Summary
Inflammatory bowel diseases (IBD), including Crohn’s disease (CD), are genetically linked to host pathways that implicate an underlying role for aberrant immune responses to intestinal microbiota. However, patterns of gut microbiome dysbiosis in IBD patients are inconsistent among published studies. Using samples from multiple gastrointestinal locations collected prior to treatment in new-onset cases, we studied the microbiome in the largest pediatric CD cohort to date. An axis defined by an increased abundance in bacteria which include Enterobacteriaceae, Pasteurellacaea, Veillonellaceae, and Fusobacteriaceae, and decreased abundance in Erysipelotrichales, Bacteroidales, and Clostridiales, correlates strongly with disease status. Microbiome comparison between CD patients with and without antibiotic exposure indicates that antibiotic use amplifies the microbial dysbiosis associated with CD. Comparing the microbial signatures between the ileum, rectum, and fecal samples indicates that at this early stage of disease, assessing the rectal mucosa-associated microbiome offers unique potential for convenient and early diagnosis of CD.
Introduction
Inflammatory bowel disease (IBD) is a complex disease in which genetic and environmental circuits establish and contribute to disease pathogenesis. Recent large-scale genome-wide association studies link IBD to host-microbe pathways central to sensing/signaling and mucosa-initiated effector responses (Jostins et al., 2012). Studies of the intestinal gut microbiota imply that an unbalanced microbial community composition is associated with a dysregulated immune response (Khor et al., 2011). The microbiome thus likely plays a role in the pathogenesis of IBD (Manichanh et al., 2012), but this role remains poorly understood. Previous studies characterized patients with established disease, but the use of small cohorts resulted in a lack of statistical power to accommodate diverse clinical covariates (Papa et al., 2012), and results of these studies were likely affected by the application of treatments (Morgan et al., 2012). The existing new-onset studies that examine the fecal microbiome (Kaakoush et al., 2012) detected a disease signal; however, because fecal bacterial ecosystems differ from those in the intestinal mucosa (Momozawa et al., 2011), studies of strictly fecal communities may face limitations in identifying microbes more directly involved in disease initiation or progression.
To improve our understanding of how the microbiota contributes to the inflammatory cascade of Crohn’s disease (CD) pathogenesis, we performed a study that addresses several important limitations of previous work. We applied a standardized approach to a large, multicenter cohort of new-onset CD, collecting samples before treatment initiation, and including subjects representing the variety of disease phenotypes with respect to location, severity, and behavior. Here we report on 668 patients that include those with CD and non-IBD controls (Table S1A), representing the largest single cohort microbiome study related to new-onset IBD to date as well as representing the largest characterization of mucosal-associated microbiota in non-IBD subjects. We used a combination of next-generation sequencing to deeply characterize the disease-associated microbiota, and a well-established multivariate analysis method to account for a wide range of demographic and clinical covariates (e.g., age, gender, race, disease severity, behavior, and location) (Morgan et al., 2012). The strength of this study lies in the sampling prior to treatment, the size of the cohort, and the concurrent sampling of different sites, including multiple mucosal tissue sites, and the luminal content as stool samples. Finally, we combined two additional cohorts with the RISK cohort, resulting in a total of 1,742 samples from pediatric or adult patients, with either new-onset or established disease, for which tissue biopsies and/or fecal samples were processed through a uniform sequencing and analysis approach. This multi-cohort study allows us to position the unique RISK cohort in the context of a comprehensively defined diversity landscape of IBD, and to identify robust and generally applicable biomarkers.
Results
A unique treatment-naïve inception cohort for pediatric CD
We studied the mucosal- and lumen-associated microbiota in a large, well-characterized inception cohort for CD in children. We included subjects from 3 to 17 years of age with a well-established diagnosis of CD (n = 447), and control subjects (n = 221) with non-inflammatory conditions, for example presenting with abdominal pain and diarrhea (Table S1A). Mucosal tissue biopsies (terminal ileum and rectum) and serum samples were collected as part of the diagnostic colonoscopic examination prior to the initiation of treatment. A subset of the enrolled patients (n = 233) also provided a fecal sample prior to treatment start. The diagnosis and disease categorization was confirmed after a minimum of six months follow-up, and was based on a combination of endoscopic, histological, and radiological investigations. A total of 1,321 samples, including 630 ileal and 387 rectal tissue biopsies and 304 stool samples, were submitted for microbiome profiling using 16S rRNA gene sequencing on the Illumina MiSeq platform (version 2) with 175 bp paired-end reads. After quality filtering and assembling overlapping paired-end reads, more than 45.5 million sequences were retained (mean of 29,915 sequences per sample), providing the most in-depth characterization of treatment-naïve CD associated communities to date.
The key players of the microbial dysbiosis in new-onset pediatric CD
An unweighted UniFrac-based comparison of the mucosal-associated microbiota from patients with new-onset CD and controls indicated that the overall diversity in microbial composition was mainly differentiated by sample type and microbial diversity, but disease phenotype was not strong enough to differentiate patients (Figure 1A). Instead, complex microbial communities from samples with multiple clinical covariates are best explored by multivariate association tests at the level of specific microbial community members. We identified microbial organisms that reached statistically significant association with subjects’ disease phenotype using the MaAsLin pipeline, which identifies significant associations of the microbiota with multiple, potentially confounded sample variables (see Experimental Procedures). This has the benefit of testing for disease characteristics, while controlling for several known or potential confounding variables, such as past antibiotic use, age, gender, and race. Correction for other factors that typically have a significant impact on the microbial composition, including treatment and disease duration, was not necessary because all samples were collected prior to treatment and at standard intestinal sites regardless of the segments involved in the disease.
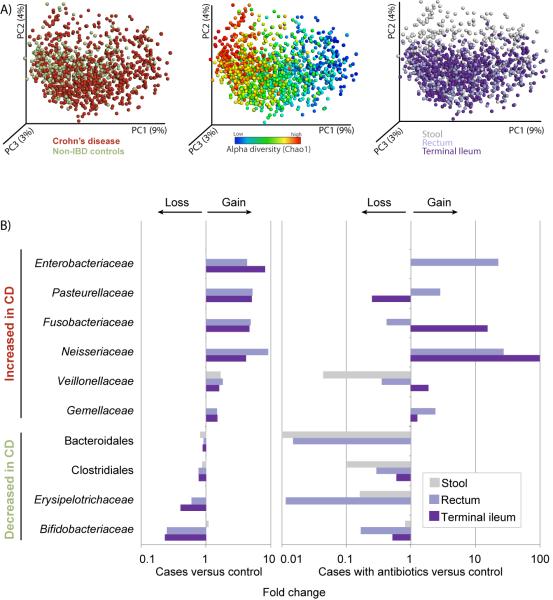
Figure 1. Most differential taxa in pediatric CD.
(A) A set of principal coordinate plots of the unweighted UniFrac distance, with each sample colored either by the disease phenotype (left), alpha diversity (middle), or sample type (right). PC1, PC2, and PC3 represent the top three principal coordinates that captured most of the diversity, with the fraction of diversity captured by that coordinate shown in percent. (B) Differences in abundance are shown for the taxonomic biomarkers that were detected using a multivariate statistical approach (see Experimental Procedures and Table S2). The fold change for each taxon was calculated by dividing the mean abundance in the cases by that of the controls. Several taxonomic biomarkers measured at both the ileal and rectal sites were found to be significantly correlated with disease phenotype; however, most of that microbial signal was lost in the stool samples. The fraction of patients that were on antibiotics during sample collection was considered as an individual subtype, due to the large confounding impact antibiotic exposure causes on the microbial composition (see Table S2). The left shows cases without antibiotic treatment, and the right includes the fraction of cases (10%) that were under antibiotic pressure at sampling. The taxa at the top are increased in disease state, whereas the taxa at the bottom follow an opposite trend. Apparent missing bars are cases in which there is no difference, or fold change equals 1. Use of antibiotics does impact the microbial composition by tipping the microbial community further towards a dysbiotic state, and has a differential impact on the taxa, depending on organism and sampling site. Related to Figure S1, Table S1.
Biomarker detection analysis of mucosal-associated microbiome showed that inflammatory conditions were most strongly associated with an overall drop in species richness and an alteration in the abundance of several taxa (Figure 1B, Table S2A). Several of these taxa have been reported in previous studies (Papa et al., 2012; Morgan et al., 2012), including Enterobacteriaceae, Bacteroidales, and Clostridiales. However, we were able to identify additional taxa as significant biomarkers for disease. Most noticeably, we detected positive correlations between CD and abundances of Pasteurellaceae (Haemophilus sp.), Veillonellaceae, Neisseriaceae, and Fusobacteriaceae. Fusobacterium has previously been suggested as a biomarker for IBD (Strauss et al., 2011), and was also recently shown to promote a beneficial microenvironment for the progression of colorectal carcinoma (Kostic et al., 2012), a long-term complication of IBD. Subsampling the dataset to either smaller sample sizes or lower sequencing depths indicated that sample size contributes more substantially to the increased statistical power, highlighting the importance of sampling a large cohort. Lowering the number of sequences to 300 reads/sample (~1% of the data) did not affect our ability to detect these taxa (results not shown), but reducing to half of the samples or lower did begin to affect the recovery of some taxa (Figure S1A). Interestingly, few of these taxa were present at a higher abundance in patients under the age of 10, and were thus negatively correlated with age, including Pasteurellaceae and Neisseriaceae (Table S2A, Figure S1B).
An increased level of Bacteroides and Clostridiales was maintained in non-CD patients relative to those with CD (Table S2A). Specific negative associations with CD were detected for several genera, including Bacteroides, Faecalibacterium, Roseburia, Blautia, Ruminococcus, Coprococcus, and a number of taxa within the families of Ruminococcaceae and Lachnospiraceae. A well-described anti-inflammatory organism that is considered to be a sensor and marker of health is Faecalibacterium prausnitzii (Sokol et al., 2008). Reduced ileal abundance of F. prausnitzii has been associated with a higher rate of endoscopic recurrence of inflammation six months after ileo-cecal resection.
New-onset mucosal-associated dysbiosis is only weakly reflected in stool
The imbalance in the microbial community network was only observed in the microbiome profiles obtained from tissue samples, and was not seen in the stool samples collected at the time of the diagnosis (Figure 1B). This was confirmed by performing separate biomarker detection on the stool samples of CD patients and controls with non-inflammatory conditions, resulting in only a short list of taxa significantly associated with disease. This included a gain in Streptococcus and a loss in a few taxa belonging to the order of Clostridiales, including Dorea, Blautia, and Ruminococcus (Table S2B). With a mean abundance far below 0.1%, all of these taxa were minimally contributing to the overall shift. Consequently, we further investigated the precise differences between the mucosal tissue and stool samples within patients with new-onset CD (Figure S1C, Table S2C). Of the four above-mentioned taxonomic groups that were decreased in CD, all except Bacteroidales were found to be significantly increased in stool samples, along with Lactobacillus, Enterococcus, and Streptococcus. In addition, the levels of Fusobacteriaceae and Neisseriaceae were reduced, but no significant differences were noted for any of the other organisms typically associated with inflammatory conditions. Based on these observations, we can infer that the microbial balance is less shifted towards a dysbiotic state in the lumen despite the disease, explaining the lack of a biomarker signal and emphasizing the need to examine tissue biopsies in addition to stool samples in order to gain a better understanding of possible mechanisms.
Antibiotic exposure amplifies the microbial dysbiosis
Antibiotic usage has previously been linked to substantial taxonomic changes in the gastrointestinal microbial composition (Dethlefsen et al., 2008; Antonopoulos et al., 2009; Manichanh et al., 2010). Here, a small subset of the CD patients (n = 57 / 447, 13%) was on antibiotics during sample collection, allowing a comparison between the microbiome in CD patients with and without antibiotic exposure. Although a weak effect on disease severity (PCDAI) and overall species diversity between the patient groups with and without antibiotics was found (p = 0.043 and 0.02, respectively; Student’s t-test), we observed a strong effect on the microbial composition, and exposure to antibiotics generally amplified the dysbiosis. A more extreme impact was seen on the abundance levels of the phyla increased in non-inflammatory conditions, including Bacteroides, Clostridiales, and Erysipelotrichaceae, which was most pronounced in rectal biopsy and stool samples, with a differential effect depending on taxa and sample type, e.g., a ten-fold increase in Fusobacteriaceae in the ileum and Enterobacteriaceae in the rectum (Figure 1B). The Pasteurellaceae were suppressed in the ileum, whereas the Veillonellaceae were decreased in the rectum and stool. We note that excluding samples from subjects with antibiotics exposure during sampling does not change the key players of the dysbiotic state outlined above (Table S2A’). These results do not provide any causative explanation, but are relevant in the context of previously described associations between higher antibiotic exposure and the diagnosis of CD (Hviid et al., 2011). We hypothesize that the use of antibiotics has the potential to impact the overall community structure and increase the potential for exposure to dysbiosis.
The functional dysbiosis at the mucosal tissue sites reflects that of established disease
Shotgun-sequencing for metagenomics would provide the greatest precision of microbial community assays and direct evidence of microbial function. However, nucleotide extracts of mucosal tissue samples consist of extremely high fractions (>99%) of host-derived nucleotides. In the absence of methods to efficiently dissociate the microbial from the host fraction, no cost-effective shotgun sequencing of the microbial communities can be performed. Therefore, we predicted the functional composition of these mucosal-associated microbiota using PICRUSt (Langille et al., 2013) (Table S2D). This algorithm estimates the functional potential of microbial communities given a marker gene survey and the set of currently sequenced reference genomes with an accuracy of 80-90% on human gut communities. The functional changes in the samples of new-onset CD patients included a loss in basic biosynthesis (related to reductions in Bacteroides and Clostridia) and a switch towards pathobiont-like auxotrophy (increase in aerobic or aerotolerant taxa, i.e., Proteobacteria and Pasteurellaceae). Further, biomethanation was replaced by acetogenesis in order to reduce accumulated hydrogen. An increased disease severity amplified the disease signal of oxidative stress and auxotrophy further. Interestingly, components of the benzoate metabolic pathway were associated with disease (Aminobenzoate degradation) and disease severity (Fluorobenzoate degradation). Intermediaries of benzoate metabolism are known to influence microbial dysbiosis as a stress response (Eloe-Fadrosh and Rasko, 2013) and have the ability to promote Enterobacteriacea growth and virulence (Freestone et al., 2007). Antibiotic exposure had several overlapping effects on the functional composition of the gut microbiota and took up one third of all significant associations, including a unique series of pathways related to xenobiotic metabolism (degradation of aminobenzoate, styrene, chloroalkene, toluene, benzoate, etc.).
Describing disease status with the Microbial Dysbiosis index
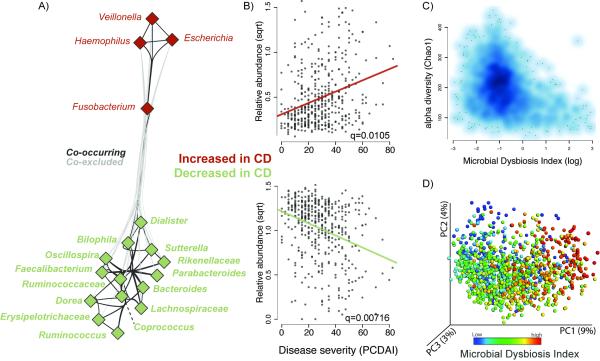
We inferred a taxon-taxon interaction network for the ileal samples (see Experimental Procedures) and found novel relationships among disease-associated organisms mentioned above (Figure 2A, Table S2E). Out of all significant interactions found, 52% supported a cooccurrence within the two groups of taxa that behave similarly with respect to disease, i.e., increase or decrease in CD respectively, and 30% supported a strong co-exclusion between these two groups. Importantly, the taxa within the families Enterobacteriaceae, Fusobacteriaceae, Pasteurellaceae, and Veillonellaceae were often found together, as well as different taxa within the Clostridia or Clostridia and Bacteroidetes teaming up with one another. These observations led us to calculate the log of [total abundance in organisms increased in CD] over [total abundance of organisms decreased in CD] for all samples, hereafter referred to as the Microbial Dysbiosis index (MD-index). This MD-index, derived on disease phenotype, showed a strong positive correlation with clinical disease severity (PCDAI) (Figure 2B) and negative correlation with species richness (Figure 2C), demonstrating that a severe disease state manifests a strongly reduced species diversity in favor of a more extreme dysbiosis. Further, this index was a straightforward feature capturing the overall beta-diversity, resulting in a clear gradient by which samples group across all sample types (Figure 2D). This gradient reflects shifts in both groups of organisms, those increased and decreased with disease (Figure S2B). Lastly, the MD-index was also significantly higher in those patients positive for two or more microbial or cytokine serological markers (p < 0.0001, Student’s t-test). Since serologic markers are increasingly being used to help differentiate IBD disease phenotypes, such an association might indicate a potential link between the gut microbial biomarkers and the presence of these serologic biomarkers. However, with the data collected for this cohort, no specific correlations were found.
Figure 2. The Microbial Dysbiosis index characterizes CD severity.
(A) A correlation network was inferred for the ileal microbiota compositions using CCREPE with a checkerboard score, indicating a strong co-occurrence between taxa of the same disease-associated behavior and a co-exclusion between taxa of a different behavior. Nodes represent the different taxa, and color corresponds to their behavior in disease, with green for those decreased in CD and red for those increased in CD. Edges between nodes represent correlations between the nodes they connect, with edge colors of dark and light grey indicating positive and negative correlations, respectively. For clarity, only edges corresponding to correlations whose similarity was less than 0.3 are shown. (B) Scatterplot of the arcsine square root transformed abundances of all summed abundances for the taxa increased (top) or decreased (bottom) in CD, versus the pediatric CD activity index (PCDAI (Hyams et al., 1991)) as a measure for disease severity. (C) Scatter density plot of the species richness (Chao1, (Chao et al., 2006)) versus the Microbial Dysbiosis index (MD-index) for each sample. The increase in blue color (white to dark blue) reflects the density of the scatter plot. The MD-index is defined as the log of [total abundance in organisms increased in CD] over [total abundance of organisms decreased in CD] (organisms listed in Fig 1A), and is intended as an overall summary statistic for the microbial dysbiosis described in more detail in panel A. In samples with a high MD-index (> 1), a strong reduction in the species richness was observed. (D) A principal coordinate plot of the unweighted UniFrac distance, colored by the MD-index. Sqrt, square root.Related to Figure S2, Table S2.
CD is characterized by inflammation spanning multiple tissue layers, with deep ulcer formation linked with worse long-term disease outcomes. The RISK cohort measured deep ulceration during the diagnostic colonoscopy, presenting us with the opportunity to examine a link between the gut microbiota and mucosal ulceration. The recorded prevalence of any deep ulcers (ileum or colon) with CD patients amounted to 42% (Table S1A). In those patients, we observed increased levels of Pasteurellacaea and Veillonellaceae (p < 0.01, FDR corrected p < 0.15) and Rothia mucilaginosa (p = 0.0004, FDR corrected p = 0.02). In addition, an association between the KEGG pathway for pathogenic Escherichia coli infection was positively associated with ulcer formation (Table S2D). Further experimental study will be needed to determine whether any of these organisms are causally involved in ulceration in IBD patients, or merely adapted to live in this affected environment.
Shotgun metagenome-based identification of microbial biomarkers
Most published studies of the microbiome in IBD so far have used a 16S rRNA gene-based approach, and are thereby limited in characterizing the microbiota to a resolution at the family/genus level. To study the microbiota at a higher resolution, a subset of 43 stool samples (10 controls and 33 subjects) were shotgun-sequenced for metagenomics using the Illumina HiSeq2000 platform (mean 13.3 gigabases (Gb) and s.d. 2.5 Gb per sample, paired-end reads, fragment insert size 180b). Metagenomic data were filtered for human and low-quality reads, and further analyzed as described in Experimental Procedures. As indicated above, the stool samples of patients do not reflect the dysbiosis in a similar way as the mucosal tissue samples, a finding that we confirmed at both the taxonomic and functional levels with these data (results not shown). Nevertheless, mucosal-associated organisms were not restricted to any particular intestinal location, and were readily observed in all sample types, although at lower abundances. Therefore, by adding metagenomics data on the subset of stool samples, we were able to profile the composition of microbial communities at a finer taxonomic resolution (Table S2F). The dominant species increased in CD were Escherichia coli, Fusobacterium nucleatum, Haemophilus parainfluenzae (Pasteurellaceae), Veillonella parvula, Eikenella corrodens (Neisseriaceae), and Gemella moribillum. The dominant species decreased in CD were Bacteroides vulgatus, Bacteroides caccae, Bifidobacterium bifidum, Bifidobacterium longum, Bifidobacterium adolescentis, Bifidobacterium dentum, Blautia hansenii, Ruminococcus gnavus, Clostridium nexile, Faecalibacterium prausnitzii, Ruminoccus torques, Clostridium bolteae, Eubacterium rectale, Roseburia intestinalis, and Coprococcus comes.
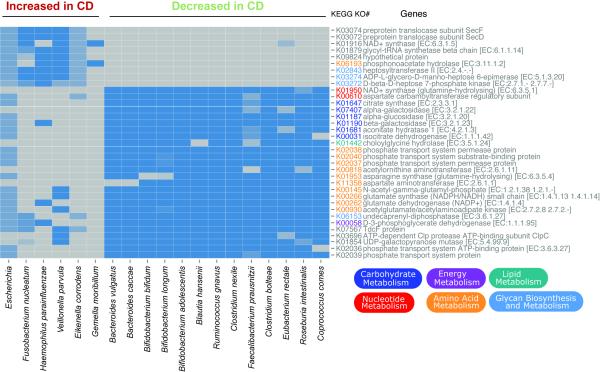
This detailed information and availability of genomic data will be useful in further functional characterization of these organisms and their roles in disease pathogenesis. In a first exploration, we performed a comparison between representative reference genomes for each of these species, generating a view of the differential KEGG pathways (Figure 3). The species increased in CD uniquely contributed pathway components of glycerophospholipid and lipopolysaccharide metabolism, found to instigate inflammation (Morita et al., 1999), and phosphonoacetate hydrolase, providing access to a novel carbon and phosphate source not accessible to most other organisms (Kim et al., 2011). Interestingly, the latter is a zinc-dependent enzyme that might contribute to a mineral deficiency common in newly diagnosed IBD patients. The pathway components unique to the species decreased in CD contribute to the bile acid and amino acid biosynthesis pathways, including connections between amino acid metabolism and energy, carbohydrate, or nucleotide metabolism. Collectively, these provide access to complex carbohydrates, and the break at the alpha-ketoglutarate step of the TCA cycle is indicative of a true anaerobic lifestyle, as indicated previously (Morgan et al., 2012).
Figure 3. Comparative genomics of CD biomarkers.
The KEGG metabolic pathways that differentiate the species by behavior in disease state are shown as a heatmap. A selection of reference genomes that are representative for the species increased or decreased with disease were obtained from IMG (JGI), and biomarker detection was performed on their gene content at the level of KEGG pathways. Several were statistically significant (Wilcoxon, p < 10e-8) and are visualized here. Related to Table S2.
Biopsy-associated microbiome can diagnose CD
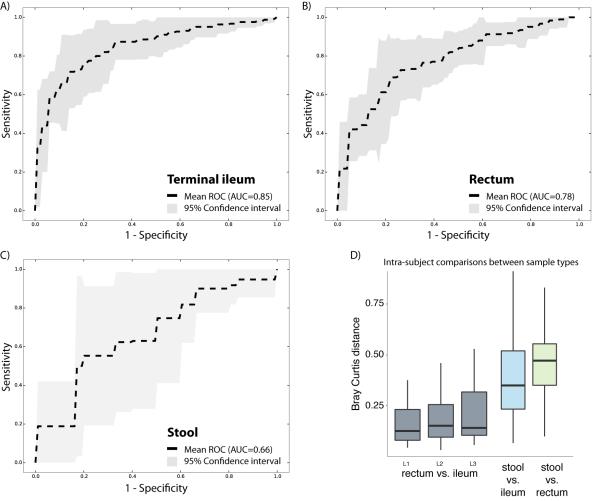
Several recent studies have explored the potential for identifying disease states based on the host-associated microbial composition, including skin swabs for psoriasis (Statnikov et al., 2013), and fecal samples for obesity (Le Chatelier et al., 2013), autism (Hsiao et al., 2013), or IBD (Papa et al., 2012). Here, we evaluated how the microbiome composition in three different sample types performed for classifying subjects by CD state using a receiver operating characteristic (ROC) analysis. We included a total of 425 tissue biopsies of the ileum, 300 of the rectum, and 199 stool samples in three independent analyses (Figure 4A-C). Microbiome profiles were collapsed to the genus-level abundances and normalized (see Experimental Procedures). The best performance was obtained by the ileal samples (AUC = 0.85), which was closely followed by the rectal biopsies (AUC = 0.78), both with a narrow confidence interval. The stool samples, however, performed less well (AUC = 0.66) and had also a low consistency (broader confidence interval). A previous study was able to get a higher performance with stool samples for disease classification in an IBD cohort (Papa et al., 2012), but their patient cohort distinguishes itself from RISK by the fact that subjects had a mean disease duration of 34.8 months, and RISK cohort is entirely new-onset, with samples only taken at the time of diagnosis. This finding is consistent with the biomarker detection analysis that indicated that several taxonomic groups were increased or decreased between cases and controls when comparing mucosal-associated microbiome profiles, but not in stool samples (Figure 1). Interestingly, classification of subjects by disease state was not affected by disease location. In this cohort, 22% had disease confined to the ileum, 25% to the colon, and 53% had ileocolonic disease. Samples from both tissue biopsy locations could classify subjects, even if disease was confined to the other location. In fact, microbiome composition between the different biopsy sites was found to be far less different than between tissue and stool, for all three disease sub-phenotypes (Figure 4D).
Figure 4. Disease classification performs well on biopsy-associated microbiome profiles.
(A-C) For each of the three sample types, including terminal ileum biopsy (A), rectum biopsy (B), and stool sample (C), we evaluated the accuracy of disease classification using L1 penalized logistic regression with ROC curves representing the results. Dashed lines show the mean performance obtained when genus-level features were used, and the surrounding grey area is the 95% confidence interval. Terminal ileum biopsies performed best (AUC = 0.85), closely matched by the rectum biopsies (AUC = 0.78). However, the classifier based on the stool samples collected at the time of the diagnosis performs less well (AUC = 0.66), and with low consistency (large confidence interval). (D) The intra-subject diversity in microbiome composition was determined for all pairwise sample type combinations. Both biopsy samples were found to be highly similar, whereas the stool sample was quite diverse. Further, we also compared whether disease location would impact the intra-subject diversity between the two tissue biopsy locations. The location of the disease, ileal (L1), colonic (L2), or ileocolonic (L3), did not significantly disrupt the similarity between the two intra-subject mucosal-associated microbiota. Also, no biomarker was detected allowing us to distinguish these disease sub-phenotypes. Related to Figure S3.
Further, this cohort presented the opportunity to derive a predictive model for future disease outcome, as patients with a positive diagnosis for CD were re-examined at a 6- and 12-month follow-up, at which point the disease activity index (PCDAI) was determined. For all patients with such follow-up data available (n = 305), 7.5% had an increased disease severity 6 months after diagnosis, and for 22% the severity reduced from severe (PCDAI > 30) to remission (PCDAI < 10). No tissue samples were collected at later time points, and thus no follow-up mucosal microbiome profiles exist. However, we could evaluate a model for predicting whether a patient will develop an exacerbated or reduced disease severity over the next 6 months, using the microbiome and clinical covariates collected at the time of onset of disease. Using a random forests classifier trained on 90% of the data, we found we were able to predict high 6-month PCDAI (≥ 10) in the remaining 10% of the data with 67.0% accuracy, a 14% improvement over the predictive accuracy of a model trained only on clinical covariates (52.9 +/- 0.4%) (Figure S3A). Although the absolute level of accuracy is modest, the performance gain driven by the microbiome is a direct and unbiased demonstration of the utility of microbiome features for predicting clinical outcomes. To determine how influential a given feature was in building the predictive model, we tested the decrease in accuracy of the model when that feature was removed. The most influential features for predicting future PCDAI according to this test were age of onset, PCDAI at diagnosis, and levels of disease-associated organisms, including Enterobacteriaceae, Fusobacterium, and Haemophilus. Age of onset and levels of Enterobacteriaceae were negatively correlated with future PCDAI, and PCDAI at diagnosis and levels of Fusobacterium and Haemophilus were positively correlated with future PCDAI (Figure S3B).
Comparing pediatric CD with other adult established disease cohorts
To position the above findings in the context of other IBD microbiome studies, we resequenced samples from a previously published study (Morgan et al., 2012) and included samples from several other cohorts, using the same sequencing approach. Combining all of these samples resulted in data from more than 1,500 subjects, of whom 46% had CD, 31% had ulcerative colitis (UC), and 19% were non-IBD controls, and included both tissue biopsies (88%) and stool samples (12%) (Table S1B). Two important differences between the RISK cohort participants and the other cohorts were (i) the lower age range (13 yrs, s.d. 3, versus 41 yrs, s.d. 15), and (ii) the fact that RISK exclusively enrolled patients with new-onset disease, whereas the mean disease duration at time of sampling in the other cohorts was 7 years since diagnosis (s.d. 11, range 0 to 62). The combined dataset consisted of nearly 60 million paired-end 16S reads, with a mean of 23,620 filtered sequences per sample.
To our knowledge, this is the largest uniformly generated microbiome dataset for CD specifically, but also IBD in general, revealing an extensive landscape of microbial diversity across a wide range of subjects and disease phenotypes. We therefore took the opportunity to examine the effect size of disease phenotype on the microbiome composition relative to the effects introduced by cohort, age, gender, treatment, and biopsy location. We surveyed effects on the biodiversity as a whole, and on the organisms contributing to the dysbiosis specifically, and used a linear mixed effects model including cohort and subject as random factors (see Experimental Procedures). The results revealed that inter-individual variation, cohort and sample type had significant effects on the overall microbial community composition, and that variation introduced by disease phenotype and treatment were hidden underneath those primary factors (Table S2G). However, within specific sample types (e.g., terminal ileum), disease can explain up to 10% of the variation seen in subsets of key dysbiotic taxa, in which the effects of having CD versus control do surface. The earlier described impact of antibiotics on the microbial community network was one of the stronger signals across all cohorts, and found in almost all included sample types. Interestingly, the signal was stronger in the mucosal-associated microbiome profiles than in the stool samples (Figure S2A). In contrast, the impact of antibiotics on overall microbial diversity was stronger in stool than in mucosal samples, again indicating that conclusions may differ based on which sample types are studied. Taken together, this advocates for a need to standardize sample collection, include diverse sample types to represent different gastrointestinal compartments, and account for treatment effect in order to get the most statistical power to study the role of the microbiome in diseases.
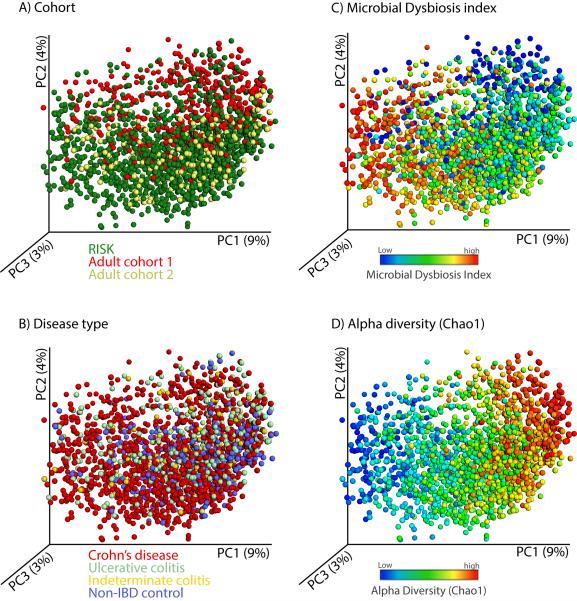
Lastly, we aimed to determine the effect on individual taxa using a biomarker detection approach and compare the results across the different cohorts and disease phenotypes. Most of the organisms that were found to be increased in CD were also found to be significantly correlated with UC, but those taxa that were negatively associated with CD were not found to be significant in association with UC (Table S2H). When using a weighted UniFrac distance calculation and a PCoA visualization (Figure 5), the clustering by overall microbial community composition was strongly affected by cohort (Figure 5B) and different disease subgroups significantly overlapped (Figure 5A). However, the MD-index (Figure 5C) and species richness (Figure 5D) still explained the first principal coordinate even when the different cohorts were combined and demonstrate that the specific disease-associated taxa were consistent.
Figure 5. A view of the microbial composition across different IBD cohorts.
We combined microbial profiles obtained for 1,742 subjects from three different IBD cohorts and generated a set of principal coordinate plots of the unweighted UniFrac distance, where each sample was colored by (A) cohort, (B) disease type, (C) MD-index, or (D) species richness (Chao1). From this combined view, it is clear that the first principal coordinate (PC1) stratifies the samples by species richness, which is negatively correlated with MD-index, and that the second principal coordinate (PC2) is largely affected by cohort. Disease phenotype is no obvious driver for sample clustering.
Discussion
Our results on the microbiome at the onset of CD have identified the key constituents of the complex gut microbial community that define a mucosal surface in homeostasis or dysbiosis. Our work demonstrates that the creation of such a large multi-center cohort increases the resolution and statistical power for studying the role of the microbiome in disease. Several of the taxa we identified were only reliably associated with disease phenotype when using several hundreds of samples. Achieving similar sample sizes by combining independent cohorts for a cross-study comparison limits the study of parameters with large effect size that surpass the bias introduced by differences in collection and sample handling (Lozupone et al., 2013). Capturing microbial shifts in their full complexity, including taxa with smaller shifts in relative abundance comparing cases versus controls, require these large, optimal study designs.
Several of the organisms identified in this study are known to reside at the inflamed mucosa with the potential to exacerbate inflammation and/or invade intestinal epithelial cells, including strains of Escherichia (Rolhion and Darfeuille-Michaud, 2007) and Fusobacterium (Strauss et al., 2011). Others, such as Haemophilus and Veillonella, have recently been reported to contribute to oral dysbiosis in IBD patients (Said et al., 2013). Haemophilus spp., like the Enterobacteriaceae, are well adapted to survive in oxidative stress environments and intensify oxidative stress in airway infections (Harrison et al., 2012). Interestingly, Veillonella spp. are closely related to the Clostridiales, who are otherwise considered to be beneficial to the host (Furusawa et al., 2013). Prior literature, however, indicates that Veillonella produce lipopolysaccharides (Gupta, 2011), a gene cluster they might have gained through horizontal gene transfer (Michael Fischbach, personal communication). Rothia mucilaginosa, here reported at increased levels in patients with intestinal ulcer formation, has been found to act as an opportunistic pathogen in immunocompromised patients (Chavan et al., 2013) and has a genomic content that is well adapted to live within the microaerophilic surface of the mucus layer in cystic fibrosis lungs (Lim et al., 2013). Most of these organisms are relatively rare in the colon, and typically part of the normal human oral and upper respiratory tract microbiota, but could become opportunistic colonizers in conditions of altered mucosal changes in tissue oxygenation and disruption of mucosal barrier function.
We observed that both the ileal and rectal biopsy have similar discriminatory power for classifying disease, regardless of the disease location. This creates the opportunity to use a minimally invasive sampling approach that avoids bowel preparation prior to the colonoscopy, and to perform dense sampling of the mucosal-associated microbiome to monitor the response to treatment and potentially predict changes in disease flares. Being able to account more readily for the microbiota in larger cohort sizes will be of value in defining disease sub-phenotypes and tracking treatment effects in clinical trials, something that is currently not annotated. Understanding the microbial communities of the small intestine remains of tremendous value. Mucosal-associated microbes are uniquely positioned to influence the immune system (Belkaid and Naik, 2013); particularly, the porous mucus layer in the ileum has been shown to educate the immune system to develop tolerance towards commensals (Shan et al., 2013).
Large-scale collection of stool samples would be an even less invasive approach, but for this cohort did not reflect the mucosal dysbiosis, in contrast to an earlier study (Papa et al., 2012). The main difference between the two cohorts is that new-onset patients were sampled at the time of diagnosis. The earlier study included samples of patients with established disease (mean of 3 years at time of sampling) and a treatment history. We observed that the microbial community associated with the inflamed epithelium had an increased level of aerobic and facultative anaerobes (e.g., Protebacteria), whereas obligate anaerobes prevailed in the feces (e.g., Bacteroides and Clostridiales). The microbial community in stool from patients with established disease also consists of less anaerobes (Papa et al., 2012). This is consistent with a recent observation that the oxygen level in the lumen increases with intestinal inflammation to a level that gut microbiota start to shift towards an aerotolerant composition in response to an oxidative stress (Mimouna et al., 2011). In order to validate this observation, future work will need to consistently capture samples from patients across a wide range of disease durations.
Another factor affecting the gut microbial composition is the use of antibiotics, as shown in a subset of the RISK cohort patients. Previously, antibiotics have been claimed to provide benefits for CD patients as a first-line therapy. However, we question this practice based on our observation that the microbial network appears more dysbiotic in the context of antibiotic exposure. Loss of protective microbes has the potential of triggering a proliferation of less beneficial taxa (Looft and Allen, 2012), exacerbating the inflammation. Similarly, changing dietary patterns can introduce such shifts as quickly (Wu et al., 2011), particularly in those individuals with reduced microbial complexity (Fang and Evans, 2013). For example, the vitamin D pathway has importance in gut homeostasis and in signaling between the microbiota and the host immune system, and may thus have implications for the development, severity, and management of inflammation (Mouli and Ananthakrishnan, 2014).
The data presented here provide a unique framework for understanding the microbial dysbiosis in new-onset CD. This will further develop principles that are likely to govern therapeutics in IBD, but will need to be carefully thought through (Fischbach et al., 2013). These include in particular those efforts that aim to shift the microbiome following a path that is based on the successful principles applied to recurrent Clostridium difficile infections, as these are unlikely to be directly applicable to the multifactorial disease pathogenesis of IBD.
Experimental Procedures
Study population and sample collection
A total of 447 children and adolescents (< 17 years) with newly diagnosed CD and a control population composed of 221 subjects with non-inflammatory conditions of the gastrointestinal tract were enrolled to the RISK study in 28 participating pediatric gastroenterology centers in North America between November 2008 and January 2012 (Table S1A). Biopsies were taken from the terminal ileum and rectum using standard endoscopic forceps, and placed into a sterile cryovial with RNAlater (Qiagen) on ice in the Endoscopy Suite. Nucleotides were isolated from these biopsies using the Qiagen AllPrep Mini Kit.
16S rRNA gene sequencing
The 16S gene dataset consists of sequences targeting the V4 variable region. Detailed protocols used for 16S amplification and sequencing are as previously described (Caporaso et al., 2012). Sequencing was performed on the Illumina MiSeq platform according to the manufacturer’s specifications with addition of 5% PhiX, and generating paired-end reads of 175b in length in each direction. The overlapping paired-end reads were stitched together (approximately 97bp overlap), and further processed in a data curation pipeline implemented in QIIME 1.7.0 as pick_closed_reference_otus.py (Caporaso et al., 2010). All 16S rRNA sequences have been deposited at the National Center for Biotechnology Information as two BioProjects with ID PRJNA237362 and PRJNA205152, and are also available in a variety of tables through www.microbio.me/qiime/under Study ID 2516.
Shotgun metagenomic sequencing
Metagenomic data production and processing were performed as described previously (HMP Consortium, 2012). In brief, library construction was performed on the Illumina HiSeq 2000 platform, targeting 7 Gb of sequence per sample with 101bp, paired-end reads. Species abundances were calculated with MetaPhlAn 1.7.7 (Segata et al., 2012), following Bowtie 2-2.1.0 alignment (Langmead and Salzberg, 2012) to the MetaPhlAn 1.0 unique marker database.
Statistical analysis
Association testing of all covariates versus all taxa was performed by regressing the relative abundance of each taxon on these linear clinical covariates: subject, diagnosis, ulcering, ileal involvement, PCDAI, biopsy location, age, gender, race, antibiotic exposure, with subject as a random variable, using the MaAsLin algorithm with default parameters (Morgan et al., 2012).
Correlation network
We extracted the subnetworks of microbial interactions at the terminal ileum from subjects free of any antibiotics pressure, using CCREPE (Compositionality Corrected by REnormalization and PErmutation). This is a statistical methodology for co-variation analysis in compositional data developed on top of previously published work (Faust et al., 2012), using the NC-score, a similarity measure specifically designed to detect association patterns in the human microbiome and other microbial communities (see Supplemental Information).
ROC analysis
ROC curves were constructed to evaluate the performance of sparse logistic regression classifier (using L1 penalization) aiming to identify the IBD status of a subject based on his or her microbiome profile. We have checked the performance for classifier trained by samples from three different sites (ileum, rectum, and stool). Five ROC curves were gained per site using 5-fold cross-validation. A mean ROC curve was then given by averaging over all 5 individual fold ROC curves and an approximated 95% pointwise confidence interval was also constructed by using normal approximation and the sample means and variances.
Supplementary Material
Highlights.
Microbiomes from multiple GI locations in new-onset Crohn’s disease (CD) cases analyzed
Co-occurring and co-excluded CD-associated organisms identified
Rectal mucosa-associated, but not fecal, microbiome is a robust disease predictor
Antibiotics amplify the dysbiosis associated with CD
Acknowledgments
We thank the patients who donated samples for this study, the health professionals who collected them, and the Crohn’s and Colitis Foundation of America for supporting the RISK cohort. We thank Douglas Wendel, Gail Ackermann, Tim Vigers, and Tim L. Tickle for their valuable input and helpful discussions. We thank participating clinicians Marla Dubinsky, Joshua Noe, Scott Snapper, Richard Kellermayer, Michael Kappleman, Anthony Otley, Mirian Pfefferkorn, Stanley Cohen, Stephen Guthery, Neal LeLeiko, Maria Oliva-Hemker, David Keljo, Dedrick Moulton, Barbara Kircshner, Ashish Patel, David Ziring, Jonathan Evans, Jonah Essers, Bruce Aronow, and MiOk Kim. Work was supported by grants from the Crohn’s and Colitis Foundation of America, ARO grant W911NF-11-1-0473 (C.H.), NIH grants U54 DE023798, R01 HG005969 (C.H.), and R01 DK092405 (R.J.X., C.H., D.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol. 2013;14:646–653. doi: 10.1038/ni.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A, Chazdon RL, Colwell RK, Shen TJ. Abundance-based similarity indices and their estimation when there are unseen species in samples. Biometrics. 2006;62:361–371. doi: 10.1111/j.1541-0420.2005.00489.x. [DOI] [PubMed] [Google Scholar]
- Chavan RS, Pannaraj PS, Luna RA, Szabo S, Adesina A, Versalovic J, Krance RA, Kennedy-Nasser AA. Significant morbidity and mortality attributable to rothia mucilaginosa infections in children with hematological malignancies or following hematopoietic stem cell transplantation. Pediatr Hematol Oncol. 2013;30:445–454. doi: 10.3109/08880018.2013.783893. [DOI] [PubMed] [Google Scholar]
- HMP Consortium A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloe-Fadrosh EA, Rasko DA. The human microbiome: from symbiosis to pathogenesis. Annu Rev Med. 2013;64:145–163. doi: 10.1146/annurev-med-010312-133513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Evans RM. Microbiology: Wealth management in the gut. Nature. 2013;500:538–539. doi: 10.1038/500538a. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Bluestone JA, Lim WA. Cell-based therapeutics: the next pillar of medicine. Sci Transl Med. 2013;5:179ps177. doi: 10.1126/scitranslmed.3005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freestone PP, Walton NJ, Haigh RD, Lyte M. Influence of dietary catechols on the growth of enteropathogenic bacteria. Int J Food Microbiol. 2007;119:159–169. doi: 10.1016/j.ijfoodmicro.2007.07.039. [DOI] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Origin of diderm (Gram-negative) bacteria: antibiotic selection pressure rather than endosymbiosis likely led to the evolution of bacterial cells with two membranes. Antonie Van Leeuwenhoek. 2011;100:171–182. doi: 10.1007/s10482-011-9616-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison A, Bakaletz LO, Munson RS., Jr. Haemophilus influenzae and oxidative stress. Front Cell Infect Microbiol. 2012;2:40. doi: 10.3389/fcimb.2012.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou JK, Lee D, Lewis J. Diet and Inflammatory Bowel Disease: Review of Patient-Targeted Recommendations. Clin Gastroenterol Hepatol. 2013 doi: 10.1016/j.cgh.2013.09.063. S1542-3565(13)01512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hviid A, Svanstrom H, Frisch M. Antibiotic use and inflammatory bowel diseases in childhood. Gut. 2011;60:49–54. doi: 10.1136/gut.2010.219683. [DOI] [PubMed] [Google Scholar]
- Hyams JS, Ferry GD, Mandel FS, Gryboski JD, Kibort PM, Kirschner BS, Griffiths AM, Katz AJ, Grand RJ, Boyle JT, et al. Development and validation of a pediatric Crohn’s disease activity index. J Pediatr Gastroenterol Nutr. 1991;12:439–447. [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush NO, Day AS, Huinao KD, Leach ST, Lemberg DA, Dowd SE, Mitchell HM. Microbial dysbiosis in pediatric patients with Crohn’s disease. J Clin Microbiol. 2012;50:3258–3266. doi: 10.1128/JCM.01396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Benning MM, OkLee S, Quinn J, Martin BM, Holden HM, Dunaway-Mariano D. Divergence of chemical function in the alkaline phosphatase superfamily: structure and mechanism of the P-C bond cleaving enzyme phosphonoacetate hydrolase. Biochemistry. 2011;50:3481–3494. doi: 10.1021/bi200165h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- Lim YW, Schmieder R, Haynes M, Furlan M, Matthews TD, Whiteson K, Poole SJ, Hayes CS, Low DA, Maughan H, et al. Mechanistic model of Rothia mucilaginosa adaptation toward persistence in the CF lung, based on a genome reconstructed from metagenomic data. PLoS One. 2013;8:e64285. doi: 10.1371/journal.pone.0064285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looft T, Allen HK. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes. 2012;3:463–467. doi: 10.4161/gmic.21288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh J, Gonzalez A, Ackermann G, Wendel D, Vazquez-Baeza Y, Jansson JK, Gordon JI, Knight R. Meta-analyses of studies of the human microbiota. Genome Res. 2013;23:1704–1714. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Reeder J, Gibert P, Varela E, Llopis M, Antolin M, Guigo R, Knight R, Guarner F. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 2010;20:1411–1419. doi: 10.1101/gr.107987.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimouna S, Goncalves D, Barnich N, Darfeuille-Michaud A, Hofman P, Vouret-Craviari V. Crohn disease-associated Escherichia coli promote gastrointestinal inflammatory disorders by activation of HIF-dependent responses. Gut Microbes. 2011;2:335–346. doi: 10.4161/gmic.18771. [DOI] [PubMed] [Google Scholar]
- Momozawa Y, Deffontaine V, Louis E, Medrano JF. Characterization of bacteria in biopsies of colon and stools by high throughput sequencing of the V2 region of bacterial 16S rRNA gene in human. PLoS One. 2011;6:e16952. doi: 10.1371/journal.pone.0016952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita H, Nakanishi K, Dohi T, Yasugi E, Oshima M. Phospholipid turnover in the inflamed intestinal mucosa: arachidonic acid-rich phosphatidyl/plasmenyl-ethanolamine in the mucosa in inflammatory bowel disease. J Gastroenterol. 1999;34:46–53. doi: 10.1007/s005350050215. [DOI] [PubMed] [Google Scholar]
- Mouli VP, Ananthakrishnan AN. Review article: vitamin D and inflammatory bowel diseases. Aliment Pharmacol Ther. 2014;39:125–136. doi: 10.1111/apt.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa E, Docktor M, Smillie C, Weber S, Preheim SP, Gevers D, Giannoukos G, Ciulla D, Tabbaa D, Ingram J, et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One. 2012;7:e39242. doi: 10.1371/journal.pone.0039242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath HC, Herfarth HH, Ikeda JS, Grenther WB, Hamm TE, Jr., Balish E, Taurog JD, Hammer RE, Wilson KH, Sartor RB. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J Clin Invest. 1996;98:945–953. doi: 10.1172/JCI118878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolhion N, Darfeuille-Michaud A. Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:1277–1283. doi: 10.1002/ibd.20176. [DOI] [PubMed] [Google Scholar]
- Said HS, Suda W, Nakagome S, Chinen H, Oshima K, Kim S, Kimura R, Iraha A, Ishida H, Fujita J, et al. Dysbiosis of Salivary Microbiota in Inflammatory Bowel Disease and Its Association With Oral Immunological Biomarkers. DNA Res. 2013 Sep 7; doi: 10.1093/dnares/dst037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan M, Gentile M, Yeiser JR, Walland AC, Bornstein VU, Chen K, He B, Cassis L, Bigas A, Cols M, et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 2013;342:447–453. doi: 10.1126/science.1237910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statnikov A, Alekseyenko AV, Li Z, Henaff M, Perez-Perez GI, Blaser MJ, Aliferis CF. Microbiomic signatures of psoriasis: feasibility and methodology comparison. Sci Rep. 2013;3:2620. doi: 10.1038/srep02620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, Lynch T, Allen-Vercoe E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.