Abstract
Bacteroides fragilis, an opportunistic pathogen of humans, is a leading cause of bacteraemias and anaerobic abscesses which are often treated with metronidazole, a drug which damages DNA. This study investigated the responses of the B. fragilis recA three gene operon to the stress experienced during metronidazole treatment and exposure to reactive oxygen species simulating those generated by the host immune system during infection. A transcriptionally regulated response was observed using quantitative RT-PCR after metronidazole and hydrogen peroxide treatment, with all three genes being upregulated under stress conditions. In vivo and in vitro analysis of the functional role of the second gene of the operon was done using heterologous complementation and protein expression (in Escherichia coli), with subsequent biochemical assay. This gene encoded a functional bacterioferritin co-migratory protein (BCP) which was thiol-specific and had antioxidant properties, including protection of the glutamine synthetase III enzyme. This in vitro data supports the hypothesis that the genes of the operon may be involved in protection of the bacteria from the oxidative burst during tissue invasion and may play a significant role in bacterial survival and metronidazole resistance during treatment of B. fragilis infections.
Keywords: Bacteroides fragilis, Bacterioferritin co-migratory protein, recA
1. Introduction
The Bacteroides genus is one of 5 predominant groups of bacteria in the intestinal microbiome, accounting for around 30% of gut microbes [1; 2]. Bacteroides fragilis, a non-spore forming, Gram-negative, anaerobic rod represents only around 0.5% of the Bacteroides in the gut lumen where it grows as a commensal; it is, however, a virulent opportunistic pathogen [2]. It is isolated from the majority of clinical cases of bacterial septicaemia resulting from intestinal ruptures or surgeries and forms abscesses in the abdomen, pelvis, lungs and brain [3]. In order for B. fragilis to colonise the abdominal cavity, the cell has to survive high oxygen levels and the initial host immune onslaught. B. fragilis has been shown, in vitro, to have an extensive, complex and co-ordinated response to oxidative stress that involves at least 3 independent regulons, 28 proteins and alterations to its physiology at the metabolic level [3; 4; 5]. These genes may be transcriptionally responsive to oxygen, hydrogen peroxide or both [3]. In a second paper by this group, up to 45% of the transcriptome was shown to be alternatively regulated in response to oxidative stress [6]. Previous research by our group showed a link between the presence of the RecA protein and survival of B. fragilis cells in the presence of reactive nitrogen species (RNS) and reactive oxygen species (ROS) [7]. These observations suggest that the RecA protein from B. fragilis may also be important for enabling cell survival in the presence of the oxygen radicals associated with the innate immune response.
The B. fragilis recA gene was previously observed by RT-PCR to be transcribed, under normal growth conditions, on the same RNA transcript as two upstream open reading frames encoding a putative bacterioferritin co-migratory protein (BCP) and a putative saccharopine dehydrogenase (SDH) [7]. BCP proteins belong to the thiol-specific antioxidant (TSA) protein family [8]. These proteins are found in several bacteria where they catalyse the reduction of hydrogen peroxide and organic hydroperoxides [8; 9], thereby preventing free radical formation and the resultant cellular oxidation damage. B. fragilis has KatA (catalase), AhpC (alkyl hydroperoxidase) and six other Tpx (thioredoxin peroxidase) proteins which can serve this protective function [10]. It is not known whether its bcp gene product may act in a similar way. The role of the sdh gene product is also not clearly understood in B. fragilis.
In this study, the functions of the B. fragilis BCP were investigated. The ability of the annotated bcp gene to complement an Escherichia coli bcp− strain (KD2301) functionally was evaluated, and the protein was also heterologously expressed and biochemically assayed for substrate preference, thiol peroxidase dependence and protective properties for the vital glutamine synthetase enzyme from B. fragilis. The transcriptional response of the bcp gene to exposure to metronidazole and hydrogen peroxide was measured along with the recA and sdh genes using quantitative RT-PCR methods (qPCR).
2. Methods and materials
2.1. Bacterial strains, plasmids and growth conditions
B. fragilis 638R was grown in supplemented brain heart infusion broth (BHISB) or on plates (BHISA) at 37°C under anaerobic conditions [11]. All bacterial strains are described in Table 1. E. coli strains were grown in LB broth and plated on LB agar with appropriate antibiotic selection. E. coli KD2301 was grown with kanamycin (10 μg/ml) [12]. E. coli BL21DE3 was grown with no selection, while E. coli BL21DE3 and KD2301 strains expressing the pET22b1247pro plasmid were grown with ampicillin (100 μg/ml). All E.coli growth was under aerobic conditions at 30°C.
Table 1.
Strains and plasmids used in this study
| Strains/ plasmids | Relevant characteristic/use* | Source/reference |
|---|---|---|
| B. fragilis | ||
| 638R | Clinical strain, RifRGentR | 39 |
| E. coli | ||
| BL21DE3 | E. coli B strain with DE3, a λ prophage carrying the T7 RNA polymerase gene and lacIq | 40 |
| BL21DE3(pET22b(+)) | BL21DE3 with empty pET22b(+) vector | This study |
| BL21DE3 (pET22b1247pro) | BL21DE3 derivative with pET vector expressing full-length B. fragilis BCP | This study |
| KD2301 | BCP deficient E.coli mutant derived from BL21DE3 | 12 |
| KD2301(pET22b1247pro) | KD2301 derivative expressing full-length B. fragilis BCP | This study |
| Plasmids | ||
| pET22b(+) | pelB coding sequence, His-tag coding C-terminal | Novagen |
| pET22b1247pro | pET22b(+) plasmid with full-length BCP protein under IPTG induction | This study |
Rif=rifampicin, Gent=gentamycin; R=resistant
2.2. Bioinformatic analysis
Protein and DNA sequences were obtained from the National Centre for Biotechnology Information (www.ncbi.nih.gov). BLAST 2.2.17 [13] was used to calculate the predicted percentage identity between protein sequences for the CDS from B. fragilis 638R (NC_016776.1) for the 3 ORFs BF638R1245, BF638R1246/7 and BF638R1248 that make up the three gene cluster. Conserved domains database (CDD) [14] searches were used to identify conserved domains in the protein sequences. KEGG analysis [15, 16] was undertaken to establish whether the other enzymes in the metabolic pathways associated with the CDD protein domain searches were present in B. fragilis.
2.3 RNA isolation and northern blot analysis
RNA isolation and northern blot analysis were performed essentially as previously described [17]. The RNA was purified using the hot-phenol method except that chloramphenicol was not added to the culture prior to harvesting. For the northern blots, RNA (50 μg) was electrophoresed on 1% agarose gels containing, 1X MOPS (40 mM 3-[N-morpholino] propanesulphonic acid) and 2.2 M formaldehyde, and transferred to a nylon membrane. 32P labelled probes were added to the membrane and allowed to hybridise overnight at 42° C. Membranes were washed in decreasing concentrations of SSPE (5X – 0.1X) until low background radiation had been reached. DNA probes were labelled by random oligonucleotide priming with the incorporation of 32P-dCTP. The recA and bcp probes were derived from PCR fragments that encompassed the central portion of the respective gene. At least 106 cpm of labelled probe /ml of hybridisation solution were added for all hybridisations.
2.4 Quantitative RT-PCR
2.4.1. Sample preparation and storage, and primer design
B. fragilis 638R was grown to mid-log phase (OD600=0.6) and then half of the culture was exposed to either 100 μM H2O2 or 1 μg/ml metronidazole. The other half of the culture was used as the uninduced control. Samples of 100 ml were taken for each treatment at time points 0, 15, 30 and 60 min. Three biological replicates were performed and each separated into three technical repeats for RNA extraction. RNA was extracted using the hot phenol method of Aiba et al. [18] with the following modifications: after 16 h precipitation of the RNA at −20°C, a DNase1 treatment was performed at 37°C for 3 h. Purification and final RNA precipitation were done using the Qiagen Total RNA kit (WhiteSci). DNA contamination and integrity of the RNA were evaluated by standard PCR of the 16S rRNA gene using the universal F27/R5 primer pair combination and the RNA preparation as template [7] (Table 2). cDNA conversion was undertaken using the First Strand cDNA synthesis kit (Fermentas) as per the manufacturer's instructions. Primers generating products of 100 bp were designed (Table 2) using Beacon primer design (Premier Biosoft). These were synthesised and purified using HPLC methods (University of Cape Town Oligo Synthesis Service) and their site specificity was tested using BLAST [13] as well as by standard PCR methods.
Table 2.
Primers used in this study
| Primer | Sequence (5' – 3') | Characteristics/use | Source |
|---|---|---|---|
|
| |||
| F27 | AGAGTTTGATCITGGCTCAG | 16S rRNA primers | 41 |
| R5 | ACGGITACCTTGTTACGACTT | ||
|
| |||
| 12F | ATATGCCTCTTCGTGACTAC | qPCR SDH primer | This study |
| 12R | GGATTAGGTAACACGGCTT | ||
|
| |||
| 13F | AGCGTGTTCTTTGGTCTTTAC | qPCR BCP primer | This study |
| 13R | CCATACTCCAAATTGCTCTAC | ||
|
| |||
| 14F | CAGTCAAGGCGGCTACAGAG | qPCR RecA primer | This study |
| 14R | CAGTTTAGCCGCATAGAAGC | ||
|
| |||
| 15F | ACACGGTCCAAACTCCTAC | qPCR 16S rRNA | This study |
| 15R | GTGAAGGATGAAGGCTCTAT | primer | |
|
| |||
| BF1247proF | CATCAACCATGGATGTAGGAGATAAAGC Ncol | Full-length primers for bcp gene used in protein expression | This study |
| BF1247proR | CGATCTCGAGAATTTGTAGAGCG Xhol | ||
2.4.2 qPCR reaction conditions, controls and optimisation
The design and implementation of the qPCR experiment were done according to the relevant MIQE guidelines [19] using the Rotogene 6000 (Corbet) 96 tube rotor. The final cycle conditions were as follows: 95°C for 10 min, 95°C for 15 s, 50°C for 20 s, and 72°C for 20 s for 45 cycles. The final reaction mix was as follows: 0.25 μl SYBR green (Celtic Diagnotstics), 1 μl cDNA template, 0.1 μl forward primer (0.5 μM), 0.1 μl reverse primer(0.5μM), 6.2 μl Sensimix (Celtic Diagnostics), and 4.85 μl MilliQ water to give a final reaction volume of 12.5 μl.
Three biological samples of each set of conditions were tested in technical triplicate. The standard curves were created by mixing the cDNA from all three technical repeats at each time point under each condition, and then a dilution series from 100–10−8 was made with sterile MilliQ water in technical duplicate for each primer pair. The controls for each run included a no template control, an RNA template control, and a genomic DNA (gDNA) control.
2.4.3. qPCR data analysis
Rotor-gene 6000 series software 1.7 was used for the primary portions of data analysis. Using the standard curve, the relative cDNA concentration was determined for each sample, at each time point, and with each primer pair. The 16s rRNA gene was used as the internal standard. A mean value for each biological repeat was established for each time point. These values were then normalised against the calibrated 16S rRNA value [19]. The relative abundance of each gene at each time point was calculated and compared to the uninduced (T0) value. The relative increases in these values were then evaluated for statistical significance [20].
2.5 Heterologous BCP expression and purification
BCP expression was undertaken using the pET expression system (Novagen) and plasmids were expressed in E. coli BL21DE3. Full-length sequence from BF638R1246/7 was obtained by PCR using BF1247proF and BF1247proR (Table 2). The PCR was carried out using Kappa Ready Mix according to the following parameters: initial denaturation of 95°C for 5 min, then 25 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s and elongation at 72°C for 3 min. A final elongation step was carried out at 72°C for 5 min. The full-length bcp gene was directionally cloned into the pET22b (+) (Novagen) vector. Cells were grown to an OD600 of 0.4 and induced with 1 mM IPTG for 3 h. The soluble cell-free extract was passed through a nickel-affinity chromatography column (His-Select Nickel Affinity gel, Sigma). The column with bound protein was sequentially washed according to the manufacturer's instructions with an additional 30 mM imidazole (50 mM NaH2PO4, 300 mM NaCl, 30 mM imidazole) wash prior to elution. The bound, purified protein was eluted using 250 mM imidazole (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole) and the purity of the BCP protein was assessed using both SDS-PAGE and His-tag western blots. Purified B. fragilis BCP was used for all of the assays. Pierce Zeba Spin desalting columns (Thermo Scientific) were used to exchange the 250 mM imidazole elution buffer for the assay buffer of 50 mM Hepes (Sigma).
2.6. Heterologous complementation of E. coli bcp− with the cloned B. fragilis bcp gene
The E. coli KD2301 [pET22b (+)] (bcp− mutant), E. coli BL21DE3 [pET22b (+)] (wild type) and E. coli KD2301 (pET22b1247pro) (complemented bcp– mutant) strains were used. The complementation of all E. coli strains was accomplished by transforming competent E. coli cells with the pET22b1247pro plasmid which expresses the full-length BCP protein from B. fragilis. Transformed cells were grown in Luria broth with 10 μg/ml ampicillin to an OD600 of 0.4 and 1 mM IPTG was added to all cultures. Cells were then grown until cultures reached an OD600 of 0.8. One millilitre of the culture was removed, centrifuged and the pellet resuspended in PBS pH 7.4. H2O2 (Sigma-Aldrich) was then added to a final concentration of 100 μM. Cells were sampled at 5 min intervals over a 15 min time period, diluted in sterile distilled water (10−1 to 10−8) and plated on Luria agar without antibiotic. The plates were incubated aerobically at 30°C for 1 day and the surviving fraction of cells was calculated for each time point. All experiments were done in biological triplicate and technical duplicate. Statistical significance was determined using the student t-test for statistical significance at p <0.05.
2.7. Biochemical assays
2.7.1. Thiol-dependent peroxidase activity of BCP
The thiol-dependent peroxidase activity of purified BCP was evaluated using the method of Jeong et al. [12] (thioredoxin + thioredoxin reductase + BCP + NADPH + substrate) with the following modifications: 8 μM of purified BCP was used in all experiments; 150 μM t-butyl hydroperoxide, 100 μM H2O2, and 100 μM linoleic acid hydroperoxide were used as substrate. Recombinant thioredoxin 1 (Trn1) and thioredoxin reductase 1 (Trx1) (Sigma) were added at published concentrations [12]. Activity of BCP was observed as a decrease in total peroxide concentration. Fox 1, a redox-sensitive indicator, was used to spectrophotometrically measure the total peroxide in each reaction as described previously [12; 21].
2.7.2. Antioxidant characteristics of the B. fragilis BCP
The antioxidant properties of the BCP were tested using partially purified glutamine synthetase III (GSIII) from B. fragilis as the target enzyme. Purification of GSIII was done according to the method of van Rooyen et al. [22]. The design of the antioxidant assay was based on the work of Jeong et al. [12] with the following modifications: 8 μM of purified BCP protein was used in all experiments; 100 μM H2O2, Trn1 (Sigma), and Trx1 (Sigma) were added at published concentrations [12]. GSIII was exposed to H2O2 in the presence of BCP alone (with NADPH), BCP with both thioredoxin and thioredoxin reductase (with NADPH), or with just thioredoxin and thioredoxin reductase (with NADPH) and assayed for activity. This was compared to GSIII activity under the same conditions but in the absence of H2O2. The activity of GSIII was assayed every 10 min for 30 min using the γ- glutamyltransferase assay [23].
3. Results and discussion
3.1. Bioinformatics
Our previous RT-PCR studies confirmed that the 3-gene cluster (recA and the two upstream ORFs) was transcribed under normal growth conditions as a single transcript [7]. Bioinformatic analysis of the two upstream genes in the recA operon was therefore undertaken using protein domain homology and KEGG analysis in order to achieve a better understanding of their potential functional roles in association with recA (Fig. 1 and 2).
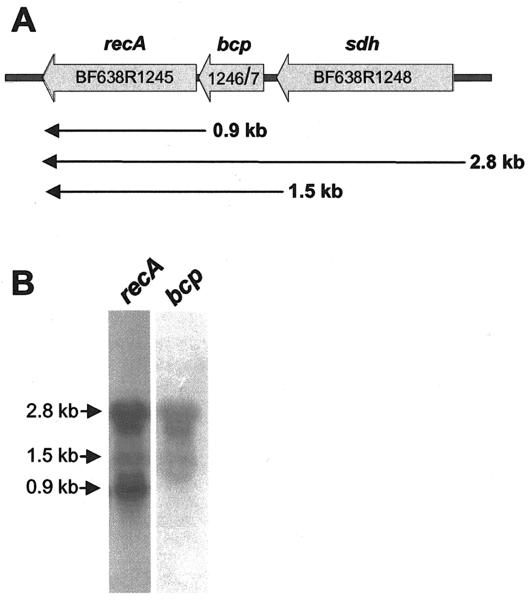
Fig. 1. Genetic and transcriptional organization of the recA, bcp, sdh locus.
A) Genetic map showing a 3.2 kb region of the BF638R chromosome containing the location of the recA, bcp and sdh genes (thick arrows). The map is drawn to scale and the thin arrows under the map indicate the approximate size of the mRNA transcripts identified by northern hybridization. B) Northern hybridization analyses showing results using either a recA or bcp probe. The location of the 2.8, 1.5 and 0.9 kb mRNA species is marked at the edge of the autoradiographs. RNA was obtained from mid-logarithmic phase cells cultured in BHISB.
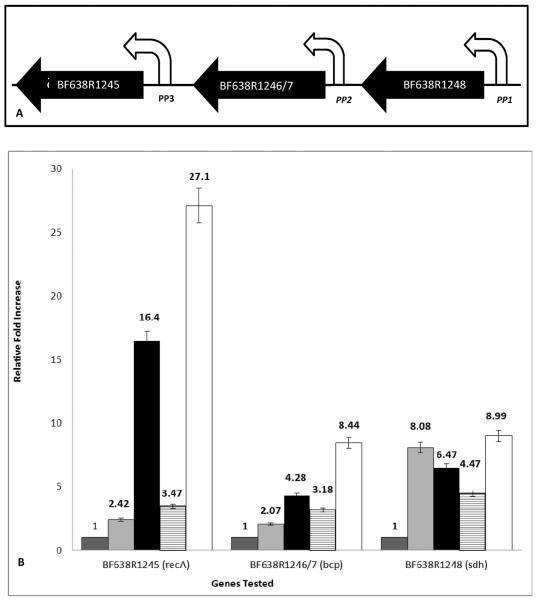
Fig. 2. (A) Schematic representation of the three genes making up the recA operon of B. fragilis.
BF638R1245 (recA); BF638R1246/7 (bcp); and BF638R1248 (sdh). White arrows indicate the promoters for each gene and the transcription direction. Putative promoters are shown in italics. (B) Relative fold increase in transcription of the three genes in the recA operon of B. fragilis. Transcription of BF638R1245; BF638R1246/7 and BF638R1248 was evaluated over time either being untreated (dark grey), after the addition of 1 μg/ml of metronidazole for 30 min (light grey) or 60 min (black), or after exposure to 100 μM H2O2 for 15 min (horizontal bars) or 30 min (white). Error bars represent the standard error of the mean.
A predicted role for the protein encoded by the BF638R1246/7 CDS was determined by doing a protein BLAST. This showed that it had amino acid homology to a large number of proteins belonging to the thioredoxin-like superfamily. Conserved domain database searches identified a conserved PRX-BCP domain as well as other typical thioredoxin domains corresponding to all the major subgroups of the TSA peroxiredoxin superfamily of proteins. This suggested a conservation of the functional role of this protein to those predicted by the presence of the PRX catalytic site. The B. fragilis BCP shows extremely high amino acid homology to the described BCP proteins of other anaerobic bacteria, including Prevotella spp. (100%) and Porphyromonas spp. (100%). Homology to the AhpC peroxidase domains and the alkylhydroperoxide reductase protein supports a similar overlapping role for the B. fragilis BCP with that of the H. pylori BCP [24].
The members of the thiol-specific antioxidant (TSA) protein family are fairly ubiquitous and catalyse the reduction of organic hydroperoxides and H2O2 [8; 9]. This reduction prevents free radical formation and oxidative damage to metabolic processes, cellular machinery and DNA. B. fragilis has a number of TSA proteins including AhpC and 2 thiol peroxiredoxin genes. However, the putative BCP encoded by BF638R1246/7 appears to be the sole BCP protein in this bacterium. The H. pylori BCP protein has been shown to be less specific in its substrate choice and less prevalent within the cell than its TSA counterpart, AhpC [8] and seems to act only in situations where the oxidative exposure is acute [8; 9]. In general, BCP acts to “mop up” oxidised iron by protecting the pool of reduced iron that has been released from the dehydratase iron sulphate active clusters. These clusters are associated with the Fe-S metalloenzymes that are central to anaerobic metabolism. This prevents widespread distribution of ferric iron which could be used in damaging Fenton reactions [25; 26]. The BCP protein also acts as an electron acceptor/donor accessory protein in the thioredoxin reduction of sulphur to remove oxygen radicals from the bacterial species [25; 26]. The functionality of the putative B. fragilis BCP was further investigated in this study.
Protein BLAST and conserved domain database searches of the putative protein encoded by the first gene of the operon, ORF BF638R1248, showed high amino acid similarity to the saccharopine dehydrogenases, lysine α-ketogluterate reductases and carboxynorspermidine synthase proteins found in other anaerobic bacteria, including a number of Prevotella spp. (97%), Porphyromonas spp. (98%), Clostridium spp. (95%) and Bacteroides spp. (98–100%). KEGG pathway analysis revealed a number of genes that encode putative enzymes that may be associated with lysine metabolism throughout the amino adipate pathway (Bfr0300 and Bfr01110 from the B. fragilis 638R strain). This pathway also shows homology to the eukaryote amino-adipate pathway for biosynthesis [27] which is not commonly employed by bacteria but has been described in Silicibacter pomeroyi as an alternative method for lysine degradation [28]. Homologues of the S. pomeroyi sdh gene have been identified in a number of closely related Cyanobacteria and Bacteroidetes where it often clusters with genes associated with oxidative stress response [29]. However, the exact role of the SDH protein in this response remains unclear. The sdh has never previously been observed to be associated in an operon with recA and bcp, as has been observed in B. fragilis.
3.2 Northern blot analysis
Northern hybridisation clearly shows that transcription of the 3 gene operon (Fig 1A) can be complex. The last gene in the operon is recA and when this was used as a probe, three distinct mRNA species were observed (Fig. 1B). The 2.8 kb transcript corresponds to transcription of the entire operon and a 1.5 kb mRNA is consistent with a transcript that includes only bcp and recA. In addition, a small 0.9 kb mRNA species matches the size of the recA gene alone, suggesting that this gene can be transcribed independently. Support for this was observed when the bcp gene was used as a probe. As shown in Fig. 1B, both the 2.8 and 1.5 kb mRNA species were present; however, the smallest RNA was missing, suggesting that this was in fact a recA-specific mRNA.
3.3. Quantitative qPCR
In view of the predicted links between the 3 genes of the recA operon and oxidative stress, the transcriptional responses of the recA and its associated genes were examined using qPCR following H2O2 and metronidazole (Mtz) treatment. Analysis using the Pfaffl criteria [20] revealed a statistically significant increase (more than 2-fold) in the transcriptional levels of all 3 genes under both types of inducing conditions when compared to the uninduced state of cell growth. After Mtz induction, all three genes were upregulated after 30 or 60 min of exposure as follows: sdh 8.08- and 6.47-, bcp 2.07- and 4.28- and recA 2.42- and 16.43-fold increase, respectively (Fig. 2B). After H2O2 exposure, a similar pattern of induction was observed and all 3 genes were transcriptionally upregulated at 15 and 30 min of exposure as follows: sdh 4.47-and 8.99-, bcp 3.18- and 8.44- and recA 3.47- and 27.1-fold, respectively.
Taken together with the previous RT-PCR findings [7] and northern blot analysis (Fig 1), co-transcription of the 3 genes observed in the qPCR experiment confirm that the 3 genes of the operon function together in a transcriptionally regulated manner as well as independently in response to the stressors tested. The occurrence of a recA gene in an operon has been observed in a number of bacterial species including Porphyromonas gingivalis [29], which has a large recA operon that includes bcp, vimA, E and F. This operon is crucial for survival of this pathogen in the oral cavity, especially in incidences of oxidative stress and the establishment of infection [29]. The transcriptional regulation of the B. fragilis sdh, bcp and recA observed in this study has shown that the operon is responsive to oxygen and nitrogen radicals which simulate those associated with the inflammatory response in the human host [30]. The gene products may therefore assist this bacterium in surviving a transient oxidative burst while traversing the endothelial wound and surviving in the oxygen-rich abdominal cavity before abscess formation generates an anoxic environment [2; 6; 31].
The different transcriptional levels of recA, bcp and sdh, associated with different agents of stress and also in relation to each other, are interesting and suggest that the transcription of these three genes as a single transcriptional unit may be a conditional event. This conclusion is supported by the northern blot analysis which showed multiple mRNA transcripts corresponding to differential gene expression within the operon (Fig 1B), and the fact that the degree to which each of these genes is transcribed is dependent on the inducing agent [32]. In addition, the data indicate that the recA gene, although expressed as part of an operon with the two upstream genes under normal growth conditions (Fig 1B) [7], can be induced much more strongly than the others under both stress conditions (Fig 2B). This suggests that an additional promoter-like sequence exists in the bcp/recA intergenic region. This finding is supported by the fact that the 113 bp fragment immediately upstream of recA has previously been shown to function as an active promoter, as determined by promoter fusion and primer extension analyses [33]. Northern blot analysis also showed an independent mRNA transcript corresponding to only the recA gene length. RecA can therefore be expressed from the operon promoter P1 (Fig. 2A) as well as from its own promoter P3 (Fig. 2A). To date no bioinformatic studies of this region have shown a clear promoter in regions P1 and P2, but experimental data suggest that a non-canonical promoter region could be present.
This arrangement would allow the benefits of an operon for induction under stress conditions, but allows transcription of an important cellular maintenance gene, recA, to be tightly regulated under normal cellular conditions without producing unnecessary gene products. This phenomenon, termed a “conditional operon”, has been described in Mycobacterium bovis BCG in the Rv3134c/devR/devS operon where at least 3 different promoters have been identified for controlling expression under different conditions. Rv3134c and devR are only co-transcribed under hypoxia, while devR and devS are co-transcribed under hypoxic and starvation conditions [32]. In addition, the lolA and trxB genes of the TrxB operon of B. fragilis showed differential regulation at the transcriptional level [31]. The transcriptional profiles generated in this study suggest that the B. fragilis recA gene may belong to a similar conditional operon.
3.4. Heterologous complementation of an E. coli BCP mutant
In order to better understand the role of this operon under stress conditions, the functional characterisation of one of the upstream genes, bcp, was undertaken using heterologous protein expression, biochemical assay and E. coli mutant complementation. Mutational analyses, using both an insertion and a deletion method, were attempted in B. fragilis 638R, but stable B. fragilisbcp− mutants were never isolated and were possibly lethal to the cell.
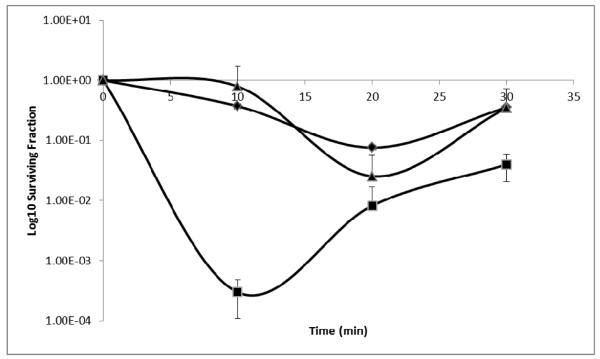
The E. coli KD2301 bcp− strain was created in a study by Jeong et al. [12] and has been used to characterise a number of BCP proteins from other bacteria including P. gingivalis [29]. BCP-deficient E. coli KD2301 showed a high degree of sensitivity to H2O2 during the first 10 min of exposure (Fig. 3), but prolonged exposure resulted in increased survival. This recovery phenomenon was reported previously in this E. coli mutant and has been attributed to the compensatory induction of other oxygen stress-responsive genes. These could include the E. coli catalase system as well as the alkyl hydroperoxidases which respond to the damage associated with H2O [12]. In this study, the E. coli bcp− mutant was complemented with the B. fragilis bcp gene. The complemented mutant strain exhibited a phenotype that was identical to that of the wild-type E. coli strain with very little decrease in survival over the first 10 min of H2O2 exposure (Fig. 3), conclusively demonstrating a functional role for the B. fragilis BCP in in vitro protection against oxygen stress, particularly during the initial stages of exposure.
Fig. 3. Heterologous complementation of an E. coli BCP mutant with BCP from B. fragilis after exposure to 100 μM hydrogen peroxide.
E. coli strains:◆BL21DE3 (wild type);▴KD2301 with pET22b1247pro (bcp complement);∎KD2301 (bcp− mutant)
3.5. Thiol-dependent peroxidase activity of B. fragilis BCP
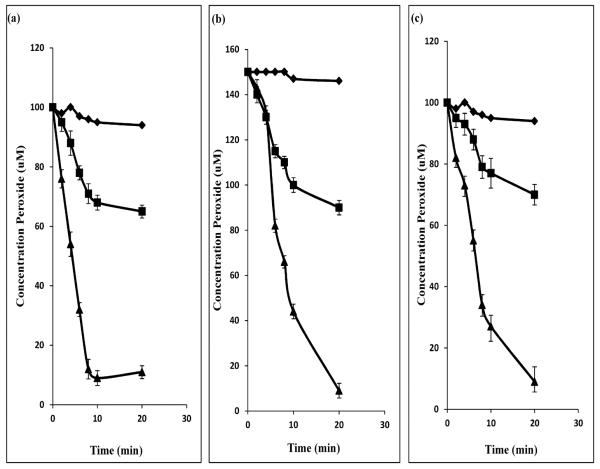
The BCP enzymes from H. pylori, E. coli, P. gingivalis and C. jejuni have been described as thiol-dependent peroxidases [8; 12; 29; 34]. They all exhibit a reliance on NADPH and require the presence of thioredoxin reductase (Trx1) and thioredoxin (Trn1) [35] for catalysis. In order to test whether the B. fragilis BCP met these criteria, an experiment was undertaken using 3 different substrates, namely, H2O2, t-butyl hydroperoxide and linoleic acid, which were treated with soybean peroxidase to produce linoleic acid hydroperoxide. BCP activity was assayed by measuring the OH− (peroxide radical) concentration of the reaction mix using the redox-sensitive Fox 1 reagent. BCP activity was measured in the presence and absence of the thioredoxins and NADPH (Fig. 4). The purified B. fragilis BCP showed activity against all 3 substrates and no preference for a single substrate could be determined. In the absence of only one of the thiolreductases, the activity against H2O2 and t-butyl hydroperoxide was greatly reduced. However, the activity against the linoleic acid was not significantly affected by the absence of either one of the thiol-reductases; only a single reductase was required (results not shown). The BCP activity observed in the absence of the thioredoxin proteins may be the result of rapid partial degradation of the peroxide substrates [36]. This suggests that there may be a substrate preference for the organic peroxides over the more complex inorganic compounds. The B. fragilis BCP showed no activity against any of the peroxide compounds in the absence of NADPH or both of the thiol reductases. It may therefore be concluded that this protein is acting in the same way as previously described BCPs and is thus a thiol-reductase-dependent peroxidase enzyme belonging to the peroxiredoxin (Prx) protein superfamily.
Fig. 4. Activity of BCP from B. fragilis using various substrates.
(a) Linoleic acid hydroperoxide 100 μM; (b) t-butyl hydroperoxide 150 μM (c) hydrogen peroxide 100 μM.∎B. fragilis BCP alone;▴Trn1 and Trx1 alone;◆Trn1, Trx1 and BCP. Trn1: thioredoxin1; Trx1: thioredoxin reductase1.
Prx proteins are strongly expressed, ubiquitous, Cys-dependent peroxidases [12; 35] having oligomeric kinetics and substrate preferences [8; 24; 35]. Since they have the ability to interact with a wide variety of substrates in terms of hydroperoxide and its reducing equivalent, BCP can remain active and retain efficacy under a wide variety of cellular conditions [35]. BCPs are more flexible in their reducing pathways than many other classes of antioxidant proteins, allowing for the high reducing capacity of this protein family, even in very unfavourable conditions [35].
The ability of BCP to recognise and act on a variety of compounds is advantageous (Fig. 4). Its small size (21 kDa) and rapid transcriptional upregulation after exposure to an oxidising agent (Fig. 2) make it an ideal candidate to act as an electron sink in cases of acute cellular exposure to oxygen stress.
3.6. The antioxidant potential of B. fragilis BCP
One of the major roles of the thiol-dependent peroxidase proteins such as BCP has been shown to be the protection of other oxygen-sensitive enzymes from oxidation. Glutamine synthetase is an example of such an oxygen-sensitive key catalytic enzyme of anaerobic metabolism. It is responsible for the synthesis of glutamine from glutamate, making it vital to cellular survival [22]. This enzyme relies on an Fe-S cluster at its catalytic centre to facilitate its metabolic function and is thus highly sensitive to oxidative damage [37] making it an ideal target for evaluating the potential of the BCP to act as an antioxidant protein.
The glnN gene encoding the B. fragilis glutamine synthetase and its product (GSIII) have previously been well characterised [22]. In this study, GSIII activity was further examined by assaying it after exposure to 100 μM H2O2 in the presence and absence of BCP. The BCP, in combination with its thioreductases, Trn1 and Trx1, was able to protect the activity of GSIII during exposure to H2O2, allowing close to 100% of GS activity to be achieved within 30 min post-exposure (Fig. 5). However, BCP alone, without the addition of the other thiol peroxidases, also maintained approximately 70–80% of the GSIII's wild type activity (Fig. 5), indicating that the BCP can act independently to protect an oxygen-sensitive enzyme from damage, possibly due to the natural degradation of the hydrogen-peroxide-producing degradation intermediates which BCP is able to degrade without the catalytic effects of the thioredoxin proteins [36]. The contribution of the thioredoxins without BCP was also evaluated and approximately 80–90% of activity was maintained. These enzymes therefore also make their own contribution to GS enzyme oxygen stress stability. An additional 10% GS activity was, however, gained by the joint activities of the BCP together with the Trn1 and Trx1. In the absence of any of these proteins, over 80% of the GSIII activity was lost following H2O2 treatment. The ability of BCP to protect the GSIII activity can be attributed to the peroxidase activity exhibited by the BCP protein in earlier experiments. A similar effect of H2O2 on GSI activity was previously seen in E. coli, although the independent activity of BCP against H2O2 is unique to this study [12].
Fig. 5. Effect of BCP on the recovery properties of the B. fragilis GSIII protein following exposure to hydrogen peroxide.
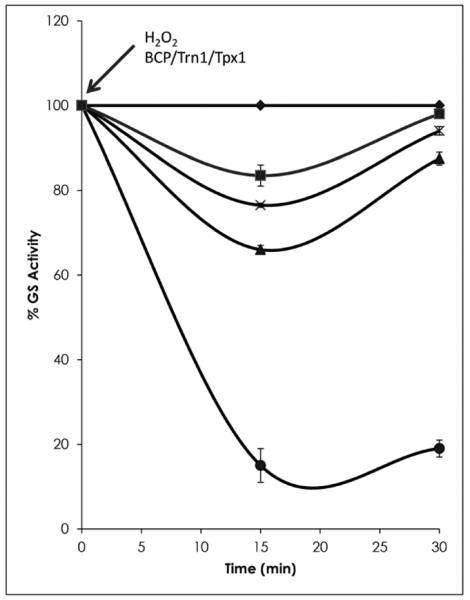
Protective properties of BCP for FeS enzymes during exposure to hydrogen peroxide demonstrated using the relative percentage activity of the B. fragilis GSIII protein.◆GS without peroxide;∎GS exposed to H2O2 in the presence of Trn1, Trx1 and BCP; x GS exposed to H2O2 in the presence of Trn1 and Trx1;▴GS exposed to H2O2 in the presence of BCP; and●GS and H2O2 only. Trn1: thioredoxin 1; Trx1: thioredoxin reductase 1 (Sigma). Error bars represent standard deviation. Arrows indicate the time of addition of various components.
The antioxidant activity of BCP in the protection of the redox-sensitive GSIII protein (Fig. 5) suggests its probable function within the in vivo environment of B. fragilis and could possibly include other Fe-S metalloenzyme targets such as PFOR [25]. This would help in the maintenance of anaerobic metabolism in the face of the oxidative burst resulting either from fluctuations within the gut microcosm or from exposure to the inflammatory response [38]. The ability of a cell to maintain metabolic function, even under severe oxidative stress, is vital for its continued survival [6; 26].
The research described in this paper, together with our previous findings on the role of the RecA protein [7], suggest that the recA operon may play a role in facilitating metabolic continuity and genomic integrity during oxidative stress by enhancing the reduction of hydroperoxides and preventing lipid oxidation and DNA damage during B. fragilis translocation through the wounded epithelium or after exposure to the high oxygen environment of the abdominal cavity. Future in vivo studies of this pathway are warranted in order to better elucidate the functional role of this novel conditional operon in the pathogenicity of B. fragilis.
Acknowledgements
S.A.N acknowledges bursary support from the National Research Foundation (NRF-DAAD) and the Carnegie Melon Foundation. This work was supported by the National Research Foundation, the Medical Research Council of South Africa, the University of Cape Town and the Carnegie Melon Foundation, as well as a Public Health Service grant AI40588 to C.J.S. from the National Institutes of Health. The authors would like to acknowledge Professor Hansel Fletcher of Loma Linda University for the use of the E. coli KD2301 strain.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Arumugam M, Raes E, Pelletier P, Le T, Yamada D, Mende G, Fernandes P, Bork, et al. Enterotypes of the human gut microbiome. Nat. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wexler H. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20(4):593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rocha E, Herren C, Smalley D, Smith C. The complex oxidative stress response of Bacteroides fragilis: the role of OxyR in control of gene expression. Anaerobe. 2003;9(4):165–173. doi: 10.1016/S1075-9964(03)00118-5. [DOI] [PubMed] [Google Scholar]
- [4].Meehan B, Malamy M. Fumarate reductase is a major contributor to the generation of reactive oxygen species in the anaerobe Bacteroides fragilis. Microbiol. 2012;158(2):539–546. doi: 10.1099/mic.0.054403-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wei Q, Le Minh P, Dötsch A, Hildebrand F, Panmanee W, Elfarash A, Schulz S, et al. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res. 2012:1–14. doi: 10.1093/nar/gks017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sund C, Rocha E, Tzianabos A, Wells W, Gee J, Reott M, O'Rourke D, et al. The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol Microbiol. 2008;67(1):129–142. doi: 10.1111/j.1365-2958.2007.06031.x. [DOI] [PubMed] [Google Scholar]
- [7].Steffens L, Nicholson S, Paul L, Nord C, Patrick S, Abratt V. Bacteroides fragilis RecA protein overexpression causes resistance to metronidazole. Res in Microbiol. 2010;161(5–3):346–354. doi: 10.1016/j.resmic.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Comtois S, Gidley M, Kelly D. Role of the thioredoxin system and the thiol-peroxidases Tpx and Bcp in mediating resistance to oxidative and nitrosative stress in Helicobacter pylori. Microbiol. 2003;149(1):121–129. doi: 10.1099/mic.0.25896-0. [DOI] [PubMed] [Google Scholar]
- [9].Lai Y, Yang J, Lin T, Lin J, Wang T. Helicobacter pylori infection and CagA protein translocation in human primary gastric epithelial cell culture. Helicobacter. 2006;11(5):451–459. doi: 10.1111/j.1523-5378.2006.00438.x. [DOI] [PubMed] [Google Scholar]
- [10].Reott M, Parker A, Rocha E, Smith C. Thioredoxins in redox maintenance and survival during oxidative stress of Bacteroides fragilis. Journal of Bacteriol. 2009;191(10):3384–3391. doi: 10.1128/JB.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Moore W, Holdeman L. Identification of anaerobic bacteria. American Journal of Clinical Nutrition. 1972;25(12):1306–1313. doi: 10.1093/ajcn/25.12.1306. [DOI] [PubMed] [Google Scholar]
- [12].Jeong W, Cha M, Kim I. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/alkyl hydroperoxide peroxidase C (AhpC) family. Journal Biological Chem. 2000;275(4):2924–2930. doi: 10.1074/jbc.275.4.2924. [DOI] [PubMed] [Google Scholar]
- [13].Altschul S, Gish W, Miller W, Myers W, Lipman J, et al. Basic local alignment search tool. Journal of Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- [14].Marchler-Bauer A, Lu S, Anderson J, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39(Database Issue):D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular datasets. Nucleic Acids Res. 2011;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rocha E, Smith C. Regulation of Bacteroides fragilis katB mRNA by Oxidative Stress and Carbon Limitation. Journal of Bacteriol. 1997;179(22):7033–7039. doi: 10.1128/jb.179.22.7033-7039.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. Journal of Biological Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- [19].Bustin S, Benes V, Garson J, Hellemans J, Huggett J, Kubista M, Mueller R, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- [20].Pfaffl M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45–e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim K, Kim I, Lee K, Rhee S, Stadtman E. The isolation and purification of a specific “protector” protein which inhibits enzyme inactivation by a thiol/Fe (III)/O2 mixed-function oxidation system. Journal of Biological Chem. 1988;263(10):4704–4711. [PubMed] [Google Scholar]
- [22].van Rooyen J, Abratt V, Belrhali H, Sewell B. Crystallization of recombinant Bacteroides fragilis glutamine synthetase (GlnN) isolated using a novel and rapid purification protocol. Protein Express Purificat. 2010;74(2):211–216. doi: 10.1016/j.pep.2010.06.007. [DOI] [PubMed] [Google Scholar]
- [23].Bender R, Janssen K, Resnick A, Blumenberg M, Foor F, Magasanik B. Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. Journal of Bacteriol. 1977;129(2):1001–1009. doi: 10.1128/jb.129.2.1001-1009.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang G, Olczak A, Walton J, Maier R. Contribution of the Helicobacter pylori thiol peroxidase bacterioferritin comigratory protein to oxidative stress resistance and host colonization. Infect and Immun. 2005;73(1):378–384. doi: 10.1128/IAI.73.1.378-384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Imlay J. Pathways of oxidative damage. Ann Rev in Microbiol. 2003;57(1):395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- [26].Imlay J. Cellular defences against superoxide and hydrogen peroxide. Annual Rev of Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nishida H, Nishiyama M, Kobashi N, Kosuge T, Hoshino T, Yamane H. A prokaryotic gene cluster involved in synthesis of lysine through the amino adipate pathway: a key to the evolution of amino acid biosynthesis Genome res. 1998;9(12):1175–1183. doi: 10.1101/gr.9.12.1175. [DOI] [PubMed] [Google Scholar]
- [28].de Mello Serrano G, Rezende e Silva Figueira T, Kiyota E, Zanata N, Arruda P. Lysine degradation through the saccharopine pathway in bacteria: LKR and SDH in bacteria and its relationship to the plant and animal enzymes. FEBS Letters. 2012;586:905–911. doi: 10.1016/j.febslet.2012.02.023. [DOI] [PubMed] [Google Scholar]
- [29].Johnson N, McKenzie R, Fletcher H. The bcp gene in the bcp-recA-vimA-vimE-vimF operon is important in oxidative stress resistance in Porphyromonas gingivalis W83. Mol Oral Microbiol. 2011;26:62–77. doi: 10.1111/j.2041-1014.2010.00596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Miller R, Britigan B. Role of oxidants in microbial pathophysiology. Clin Microbiol Rev. 1997;10(1):1–18. doi: 10.1128/cmr.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rocha E, Tzianabos A, Smith C. Thioredoxin reductase is essential for thiol/disulfide redox control and oxidative stress survival of the anaerobe Bacteroides fragilis. Journal of Bacteriol. 2007;189(22):8015–8023. doi: 10.1128/JB.00714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rodriguez J, Burbano C, Nunez C, González C, Zambrano M, Garcia M, Del Portillo P. Rv3134c/devR/devS operon of Mycobacterium bovis BCG is differentially transcribed under “in vitro” stress conditions. Tuberculosis. 2008;88(4):273–282. doi: 10.1016/j.tube.2007.11.011. [DOI] [PubMed] [Google Scholar]
- [33].Goodman H, Woods D. Molecular analysis of the Bacteroides fragilis recA gene. Gene. 1990;94(1):77–82. doi: 10.1016/0378-1119(90)90470-c. [DOI] [PubMed] [Google Scholar]
- [34].Atack J, Kelly D. Contribution of the stereospecific methionine sulphoxide reductases MsrA and MsrB to oxidative and nitrosative stress resistance in the food-borne pathogen Campylobacter jejuni. Microbiol. 2008;154(8):2219–2230. doi: 10.1099/mic.0.2008/019711-0. [DOI] [PubMed] [Google Scholar]
- [35].Reeves S, Parsonage D, Nelson K, Poole L. Kinetic and thermodynamic features reveal that E. coli BCP is an unusually versatile peroxiredoxin. Biochem. 2011;50:8970–8981. doi: 10.1021/bi200935d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Koubek E, Haggett M, Battaglia C, Khairat M, Ibne-Rasea H, Pyun J, Edwards O. Kinetics and mechanism of the spontaneous decomposition of some peroxoacids, hydrogen peroxide and t-butyl hydroperoxide. J. Am. Chem. Soc. 1968;85(15):2263–2268. [Google Scholar]
- [37].Fucci L, Oliver C, Coon M, Stadtman E. Inactivation of key metabolic enzymes by mixed-function oxidation reactions: possible implication in protein turnover and ageing. PNAS. 1983;80(6):1521–1525. doi: 10.1073/pnas.80.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mazmanian S, Round J, Kasper D. A microbial symbiosis factor prevents intestinal inflammatory disease. Nat. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- [39].Privitera G, Dublanchet, Sebald M. Transfer of multiple antibiotic resistances between species of Bacteroides fragilis. Jour of Infect Dis. 1979;139:97–101. doi: 10.1093/infdis/139.1.97. [DOI] [PubMed] [Google Scholar]
- [40].Studier F, Moffatt B. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. Journal of Mol Biol. 1986;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- [41].Chèneby D, Philippot L, Hartmann A, Hénault C, Germon J. 16S rDNA analysis for characterization of denitrifying bacteria isolated from three agricultural soils. FEMS Microbiol Ecol. 2000;34(2):121–128. doi: 10.1111/j.1574-6941.2000.tb00761.x. [DOI] [PubMed] [Google Scholar]