how dATP is able to overcome a severalfold deficit to outcompete ATP at the A-site during the S-phase of the cell cycle.
Figure 14.4.
Structure of ATP- and dATP-bound hRRM1. (A and B) ATP and dATP binding at the A-site in hRRM1 consist of a four-helix bundle. 2Fo–Fc electron density for ATP and dATP is shown which is contoured at 1σ (blue density). (C) Ribbon diagram depicting ATP-binding cones of hRRM1–TTP–ATP ternary complex (blue) and hRRM1–TTP–dATP ternary complex (magenta) were aligned to that of hRRM1–TTP complex (orange). Allosteric modulator ATP and dATP are shown in yellow and green, respectively. Reproduced with permission from Ref. 29.
2.5. Subunit oligomerization and allosteric regulation of RR
Binding of different ligands enables the formation of a biologically functional oligomer in some cases and a nonfunctional oligomer in others. In the case of RR, both the allosteric activator ATP and allosteric inhibitor dATP induce oligomerization upon binding to RR. ATP and dATP bind in a concentration-dependent manner to alter the enzyme’s oligomeric state and thereby regulate enzyme activity. Initial studies on mouse RR have shown that dATP binding to the A-site induces formation of a tetramer, which further isomerizes to an inactive form of the enzyme. ATP, in contrast, induces formation of an active hexamer upon binding to a proposed H-site.25,26 This study also examined mouse D57N RR, which differs from wild-type mouse RR in that it has lost the ability to differentiate between ATP and dATP and hence is activated by both. They argue that the D57N mutation in mouse RR produces this effect by blocking the transformation of active to inactive tetramer.25,27
Later studies on mouse RR suggest that despite the fact that ATP and dATP have opposing effects on enzyme activity, both nucleotides facilitate the formation of higher-order oligomers. Anders Hofer’s group has shown, using a technique known as gas-phase electrophoretic-mobility macromolecule analysis, that ATP/dATP-bound murine RR1 forms hexamers which can interact with dimers of the small subunit to form the enzymatically active complex α6β2. This α6β2 holoenzyme complex can exist in either an inactivated state or an activated state depending upon whether the bound nucleotide is dATP or ATP.24 Another study with the drug gemcitabine (F2CDP) which is used clinically in the treatment of various types of cancer showed that F2CDP is a substoichiometric mechanism-based inhibitor of the large subunit of both E. coli and human RR in the presence of reductant.28 In this study, F2CDP-bound RR forms a tight α2β2 complex in E. coli. Whereas human RR forms an α6β6 complex upon F2CDP binding that leads to enzyme inactivation.28 Recently, we demonstrated that dATP-induced olig-omerization is concentration dependent.29 Our studies have shown that in the absence of any effector molecules, human RR is a monomer. At 5 μm dATP, the dimer and hexamer are seen with a reduction of monomer concentration as analyzed by size-exclusion chromatography (SEC) and multiangle light scattering (MALS). Interestingly, at a physiological concentration of 20 μm dATP (the physiological concentration of dATP reported for the S-phase), most of the human RR forms a hexameric complex, with a few dimeric species as revealed by both SEC and MALS. Furthermore, it has been shown that the N-terminus is involved in hexamer formation. Upon deleting the first 74 amino acid residues, commonly known as the ATP cone, from human RR, dATP is no longer able to induce the formation of the hexamer. In addition, in the presence of human RR2, the N-terminally truncated version of hRRM1 forms an α2β2 holocomplex. This result further emphasizes the role of the A-site in oligomerization. In this study, it has been shown that the rate of dCDP and dADP formation by hRRM1 and ScRR1 is severely diminished in the dATP-induced hexamer. However, the dimeric form of eukaryotic RR is not inhibited by dATP. The above results indicate that the dATP-induced hexamerization of eukaryotic RR is a prerequisite for enzyme inhibition. It is important to mention here that these findings deviate from previous studies which reported that the dATP-induced tetramer is the least active RR form and that the hexamer is observed only at very high dATP concentration.25 The discrepancy regarding dATP-induced oligomerization in the above study is partly due to the difficulty of interpretation of dynamic light scattering data. Since dynamic light scattering cannot resolve differences in the radius of various oligomer species that are less than a factor of 4–5, the reported mammalian RR tetramer may be mixture of dimers and hexamers, which would be consistent with our results.
2.6. The X-ray structure of the dATP-induced hexamer of Class 1 RNR
To understand the structural basis of RR1 oligomerization and enzyme inactivation by dATP, we crystallized the ScRR1 hexamer complex. The complex crystallized in a hexagonal space group and diffracted to 6 Å. Although the low-resolution structure does not provide atomic detail, it is useful for indicating oligomer organization. To analyze the data, two models of ScRR1 hexamer packing were considered. In both models, ScRR1 α6 is a trimer of dimers, in which the three dimers are related to each other by a threefold axis. However, the models differ in the diameter of the central pore of the hexamer and in how dATP would mediate hexamerization. In one model, only three of the six dATP-bound ATP-binding cones participate in forming the hexamer interface, the other three free to interact with the small subunit. Hence, only three dATP molecules are present at the hexamer interface. In the other model, the interfaces that stabilize the hexamer are entirely formed by six dATP-bound ATP-binding cones. Furthermore, each of the three interfaces is formed by two dATP-bound ATP-binding cones from adjacent RR1 dimers that contact each other in an antiparallel conformation and are related by twofold symmetry29 (Fig. 14.5A).
Figure 14.5.
Subunit oligomerization packing of RR1 based on the low-resolution X-ray crystal structure of the ScRR1 hexamer. (A) ScRR1 monomers are colored in cyan and blue. ATP-binding cones are colored in red. (B) Electron micrograph of the α6–ββ′–dATP holocomplex; image showing the negative stain of holocomplex. Scale bar, 50 nm. Model of the α6•ββ′•dATP holocomplex based on cryo-EM data. Reproduced with permission from Ref. 29.
Since the hexamer model was built on the low-resolution ScRR1 hexamer structure, it was pertinent to test the hexamer interfaces of both models biochemically. Hence, site-directed mutagenesis was used to disrupt the hexamer interface predicted by each model. Interestingly, only the D16R mutation, at the N-terminus, disrupts hexamer formation in both hRRM1 and ScRR1. The results of this biochemical study validated only one model—the one in which all six ATP-binding cones participate in the hexamer interface (Fig. 14.5A).
It was hypothesized that since the D16R mutation abolishes the ability of hRRM1 to form a dATP-induced hexamer, D16R like the earlier reported mutation D57N which abolishes the ability to discriminate between ATP and dATP would prevent the allosteric inhibition of hRRM1 by dATP at physiological concentration. Interestingly, D16R hRRM1 retains the ability to reduce ADP and CDP substrates and, as expected, its activity is not inhibited by dATP. Unlike D57N, it is not activated by dATP either, however. Furthermore, D16R hRRM1 retains the ability to bind dATP, so this change is not caused by abolishing dATP binding at the A-site. Since D16R hRRM1 will still bind dATP but does not form hexamers as a result and is not inhibited by it, this supports the idea that allosteric inhibition of RR by dATP under physiological conditions requires hRRM1 to be in hexameric form.29
Our dATP-induced hexamer structure let us examine subunit packing for the first time. The cryo-EM structure showed that the dATP holoenzyme has a subunit composition of α6β2 (Fig. 14.5B and C). In a similar study, site-directed mutagenesis studies showed that the ATP-induced hexamer adopts an interface different from that of the dATP-induced hexamer. It is quite easy to envisage that there must be structural differences between the ATP-induced hexamer and the dATP-induced hexamer for one to act as a functional entity and the other to be inactive.
Recently, it has been shown by carefully examining the ATP cone of ATP- and dATP-bound Class I RNR that the largest differences between the two complexes are in the loop region that spans residues 45–52.52 This particular loop is believed to be an important part of the dimer–dimer interface in the eukaryotic hexamer. This observation led to speculation that subtle differences in the conformation of this region upon ATP and dATP binding are responsible for the different oligomeric structures of ATP-versus dATP-bound eukaryotic RR and for the different oligomerization behaviors of eukaryotic and E. coli Class 1RR.52 Rather than form hexamers as in human RR, E. coli RR appears to undergo oligomeric interconversion from α2β2 (the active oligomer for E. coli RR) to the ring-shaped α4β4 (the inactive oligomer for E. coli RR) under the influence of the allosteric effector. Moreover, this interconversion is linked with distinct subunit rearrangements upon ATP and dATP binding, which gives further insight into the molecular mechanism of the activated and inactivated states.45 Further extending the research on E. coli RR ring structure, a later study showed that the clinically approved anticancer drug gemcitabine when added to a solution of both E. coli RNR subunits induces an unusual structure known as a concatamer, in which two α4β4 rings interlock to form a unique (α4β4)2 complex.46 This study exploited a hybrid approach to structural studies by using small-angle X-ray scattering (SAXS) and electron microscopy (EM) along with X-ray crystallography to verify the formation of this unique complex. The proposed mechanism for the formation of the concatamer is that α4β4 ring formation occurs as described above and the ring subsequently opens to accommodate a second ring and then recloses. The mechanism by which the concatamer forms was further confirmed by SAXS analysis, additionally demonstrating that the structure was not a crystallography artifact. It is important to mention here that despite the structural similarity between ScRR1 and the E. coli enzyme, E. coli RR does not form a hexamer. The only complexes reported for the E. coli enzyme to this point are α2β2, α4β4, and (α4β4). It is noteworthy that the ability to form different ring structures among Class 1 RR provides to develop new strategies to inhibit the RRs.46
2.7. The oligomeric state of hRR in the presence of nonnatural ligand
As described above and in our recent study, we showed that a key to hRRM1 inhibition by the negative regulator dATP is the ability of the natural nucleotide to induce the protein to form an inactive hexamer.29 In an independent study, the dATP analog, clofarabine, was shown to inhibit hRRM1 by inducing hexamer formation.44,53 The above study indicates the importance of developing nonnatural ligands that can perturb the quaternary status of protein by inducing the inactive hexamer of RR.44,53 Recent studies by Ahmad et al. have shown that 5-NITP inhibits the activity of hRRM1. This particular nonnatural ligand was identified by in silico analysis of possible inhibitors of human RR. This analog’s structural similarities to dATP suggested that it would inhibit hRRM1 activity by binding to its allosteric sites. However, 5-NITP binds to the S-site with micromolar affinity and only weakly to the A-site as revealed by ITC studies. Furthermore, 5-NITP binding at the S-site was confirmed by the X-ray crystal structure. Thus, binding of 5-NITP does not induce formation of the inactive hexamer of hRRM1 as does dATP and may account for the moderate inhibitor potency. Regardless of its mechanism, 5-NITP produces cytostatic and cytotoxic effects against human leukemia cells by altering cell-cycle progression, probably through its action against RR.43 However, it is still important to develop nonnatural ligands that bind the A-site to force formation of an inactive hRRM1 hexamer as another way to exploit RR allosteric control in the service of anticancer and antiviral therapy.
3. TARGETING LARGE SUBUNIT OF RR
Considering the fact that RR plays an essential role in DNA synthesis and repair makes it an attractive target for cancer chemotherapy. In past few years, considerable amount of effort has been made to develop specific inhibitor against this enzyme.54 In this review, we briefly discuss about the inhibitors for cancer chemotherapy that involves the large subunit of RR. To date, four druggable sites have been identified for the large subunit of RR. As shown in Fig. 14.1, these druggable sites are the A-site, S-site, C-site, and the peptide-binding site (P-site). Among the four sites, three sites are occupied with the nucleoside and deoxynucleoside, whereas the fourth side is the P-site. Mostly nucleotide-based drugs that target RR inhibit its activity by binding at either the A-site or the C-site. Interestingly, most of the analogs that bind the A-site quite often bind the S-site. Now we briefly describe the drugs and their modes of mechanism when binding to the A-site, C-site, and P-site of RR.
Fludarabine, cladribine, and clofarabine55–58 are nucleoside analogs that target the A-site of the enzyme. These are U.S. FDA-approved clinical drugs that target the A-site of RR1. Clofarabine is the best-characterized drug among the above-mentioned nucleoside analogs and is clinically used in treating childhood leukemia.58–60 Clofarabine triphosphate is the potent inhibitor of RR (IC50=65 nM), apparently binding to allosteric site of large subunit of RR.40,61 In a recent report by Stubbe and co-workers, it was shown that clofarabine triphosphate inhibits hRRM1 with a Ki equal to 40 nM at the A-site.53 In addition, they have also shown that the clofarabine is not an irreversible inhibitor. Clofarabine triphosphate induces hexamers by binding at the A-site similar to the allosteric inhibitor dATP. Hence, it is important to mention here that nucleoside analogs that retain the ability to hexamerize the RR1 subunit similar to that of dATP are likely to be categorized as good inhibitors of RR.
Gemcitabine which is a billion dollar drug used in many chemotherapeutic regimes is a mechanism-based inhibitor that targets the C-site of RR1 has been characterized by Stubbe and co-workers.28,62 It is important to mention here that Gemcitabine is one of the few chemotherapeutic agents that are known to have any efficacy against pancreatic cancer.63,64 Gemcitabine binds at the C-site of the large subunit of RR and induces the hexamerization, α6β6 holocomplex. We were able to determine the initial interactions of F2dCDP, the diphosphate metabolite of gemcitabine, at the C-site of ScRR141 (yeast RR1). It is interesting to note that while F2dCDP differs from CDP only by substitution of two fluorines for the hydroxyl group and a hydrogen atom on the 2′ carbon of the ribose, F2CDP adopts a different conformation when binding to RR1.41 In the similar study, we have shown that differences observed between CDP and F2dCDP binding were due to unique chemical properties of the germinal fluorine atoms which are more hydrophobic and yet retain the ability to form hydrogen bonds. Our study illustrated the unique interactions that fluorine atoms can make in an instance where chemical space truly invades biological space.
Studies conducted in our lab and Cooperman’s group have suggested that P-site is an excellent drug target for anticancer therapy.32,41,65–72 Drug/molecule that targets the P-site will interfere with the quaternary structure of RR via a specific protein–protein interaction. Exploiting this particular characteristic will enable us to develop the highly specific inhibitors that do not lack the specificity associated with nucleoside analogs that target the A-, S-, and C-site of RR. The P-site comprises two well-formed subsites providing a platform for fragment-based drug design. Currently, there is no clinically used drug available that targets the P-site. However, future drugs are likely to emerge that target the P-site.
4. CONCLUSIONS
RR is a fascinating enzyme that converts ribonucleosides to deoxyribonucleosides and provides raw material for DNA synthesis and repair. Considering its important function, RR is an attractive target for cancer and antiviral therapy. Interestingly, in a recent review, it was noted that hRRM1 is a useful biomarker in cancer therapy similar to BRCA1.73 A number of clinical trials have shown that patients with tumors expressing low levels of hRRM1 have increased progression-free and overall survival rates when treated with regimens that included gemcitabine.73,74 Hence, hRR can be used in bench to bedside treatments.30 The current interest in RR research is increasing our understanding of higher-order oligomerization, enzyme turnover, and the mechanism of allosteric regulation that governs specificity. The molecular basis of activation and deactivation by ATP and dATP was partially clarified by our recent study, which provided an explanation of how dATP achieves a 100-fold higher affinity than ATP for the A-site.29 dATP achieves its high affinity by binding deeper in the pocket because it does not have an O2′ atom as does ATP. Interestingly, I18 acts as a steric gate for the O2′ atom.29 In the same study, we showed that dATP regulates RR by forming inactive hexamers, whereas the α2β2 dimer remains active. Moreover, the inactive holoenzyme complex in ScRR1 was shown to be an α6β2 oligomer, which is in agreement with a previously reported higher-order oligomer in mouse RR.24,75 Recently, using various structural and biophysical techniques, the Drenan group demonstrated that dATP inhibits E. coli RR by driving formation of the α4β4 ring-like structure.45 Furthermore, they have shown formation of the (α4β4)2 megacomplex of E. Coli RR in the presence of gemcitabine and dATP.46 It is important to mention that the difference in the high-order oligomeric ring structure between E. coli and eukaryotic RR is probably due to the difference in the N-terminal ATP-binding cones. Due to the complexity of its functions, understanding the structure, function, and regulation of RR is intellectually very satisfying. As we begin to understand more about how this complicated molecule functions, we will discover novel ways to develop chemotherapies that target hRRM1.
Acknowledgments
This research was supported by NIH grants 1R01GM100887-01 and R01CA100827-04A1. We would like to thank Dr. Cat Faber for proofreading. We also like to thank the present and past members of Dealwis lab for contributing to the work on ribonucleotide reductase.
ABBREVIATIONS
- 5-NITP
5-nitro-indolyl-2′-deoxyribose triphosphate
- ADP
adenosine diphosphate
- AMPPNP
adenosine 5-(β,γ-imido) triphosphate tetralithium salt
- CDP
cytidine diphosphate
- dGTP
deoxyguanosine triphosphate
- dNTP
deoxynucleoside triphosphate
- hRRM1
human ribonucleotide reductase
- ScRR1
Saccharomyces cerevisiae RR1
References
- 1.Hilser VJ. Biochemistry. An ensemble view of allostery. Science. 2010;327(5966):653–4. doi: 10.1126/science.1186121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. J Mol Biol. 1963;6:306–29. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 3.Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308(5727):1424–8. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 4.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 5.Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5(1):365–85. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 6.Kantrowitz ER, Lipscomb WN. Escherichia coli aspartate transcarbamylase: the relation between structure and function. Science. 1988;241(4866):669–74. doi: 10.1126/science.3041592. [DOI] [PubMed] [Google Scholar]
- 7.Perutz M. Cooperativity and allosteric regulation in proteins. Cambridge, UK: Cambridge University Press; 1990. [Google Scholar]
- 8.Kolberg M, Strand KR, Graff P, Andersson KK. Structure, function, and mechanism of ribonucleotide reductases. Biochim Biophys Acta. 2004;1699(1–2):1–34. doi: 10.1016/j.bbapap.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 10.Thelander L, Reichard P. Reduction of ribonucleotides. Annu Rev Biochem. 1979;48:133–58. doi: 10.1146/annurev.bi.48.070179.001025. [DOI] [PubMed] [Google Scholar]
- 11.Reichard P, Baldesten A, Rutberg L. Formation of deoxycytidine phosphates from cytidine phosphates in extracts from Escherichia coli. J Biol Chem. 1961;236:1150–7. [PubMed] [Google Scholar]
- 12.Brown NC, Canellakis ZN, Lundin B, Reichard P, Thelander L. Ribonucleoside diphosphate reductase. Purification of the two subunits, proteins B1 and B2. Eur J Biochem. 1969;9(4):561–73. doi: 10.1111/j.1432-1033.1969.tb00646.x. [DOI] [PubMed] [Google Scholar]
- 13.Holmgren A, Reichard P, Thelander L. Enzymatic synthesis of deoxyribonucleotides, 8. The effects of ATP and dATP in the CDP reductase system from E. coli. Proc Natl Acad Sci USA. 1965;54(3):830–6. doi: 10.1073/pnas.54.3.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore EC, Reichard P. Enzymatic synthesis of deoxyribonucleotides. VI. The cytidine diphosphate reductase system from Novikoff hepatoma. J Biol Chem. 1964;239:3453–6. [PubMed] [Google Scholar]
- 15.Blakley RL, Ghambeer RK, Nixon PF, Vitols E. The cobamide-dependent ribonucleoside triphosphate reductase of lactobacilli. Biochem Biophys Res Commun. 1965;20(4):439–45. doi: 10.1016/0006-291x(65)90597-8. [DOI] [PubMed] [Google Scholar]
- 16.Berglund O, Karlstrom O, Reichard P. A new ribonucleotide reductase system after infection with phage T4. Proc Natl Acad Sci USA. 1969;62(3):829–35. doi: 10.1073/pnas.62.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reichard P. From RNA, to DNA, why so many ribonucleotide reductases? Science. 1993;260(5115):1773–7. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- 18.Cotruvo JA, Jr, Stubbe J. Class I ribonucleotide reductases: metallocofactor assembly and repair in vitro and in vivo. Annu Rev Biochem. 2011;80:733–67. doi: 10.1146/annurev-biochem-061408-095817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Licht S, Gerfen GJ, Stubbe J. Thiyl radicals in ribonucleotide reductases. Science. 1996;271(5248):477–81. doi: 10.1126/science.271.5248.477. [DOI] [PubMed] [Google Scholar]
- 20.Jordan A, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 1998;67:71–98. doi: 10.1146/annurev.biochem.67.1.71. [DOI] [PubMed] [Google Scholar]
- 21.Reichard P. Ribonucleotide reductases: the evolution of allosteric regulation. Arch Biochem Biophys. 2002;397(2):149–55. doi: 10.1006/abbi.2001.2637. [DOI] [PubMed] [Google Scholar]
- 22.Stubbe J. Ribonucleotide reductases. Adv Enzymol Relat Areas Mol Biol. 1990;63:349–419. doi: 10.1002/9780470123096.ch6. [DOI] [PubMed] [Google Scholar]
- 23.Stubbe J, Ge J, Yee CS. The evolution of ribonucleotide reduction revisited. Trends Biochem Sci. 2001;26(2):93–9. doi: 10.1016/s0968-0004(00)01764-3. [DOI] [PubMed] [Google Scholar]
- 24.Rofougaran R, Vodnala M, Hofer A. Enzymatically active mammalian ribonucleotide reductase exists primarily as an alpha6beta2 octamer. J Biol Chem. 2006;281(38):27705–11. doi: 10.1074/jbc.M605573200. [DOI] [PubMed] [Google Scholar]
- 25.Kashlan OB, Cooperman BS. Comprehensive model for allosteric regulation of mammalian ribonucleotide reductase: refinements and consequences. Biochemistry. 2003;42 (6):1696–706. doi: 10.1021/bi020634d. [DOI] [PubMed] [Google Scholar]
- 26.Cooperman BS, Kashlan OB. A comprehensive model for the allosteric regulation of Class Ia ribonucleotide reductases. Adv Enzyme Regul. 2003;43:167–82. doi: 10.1016/s0065-2571(02)00035-3. [DOI] [PubMed] [Google Scholar]
- 27.Kashlan OB, Scott CP, Lear JD, Cooperman BS. A comprehensive model for the allosteric regulation of mammalian ribonucleotide reductase. Functional consequences of ATP- and dATP-induced oligomerization of the large subunit. Biochemistry. 2002;41:462–74. doi: 10.1021/bi011653a. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Lohman GJ, Stubbe J. Enhanced subunit interactions with gemcitabine-5′-diphosphate inhibit ribonucleotide reductases. Proc Natl Acad Sci USA. 2007;104(36):14324–9. doi: 10.1073/pnas.0706803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fairman JW, Wijerathna SR, Ahmad MF, et al. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat Struct Mol Biol. 2011;18(3):316–22. doi: 10.1038/nsmb.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wijerathna SR, Ahmad MF, Xu H, Fairman JW, Zhang A, Kaushal PS, et al. Targeting the large subunit of human ribonucleotide reductase for cancer chemotherapy. Pharmaceuticals. 2011;4:1328–54. doi: 10.3390/ph4101328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhlin U, Eklund H. Structure of ribonucleotide reductase protein R1. Nature. 1994;370 (6490):533–9. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- 32.Xu H, Faber C, Uchiki T, Fairman JW, Racca J, Dealwis C. Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation. Proc Natl Acad Sci USA. 2006;103(11):4022–7. doi: 10.1073/pnas.0600443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown NC, Reichard P. Role of effector binding in allosteric control of ribonucleoside diphosphate reductase. J Mol Biol. 1969;46(1):39–55. doi: 10.1016/0022-2836(69)90056-4. [DOI] [PubMed] [Google Scholar]
- 34.Reichard P, Eliasson R, Ingemarson R, Thelander L. Cross-talk between the allosteric effector-binding sites in mouse ribonucleotide reductase. J Biol Chem. 2000;275 (42):33021–6. doi: 10.1074/jbc.M005337200. [DOI] [PubMed] [Google Scholar]
- 35.Stubbe J, Riggs-Gelasco P. Harnessing free radicals: formation and function of the tyrosyl radical in ribonucleotide reductase. Trends Biochem Sci. 1998;23(11):438–43. doi: 10.1016/s0968-0004(98)01296-1. [DOI] [PubMed] [Google Scholar]
- 36.Jordan A, Pontis E, Atta M, et al. A second class I ribonucleotide reductase in Enterobacteriaceae: characterization of the Salmonella typhimurium enzyme. Proc Natl Acad Sci USA. 1994;91(26):12892–6. doi: 10.1073/pnas.91.26.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roshick C, Iliffe-Lee ER, McClarty G. Cloning and characterization of ribonucleotide reductase from Chlamydia trachomatis. J Biol Chem. 2000;275(48):38111–9. doi: 10.1074/jbc.M006367200. [DOI] [PubMed] [Google Scholar]
- 38.Nordlund P, Eklund H. Structure and function of the Escherichia coli ribonucleotide reductase protein R2. J Mol Biol. 1993;232(1):123–64. doi: 10.1006/jmbi.1993.1374. [DOI] [PubMed] [Google Scholar]
- 39.Eriksson M, Uhlin U, Ramaswamy S, et al. Binding of allosteric effectors to ribonucleotide reductase protein R1: reduction of active-site cysteines promotes substrate binding. Structure. 1997;5(8):1077–92. doi: 10.1016/s0969-2126(97)00259-1. [DOI] [PubMed] [Google Scholar]
- 40.Parker WB, Shaddix SC, Chang CH, et al. Effects of 2-chloro-9-(2-deoxy-2-fluoro-beta-D-arabinofuranosyl)adenine on K562 cellular metabolism and the inhibition of human ribonucleotide reductase and DNA polymerases by its 5′-triphosphate. Cancer Res. 1991;51(9):2386–94. [PubMed] [Google Scholar]
- 41.Xu H, Faber C, Uchiki T, Racca J, Dealwis C. Structures of eukaryotic ribonucleotide reductase I define gemcitabine diphosphate binding and subunit assembly. Proc Natl Acad Sci USA. 2006;103(11):4028–33. doi: 10.1073/pnas.0600440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmad MF, Kaushal PS, Wan Q, et al. Role of arginine 293 and glutamine 288 in communication between catalytic and allosteric sites in yeast ribonucleotide reductase. J Mol Biol. 2012;419(5):315–29. doi: 10.1016/j.jmb.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmad MF, Wan Q, Jha S, Motea E, Berdis A, Dealwis C. Evaluating the therapeutic potential of a non-natural nucleotide that inhibits human ribonucleotide reductase. Mol Cancer Ther. 2012;11:2077–86. doi: 10.1158/1535-7163.MCT-12-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aye Y, Brignole EJ, Long MJ, et al. Clofarabine targets the large subunit (alpha) of human ribonucleotide reductase in live cells by assembly into persistent hexamers. Chem Biol. 2012;19(7):799–805. doi: 10.1016/j.chembiol.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ando N, Brignole EJ, Zimanyi CM, et al. Structural interconversions modulate activity of Escherichia coli ribonucleotide reductase. Proc Natl Acad Sci USA. 2012;108 (52):21046–51. doi: 10.1073/pnas.1112715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zimanyi CM, Ando N, Brignole EJ, Asturias FJ, Stubbe J, Drennan CL. Tangled up in knots: structures of inactivated forms of E. coli class Ia ribonucleotide reductase. Structure. 2012;20(8):1374–83. doi: 10.1016/j.str.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larsson KM, Jordan A, Eliasson R, Reichard P, Logan DT, Nordlund P. Structural mechanism of allosteric substrate specificity regulation in a ribonucleotide reductase. Nat Struct Mol Biol. 2004;11(11):1142–9. doi: 10.1038/nsmb838. [DOI] [PubMed] [Google Scholar]
- 48.Eliasson R, Pontis E, Sun X, Reichard P. Allosteric control of the substrate specificity of the anaerobic ribonucleotide reductase from Escherichia coli. J Biol Chem. 1994;269(42):26052–7. [PubMed] [Google Scholar]
- 49.Kumar D, Viberg J, Nilsson AK, Chabes A. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 2010;38 (12):3975–83. doi: 10.1093/nar/gkq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar D, Abdulovic AL, Viberg J, Nilsson AK, Kunkel TA, Chabes A. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 2010;39(4):1360–71. doi: 10.1093/nar/gkq829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy KP, Xie D, Garcia KC, Amzel LM, Freire E. Structural energetics of peptide recognition: angiotensin II/antibody binding. Proteins. 1993;15(2):113–20. doi: 10.1002/prot.340150203. [DOI] [PubMed] [Google Scholar]
- 52.Hofer A, Crona M, Logan DT, Sjoberg BM. DNA building blocks: keeping control of manufacture. Crit Rev Biochem Mol Biol. 2012;47(1):50–63. doi: 10.3109/10409238.2011.630372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aye Y, Stubbe J. Clofarabine 5′-di and -triphosphates inhibit human ribonucleotide reductase by altering the quaternary structure of its large subunit. Proc Natl Acad Sci USA. 2011;108(24):9815–20. doi: 10.1073/pnas.1013274108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cerqueira NM, Fernandes PA, Ramos MJ. Ribonucleotide reductase: a critical enzyme for cancer chemotherapy and antiviral agents. Recent Pat Anticancer Drug Discov. 2007;2(1):11–29. doi: 10.2174/157489207779561408. [DOI] [PubMed] [Google Scholar]
- 55.Huang P, Chubb S, Plunkett W. Termination of DNA synthesis by 9-beta-D-arabinofuranosyl-2-fluoroadenine. A mechanism for cytotoxicity. J Biol Chem. 1990;265(27):16617–25. [PubMed] [Google Scholar]
- 56.Avery TL, Rehg JE, Lumm WC, Harwood FC, Santana VM, Blakley RL. Biochemical pharmacology of 2-chlorodeoxyadenosine in malignant human hematopoietic cell lines and therapeutic effects of 2-bromodeoxyadenosine in drug combinations in mice. Cancer Res. 1989;49(18):4972–8. [PubMed] [Google Scholar]
- 57.Griffig J, Koob R, Blakley RL. Mechanisms of inhibition of DNA synthesis by 2-chlorodeoxyadenosine in human lymphoblastic cells. Cancer Res. 1989;49(24 Pt 1):6923–8. [PubMed] [Google Scholar]
- 58.Parker WB, Shaddix SC, Rose LM, et al. Comparison of the mechanism of cytotoxicity of 2-chloro-9-(2-deoxy-2-fluoro-beta-D-arabinofuranosyl)adenine, 2-chloro-9-(2-deoxy-2-fluoro-beta-D-ribofuranosyl)adenine, and 2-chloro-9-(2-deoxy-2,2-difluoro-beta-D-ribofuranosyl)adenine in CEM cells. Mol Pharmacol. 1999;55 (3):515–20. [PubMed] [Google Scholar]
- 59.Faderl S, Gandhi V, Keating MJ, Jeha S, Plunkett W, Kantarjian HM. The role of clofarabine in hematologic and solid malignancies—development of a next-generation nucleoside analog. Cancer. 2005;103(10):1985–95. doi: 10.1002/cncr.21005. [DOI] [PubMed] [Google Scholar]
- 60.Gandhi V. Clofarabine: a viewpoint by Varsha Gandhi. Paediatr Drugs. 2005;7(4):265–6. [Google Scholar]
- 61.Xie C, Plunkett W. Metabolism and actions of 2-chloro-9-(2-deoxy-2-fluoro-beta-D-arabinofuranosyl)-adenine in human lymphoblastoid cells. Cancer Res. 1995;55(13):2847–52. [PubMed] [Google Scholar]
- 62.Wang J, Lohman GJ, Stubbe J. Mechanism of inactivation of human ribonucleotide reductase with p53R2 by gemcitabine 5′-diphosphate. Biochemistry. 2009;48(49):11622–9. doi: 10.1021/bi901588z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langer CJ. The role of new agents in advanced non-small-cell lung carcinoma. Curr Oncol Rep. 2000;2(1):76–89. doi: 10.1007/s11912-000-0014-z. [DOI] [PubMed] [Google Scholar]
- 64.Squadroni M, Fazio N. Chemotherapy in pancreatic adenocarcinoma. Eur Rev Med Pharmacol Sci. 2010;14(4):386–94. [PubMed] [Google Scholar]
- 65.Yang FD, Spanevello RA, Celiker I, Hirschmann R, Rubin H, Cooperman BS. The carboxyl terminus heptapeptide of the R2 subunit of mammalian ribonucleotide reductase inhibits enzyme activity and can be used to purify the R1 subunit. FEBS Lett. 1990;272(1–2):61–4. doi: 10.1016/0014-5793(90)80449-s. [DOI] [PubMed] [Google Scholar]
- 66.Fisher A, Laub PB, Cooperman BS. NMR structure of an inhibitory R2 C-terminal peptide bound to mouse ribonucleotide reductase R1 subunit. Nat Struct Biol. 1995;2 (11):951–5. doi: 10.1038/nsb1195-951. [DOI] [PubMed] [Google Scholar]
- 67.Pellegrini M, Liehr S, Fisher AL, Laub PB, Cooperman BS, Mierke DF. Structure-based optimization of peptide inhibitors of mammalian ribonucleotide reductase. Biochemistry. 2000;39(40):12210–5. doi: 10.1021/bi001323a. [DOI] [PubMed] [Google Scholar]
- 68.Fisher A, Yang FD, Rubin H, Cooperman BS. R2 C-terminal peptide inhibition of mammalian and yeast ribonucleotide reductase. J Med Chem. 1993;36(24):3859–62. doi: 10.1021/jm00076a015. [DOI] [PubMed] [Google Scholar]
- 69.Liehr S, Barbosa J, Smith AB, 3rd, Cooperman BS. Synthesis and biological activity of cyclic peptide inhibitors of ribonucleotide reductase. Org Lett. 1999;1(8):1201–4. doi: 10.1021/ol9909381. [DOI] [PubMed] [Google Scholar]
- 70.Pender BA, Wu X, Axelsen PH, Cooperman BS. Toward a rational design of peptide inhibitors of ribonucleotide reductase: structure-function and modeling studies. J Med Chem. 2001;44(1):36–46. doi: 10.1021/jm000335r. [DOI] [PubMed] [Google Scholar]
- 71.Cooperman BS, Gao Y, Tan C, Kashlan OB, Kaur J. Peptide inhibitors of mammalian ribonucleotide reductase. Adv Enzyme Regul. 2005;45:112–25. doi: 10.1016/j.advenzreg.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 72.Sun D, Xu H, Wijerathna SR, Dealwis C, Lee RE. Structure-based design, synthesis, and evaluation of 2′-(2-hydroxyethyl)-2′-deoxyadenosine and the 5′-diphosphate derivative as ribonucleotide reductase inhibitors. ChemMedChem. 2009;4(10):1649–56. doi: 10.1002/cmdc.200900236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jordheim LP, Guittet O, Lepoivre M, Galmarini CM, Dumontet C. Increased expression of the large subunit of ribonucleotide reductase is involved in resistance to gemcitabine in human mammary adenocarcinoma cells. Mol Cancer Ther. 2005;4(8):1268–76. doi: 10.1158/1535-7163.MCT-05-0121. [DOI] [PubMed] [Google Scholar]
- 74.Jordheim LP, Seve P, Tredan O, Dumontet C. The ribonucleotide reductase large subunit (RRM1) as a predictive factor in patients with cancer. Lancet Oncol. 2011;12:693–702. doi: 10.1016/S1470-2045(10)70244-8. [DOI] [PubMed] [Google Scholar]
- 75.Rofougaran R, Crona M, Vodnala M, Sjoberg BM, Hofer A. Oligomerization status directs overall activity regulation of the Escherichia coli class Ia ribonucleotide reductase. J Biol Chem. 2008;283(51):35310–8. doi: 10.1074/jbc.M806738200. [DOI] [PubMed] [Google Scholar]