Abstract
Drosophila SNF is a member of the U1A/U2B″/SNF protein family that is found in U1 and U2 snRNPs, where it binds to Stemloop II and Stemloop IV of U1 and U2 snRNA, respectively. SNF also binds to the U2A′ protein, but only in the U2 snRNP. Although previous reports have implicated U2A′ as a necessary auxiliary protein for the binding of SNF to Stemloop IV, there are no mechanisms that explain the partitioning of U2A′ to the U2 snRNP and its absence from the U1 snRNP. Using in vitro RNA binding isotherms and isothermal titration calorimetry, the thermodynamics of SNF/RNA/U2A′ ternary complex formation have now been characterized. There is a very large binding cooperativity unique to Stemloop IV that favors formation of the SLIV/SNF/U2A′ complex. The binding cooperativity, or heterotropic linkage, is interpreted with respect to linked conformational equilibria of both SNF and its RNA ligand and so represents an example of protein–RNA allostery.
The spliceosome catalyzes eukaryotic pre-mRNA splicing and is one of the most complex and dynamic macromolecular machines in the nucleus.1 At the core of this machinery are five major snRNPs [small nuclear ribonucleoproteins (U1, U2, and U4–U6 snRNPs)], which each contain a single unique snRNA and multiple associated proteins, some of which are unique to a given snRNP and others of which are shared among snRNPs. In particular, the U1 and U2 snRNPs of many metazoans have a common protein, first identified in Drosophila. This Drosophila SNF2,3 protein (for sans fille) binds to U1 snRNA Stemloop II (SLII) and U2 snRNA Stemloop IV (SLIV).4 To date, there are no data regarding the in vivo function of SNF in the snRNPs, although protein mutations result in defects to Drosophila sex determination, and genetic data show that a SNF deletion is embryonic lethal in the fly.5
SNF contains two RNA recognition motifs (RRMs), the first of which is responsible for specific binding to both RNAs.4 RRMs are the most abundant RNA binding domains in eukaryotes and are characterized by an α/β sandwich topology. A nuclear magnetic resonance (NMR) solution structure6 of SNF RRM1 shows its classic RRM fold (Figure 1A), but there are no structures of SNF in bimolecular complexes with either SLII or SLIV. However, SNF is a member of the U1A/U2B″/SNF family of RNA binding proteins, all of which contain an N-terminal RNA binding RRM. The homology between the three RRMs (∼74% identical) allows us to use existing cocrystals of human U1A RRM1 bound to SLII7 and human U2B″ RRM1 bound to SLIV8 as models for possibly analogous SNF interactions. In cocrystals of U1A RRM1 bound to SLII, and U2B″ bound to SLIV, the RNA is spread out over the surface of the four-stranded antiparallel β-sheet. In these complexes, two aromatic amino acids stack with nucleobases (Figure 1B); we anticipate that this orientation also describes SNF/RNA complexes.
Figure 1.
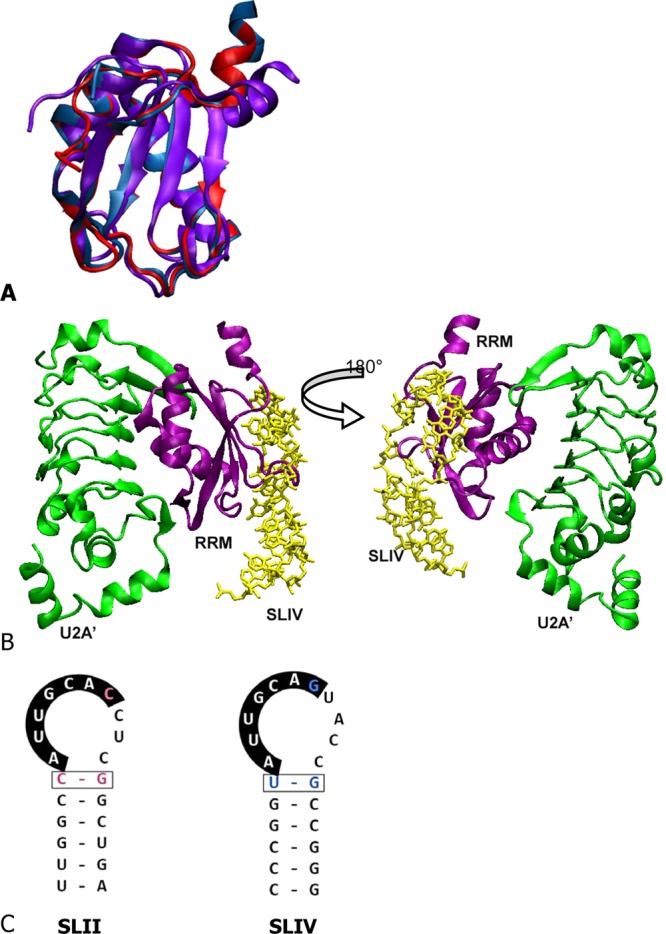
RNA and proteins. (A) Overlay of structures of the U1A RRM solution NMR structure,44 the SNF solution NMR structure6 (purple), and U2B″ from the ternary complex8(blue). (B) Cartoon representation of U2B″/SLIV/U2A′ ternary complex formation (Protein Data Bank entry 1A9N), the model of ternary complex formation for Drosophila SNF. RNA is colored yellow, RRM purple, and U2A′ green. Structures represented with VMD.45 (C) Drosophila U1 snRNA SLII and U2 snRNA SLIV sequences.
The sequences of SLII and SLIV are remarkably similar, and SNF binds to each with affinities that are uniquely dependent on salt and temperature, reflecting differences in binding mechanisms for the two RNAs.4 The RNA sequences are shown in Figure 1C; the conserved nucleobases in the loops (5′AUUGCAC/G) are primary contacts for U1A and are likely to be maintained for SNF. Binding of SNF to SLIV is complicated by the association of SNF with U2A′ protein, which is also phylogenetically conserved in metazoans.
In Drosophila, U2A′ is a 265-residue protein that contains an N-terminal leucine-rich repeat (LRR) and a C-terminal domain of ∼100 residues, predicted to be mostly disordered (using IUPRED9). In a ternary SLIV/U2B″/U2A′ crystal structure,8 the LRR domain of human U2A′ interacts with the α-helical side of human U2B″ RRM1. As the ternary complex shows, the U2B″ RRM is sandwiched between SLIV and the U2A′ LRR (Figure 1B). By analogy to the complex formed by the homologous U2B″ RRM1, SNF is thought to form a ternary complex with both SLIV and U2A′. As with the U1A/SLII cocrystal, an important caveat is that the RNA binding properties of U1A, U2B″, and SNF are quite different,4,10,11 so inferences from structural comparisons must be cautious.
Early in vitro pull-down experiments with [35S]SNF showed that it bound to Xenopus U1 snRNA.2 However, the ability of Xenopus U2 snRNA to pull down [35S]SNF was enhanced when SNF was co-incubated with in vitro-translated human U2A′ or with Drosophila nuclear extract,2 which presumably contained Drosophila U2A′. These results led to the conclusion that protein/protein interactions between U2A′ and SNF enhanced the affinity of SNF for SLIV, promoting the formation of the SLIV/SNF/U2A′ ternary complex. However, those experiments neither explained the apparent absence of a SLII/SNF/U2A′ ternary complex nor provided a mechanism that explained the formation of a SLIV/SNF/U2A′ ternary complex.
We used purified recombinant proteins and RNAs to perform in vitro experiments that compare binding in the ternary complex system (RNA/SNF/U2A′) with bimolecular binding of SNF/U2A′ and SNF/RNA complexes. Our system allows us to analyze SNF binding in terms of binding cooperativity, which we define as the degree to which binding by one ligand (RNA) affects binding of the second ligand (U2A′). Intriguingly, while the cooperativity for the SLIV/SNF/U2A′ complex is large, the cooperativity of SLII/SNF/U2A′ binding is marginal. Of most significance is the fact that the RNA-dependent thermodynamic cooperativity between protein/RNA and protein/protein interactions is sufficient to explain the characteristic partitioning behavior of U2A′ to the U2 snRNP and exclusion from the U1 snRNP. We finally describe protein/protein and protein/RNA binding in terms of allosteric models that include contributions of RNA and protein internal conformational equilibria.
Materials and Methods
Protein Constructs and Purification
Full-length SNF was purified as previously described.4
A pGEX-2T plasmid containing the gene for Drosophila U2A′ was obtained from H. Salz. The U2A′ gene was subcloned into our Ptac expression vector under an isopropyl β-d-1-thiogalactopyranoside (IPTG) inducible promoter, and the three cysteines in the protein were Quick-changed to their human sequence counterparts (C19V, C38T, and C119S) for the sake of biochemical convenience. [EMSA experiments using both constructs showed no difference in binding properties (data not shown).] The protein construct was truncated at position 180, so what we call U2A′ is the protein LRR domain. Protein expression was induced in Escherichia coli BL 21 cells at an OD of 0.8 in LB medium with 0.1 mM IPTG at 17 °C overnight to reduce the level of inclusion body formation. Cells were spun down and resuspended on ice in 30 mM sodium acetate (pH 5.3), 200 mM NaCl, 2 mM EDTA, and 8.5% sucrose. A protease inhibitor cocktail (Sigma), phenylmethanesulfonyl fluoride, and DNase II were added prior to lysis. Cells were French pressed, spun down in an ultracentrifuge, and filtered through a 0.22 μm filter, and the supernatant was loaded onto a prepacked GE Hi-Trap SP-HP cation exchange column at 4 °C. The column was washed with 50 mM NaCl and 50 mM Tris (pH 7.5). A salt gradient from 50 to 375 mM NaCl was run at a rate of 1.5 mL/min over 2.5 h. Fractions with U2A′ were collected and concentrated into 100 mM arginine, 50 mM KCl, and 10 mM cacodylate (pH 7). The arginine was necessary to maintain protein solubility at high concentrations. The concentrated protein was then run on a Superdex 75 10/300 GL (GE) gel filtration column in the same buffer, with a flow rate of 0.3 mL/min to remove impurities. The protein was eluted as a single, symmetric peak. Clean fractions were collected and concentrated to ∼100 μM for further use.
Fluorescence Titrations
For fluorescence binding experiments, we used 6-carboxyfluorescein (6-FAM) 5′-end-labeled RNAs (IDT) with sequences of 5′-6-FAM-GGGCCCGGCAUUGCACCUCGCCGGGUCC (SLII) and 5′-6-FAM-GGGCCCGGUAUUGCAGUACCGCCGGGUCC (SLIV). Loop nucleotides are underlined. These RNAs were also 3′-end-labeled (using T4 RNA ligase) with [α-32P]pCp (cytidine 3′,5′-bis-phosphate) to assess whether the 5′-fluorescein label affects RNA binding as measured by nitrocellulose filter binding experiments. Filter binding assays with FAM-RNAs and in vitro T7 RNA polymerase SLII and SLIV showed no difference in dissociation constants (data not shown).
Fluorescence experiments were performed using an SLM 8000 fluorimeter. Cuvettes and stir bars were soaked in HCl for 15 min to eliminate RNase contamination, thoroughly rinsed with RNase-free water, and then blocked for 1 h with 250 mM KCl, 10 mM potassium phosphate (pH 8), 1 mM MgCl2, and 40 μg/mL BSA. RNA stocks were diluted in water, heated to 65 °C for 5 min, and quenched on ice. A 1/10 volume of 10× buffer was added to complete RNA folding.
Fluorescence emission spectra were recorded on samples containing 10 nM RNA and variable protein concentrations (as indicated in the figures). The buffer was the same as that used for blocking. The temperature was held constant with a circulating water bath at 23 °C. Protein stocks were sufficiently concentrated that the RNA dilution was <1%. The excitation wavelength was 490 nm, and the slit widths were 8 and 2 nm for the excitation and emission monochromators, respectively. The emission wavelength was varied between 495 and 600 nm. Buffer reference spectra were subtracted from the sample spectra, and the emission intensities were normalized to the maximal intensity of the free RNA.
SNF/U2A′ titrations were performed in 250 mM KCl, 10 mM potassium phosphate (pH 8), 1 mM MgCl2, 40 μg/mL BSA, 5 mM DTT, and RNasin. Titrations were performed at 23 °C while the mixtures were being constantly stirred. For a single titration of SNF or the SNF/U2A′ complex into FAM-RNA, the cuvette and titrant concentration of FAM-RNA was held constant at 0.1 or 0.5 nM (the lower concentration was used for the highest-affinity interactions). The cuvette and titrant also contained identical concentrations of U2A′. The sample was excited at 490 nm, and the emission intensity was recorded at 520 nm (excitation and emission slit openings of 8 and 16 nm, respectively). SNF or the SNF/U2A′ complex was titrated into the RNA, and the fluorescence emission intensity was recorded for each addition of SNF. The intensity data were converted to fraction fluorescence enhancement and normalized to the maximal fluorescence enhancement. Titrations were collected at multiple concentrations of U2A′, and the data were globally fit in Scientist (Micromath) to eqs 1–4; fractional fluorescence enhancement corresponds to the fraction of RNA bound either by SNF or by the SNF/U2A′ complex. Titration series were repeated at least twice for each RNA. The parameter values listed in Table 1 represent the average of the series fits, with uncertainties that are the larger of either the propagated error or the standard deviation between measurements.
Table 1. Thermodynamic Binding Parameters for SNF, RNA, and U2A′a.
| SNF and FAM-SLII | SNF and FAM-SLIV | |
|---|---|---|
| KD,R,app (M) (1/KR) | (1.1 ± 0.5) × 10–9 | (8.3 ± 0.4) × 10–8 |
| ΔG°(R,binding) (kcal/mol) | –12.1 ± 0.3 | –9.6 ± 0.1 |
| KD,Uapp (M) (1/KU) | nab | (1.4 ± 0.2) × 10–6 |
| ΔG°(U,binding) (kcal/mol) | nab | –7.9 ± 0.1 |
| α | 2.2 ± 0.4 | 361 ± 51 |
| Δg = −RT ln(α) (kcal/mol) | –0.47 ± 0.1 | –3.5 ± 0.1 |
R is RNA. U is U2A′. SLII and SLIV are labeled with fluorescein (FAM). Binding buffer for all experiments consisted of 250 mM KCl, 10 mM potassium phosphate (pH 8.0), 1 mM MgCl2, 40 μg/mL BSA, 5 mM DTT, and RNasin at 22 °C. Parameter values reflect the average values from at least two separate data series. Uncertainties represent the larger of either the standard deviation of parameter values from different fits or the propagated error.
Data not available from this experiment.
Partitioning surfaces were simulated in Scientist based on binding parameters determined in the fluorescence experiments. For these surfaces, SLII and SLIV were considered competitive ligands for SNF. The partitioning surfaces were plotted in MatLab.
2-Aminopurine Fluorescence Experiments
2-Aminopurine (2AP) SLIV (Dharmacon) had the sequence 5′-GGCCGUAUUGCAGU-2AP-CCGCGGCC. The RNA stock was diluted to 300 nM in water, heated to 95 °C for 3 min, and quenched on ice. A concentrated buffer stock was added to bring the salt concentration to 50 mM KCl, with 10 mM cacodylate (pH 7) (the lower salt concentration prevented RNA dimerization).
Cuvettes and stir bars were washed with acid and blocked with BSA as described. The temperature was held constant with a circulating water bath at 23 °C. Protein stocks were sufficiently concentrated such that the RNA dilution was <1% upon addition. The excitation wavelength was 310 nm, and the slit widths were 8 and 2 nm for the excitation and emission monochromators, respectively. A polarizer in the emission path parallel to the monochromator gratings eliminated monochromator artifacts from Wood’s anomaly. The emission wavelength was varied between 340 and 460 nm. Buffer reference spectra were subtracted from the sample spectra, and the emission intensities were normalized to the maximal intensity of the free RNA.
Circular Dichroism (CD) Spectroscopy
CD spectra were buffer-subtracted and recorded at room temperature on a Jasco J715 instrument. RNA experiments were performed with an RNA concentration of 2 μM in 50 mM KCl and 10 mM cacodylate. Spectra were collected from 375 to 210 nm. For experiments with protein, protein was added to a concentration of 2 μM (SNF or SNF and U2A′). The hairpin RNA sequences were 5′-GGCCGCAUUGCACUCCGCGGCC (SLII) and 5′-GGCCGUAUUGCAGUACCGCGGCC (SLIV).
ITC Experiments
Protein samples were diluted from stock solutions into 100 mM arginine, 50 mM KCl, and 10 mM cacodylate (pH 7) and dialyzed in mini dialyzers (ThermoScientific, 2000 molecular weight cutoff) against that buffer. Final samples were prepared by diluting the protein solutions (SNF and U2A′) with equal volumes of the final buffer, including 5 mM BME. Samples were degassed prior to being loaded into the ITC injection syringe or cell. Titrations were performed on a NanoITC instrument (TAinstruments) and analyzed using NanoAnalyze.
Results
SNF/RNA/U2A′ Ternary Complexes
We have previously determined dissociation constants for binding of SNF to SLII and SLIV.4 In those experiments, we compared the binding of FL SNF, RRM1, and RRM2. We found that RRM2 does not bind to either SLIV or SLII or to a single-stranded random sequence RNA. We also determined that FL SNF and SNF RRM1 bind with a 1:1 stoichiometry to either hairpin. We now consider three-component systems (RNA, SNF, and U2A′) to explore possible mechanisms of U2A′ localization.
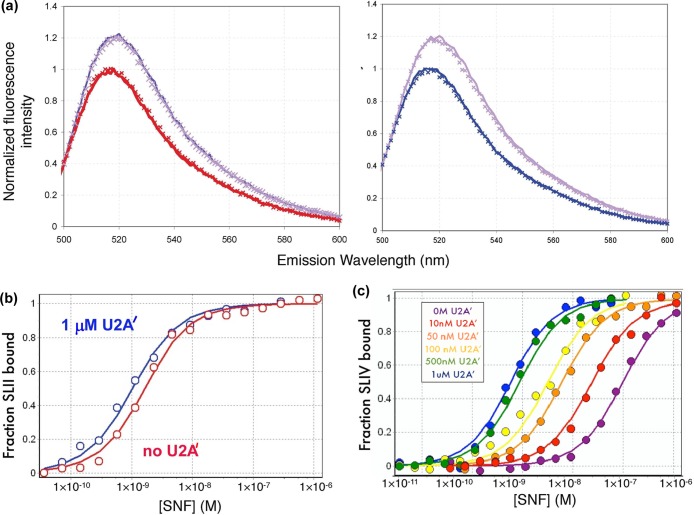
To determine the properties of formation of the RNA/SNF/U2A′ complex, the SNF/RNA binding affinity was measured at different U2A′ concentrations. Binding was monitored by fluorescence intensity changes of FAM-RNA upon addition of protein [FAM does not alter RNA binding affinity (see Materials and Methods)]. Addition of a saturating amount of SNF results in a 20% enhancement of the FAM-SLII or FAM-SLIV fluorescence intensity at 520 nm. No further change in fluorescence was observed when a large excess of U2A′ was added to the RNA alone or to the RNA bound to SNF (Figure 2a). For binding titrations, the fluorescence intensity can therefore be monitored to detect protein binding, and the enhancement is a result of binding of RNA to SNF alone or to the SNF/U2A′ complex. Representative binding curves for these experiments are shown in panels b and c of Figure 2. We observed that the presence of U2A′ imparts a marginal increase in the affinity of SNF for SLII but a very large increase in the affinity of SNF for SLIV.
Figure 2.
Binding isotherms for SLII and SLIV. (a) Fluorescence spectra of FAM-SLII (left, red) and FAM-SLIV (right, blue) increase ∼20% at 520 nm when the RNA is bound to either SNF (solid line) or SNF and U2A′ (×) under saturating conditions. Addition of 1 μM U2A′ alone did not change the FAM-RNA fluorescence (indicated by × on the RNA only spectrum). (b) Titration of SNF with or without U2A′ into fluorescein-labeled SLII. (c) Titration of SNF with or without U2A′ into fluorescein-labeled SLIV. The concentration of SLIV varied with the U2A′ concentration but was <1 nM. The SLII concentration was 0.1 nM. Conditions: 250 mM KCl, 10 mM potassium phosphate (pH 8), and 1 mM MgCl2 at 22 °C.
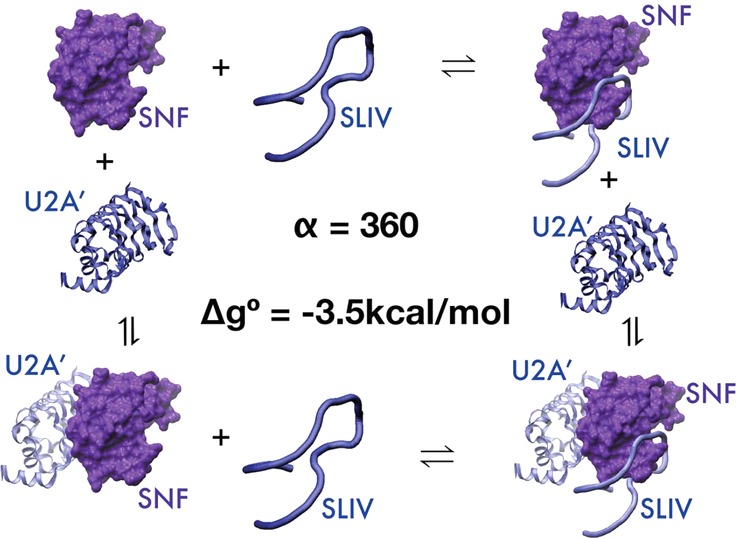
A schematic of the thermodynamic cycle for ternary complex formation is shown in Figure 3, with the right panel depicting the macromolecules. On the left, S represents the SNF protein, U represents U2A′, and R represents the RNA, either SLII or SLIV. The individual bimolecular binding events have characteristic binding parameters; KR and KU represent the bimolecular association constants for the SNF/RNA and SNF/U2A′ interactions, respectively. These binding events are also characterized by free energies of binding, ΔGRNA and ΔGU2A′, respectively. The ternary complexes can be formed by binding of U2A′ to the preformed SNF/RNA complex or by binding of RNA to the preformed SNF/U2A′ complex. These are defined by association constants KU,R and KR,U, respectively.
Figure 3.
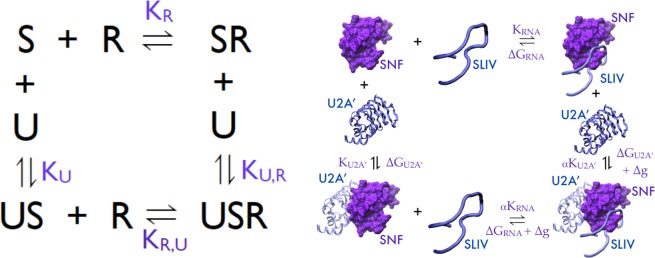
Schematics of the binding model and thermodynamic cycles for ternary complex formation. S is SNF. R is RNA. U is U2A′. The right panel gives a pictorial representation of the thermodynamic cycle. Cooperativity factor α is shown.
Consider that SNF (S) is the macromolecule that can bind two ligands, each of which binds at a single site, in the thermodynamic cycle shown in Figure 3. Conservation of energy requires that KR,U = αKR and KU,R = αKU, where α is the cooperativity parameter and describes the extent to which binding by one ligand affects binding of the second ligand. If α > 1, there is positive cooperativity between the binding events (binding by either ligand improves binding of the second ligand). When α = 1, there is no cooperativity; binding by either ligand is independent of the other. If α < 1, there is negative cooperativity in binding of the ligands. In the case of competitive ligand binding, where binding by one ligand completely precludes binding of the second ligand, α = 0. The free energy associated with cooperativity is given by Δg = −RT ln(α).
All binding data were globally fit to eqs 1–4 to obtain the two bimolecular association constants KR and KU, as well as α, the cooperativity parameter.
| 1 |
| 2 |
| 3 |
| 4 |
where FS+US is the fraction of the total RNA, bound either to SNF (S) or to the U2A′/SNF complex (US); RT, UT, and ST are the total RNA, U2A′, and SNF concentrations, respectively; R, U, and S are the concentrations of free RNA, U2A′, and SNF, respectively; α is the cooperativity parameter; and KR and KU are the bimolecular association constants for the SNF/RNA and SNF/U2A′ interactions, respectively.
We find that cooperativity of ternary complex formation depends on the RNA species bound (Table 1) [note that binding dissociation constants KD are given; KD(U,R) = 1/KU,R]. Cooperativity between U2A′ and SLII binding to SNF is only marginal (α = 2; Δg° = −0.5 kcal/mol), so it was not possible to reliably determine the bimolecular binding constant for the SNF/U2A′ interaction from these titrations. Instead, the protein/protein bimolecular binding constant was fixed to the value determined in the SLIV binding assays. For SNF binding to SLIV, the cooperativity between U2A′ and SLIV binding is very large; binding by either molecule increases the binding affinity for the other by a factor of 350 (α). Even though the apparent affinity of U2A′ for SNF in the absence of RNA is only ∼1.5 μM, the high degree of cooperativity between U2A′ and SLIV binding to SNF means that the affinity of the SLIV/SNF complex for U2A′ is 4 nM. Similarly, the apparent affinity of SNF for SLIV is shifted from 80 to 0.25 nM. Given the large cooperativity, the shift in the SLIV binding curve approaches the U2A′ saturation limit. This result is striking, corresponding to a free energy of cooperativity (Δg°) of −3.5 kcal/mol. This is a dramatic example of both the degree to which cooperativity can affect binding and of the RNA dependence of this phenomenon.
In Vivo Partitioning of Proteins in snRNPs
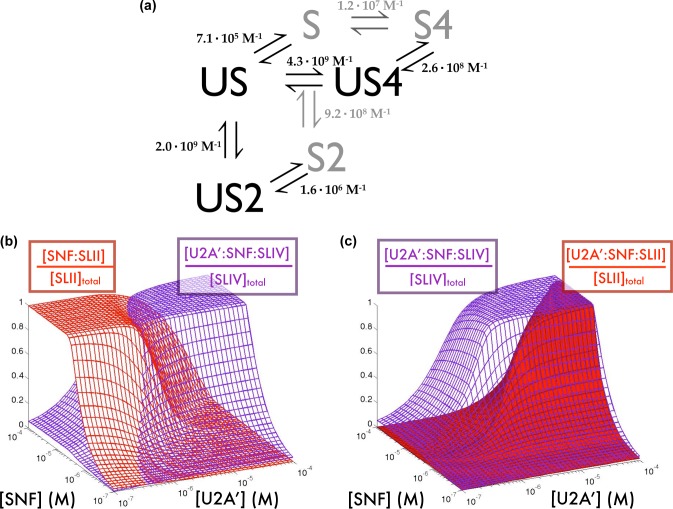
Using the experimentally determined thermodynamic parameters, we simulated the fraction of cellular U1 and U2 snRNA that would be bound by the various proteins when both proteins and both RNAs are considered simultaneously. Figure 4a shows a schematic of the two-protein, two-RNA system and all relevant binding constants. In this analysis, SLII and SLIV are considered to be competitive ligands for SNF. Panels b and c of Figure 4 and Figure 1 of the Supporting Information show the fraction of SLII and SLIV bound by SNF and U2A′ over a wide range of possible SNF and U2A′ concentrations. These simulations use the approximate cellular concentrations of U1 and U2 snRNAs of 3 and 1.5 μM, respectively.12
Figure 4.
Modeling protein distributions on snRNPs. (a) Thermodynamic model including both SLII and SLIV RNAs with binding parameters obtained from fluorescence titrations. (b) Fractions of SLII found in a bimolecular complex with SNF (red) and SLIV in a ternary complex (purple). (c) Partitioning surface showing the fraction of both SLII (red) and SLIV (purple) in ternary complexes. SLII is found primarily in the bimolecular complex and SLIV primarily in the ternary complex when [SNF] > [U2A′].
Several important observations can be made from the models of protein partitioning. First, there is a significant range of U2A′ and SNF protein concentrations for which most of SLII is found in a bimolecular complex with SNF and most of SLIV is in a ternary complex with both SNF and U2A′ (Figure 4b). Second, U2A′ partitions to the U1 snRNP only when [U2A′] > [SNF] (Figure 4c), which is generally not a condition found in cells. Even though binding of U2A′ and SLII is not negatively cooperative, the difference in the free energy of binding cooperativity (ΔΔg = 3 kcal/mol) between U2A′ and the SLII/SLIV complex binding to SNF is sufficient to effectively partition the U2A′ protein away from the U1 snRNP and into the U2 snRNP, when the concentrations of the various components are found at expected cellular levels.
Protein/Protein Interaction
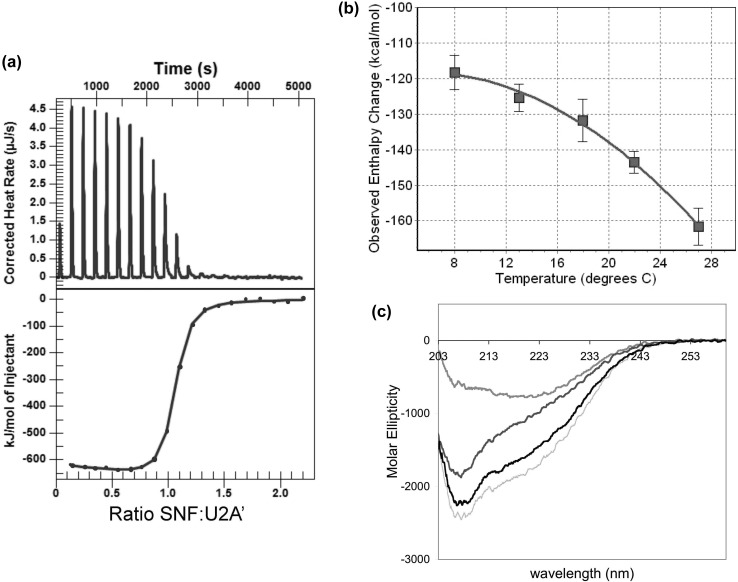
Direct measurement of the bimolecular association of U2A′ and RRMs has not been done previously. We used ITC to measure the binding thermodynamics. The titration of U2A′ with SNF shows a very large apparent enthalpy of binding (Figure 5a) that is temperature-dependent (Figure 5b), indicating a change in heat capacity (ΔCp) associated with binding. Given the nonlinearity of the temperature dependence, the data were fit to a model13 that takes into account a temperature dependence of ΔCp:
| 5 |
where T is the temperature in kelvin, TR is an arbitrary reference temperature (we chose 295 K), and ΔHR and ΔCp,R are the apparent enthalpy and heat capacity of binding at the reference temperature, respectively. Fitting the data to this model yields the following values: ΔCp,R = −3.1 ± 0.2 kcal mol–1 K–1, ΔHR = −144 ± 4 kcal/mol, and ΔΔCp = −190 ± 40 cal mol–1 K–2. To understand the origin of the large ΔCp,R and ΔH, we considered several sources that might contribute.
Figure 5.
Protein/protein interaction thermodynamics. (a) Calorimetric titration of SNF into U2A′ shows a large and negative apparent enthalpy of binding. (b) Temperature dependence of the observed enthalpy of interaction indicates a large apparent heat capacity of binding. Calorimetric titrations were conducted in 100 mM arginine, 50 mM KCl, and 10 mM cacodylate (pH 7). (c) CD spectra of SNF (gray), U2A′, and an equimolar mixture (black) show some nonadditivity in the spectra; the hashed line indicates the sum of the SNF and U2A′ spectra.
The protein/protein binding mechanism includes burial of hydrophobic surfaces. On the basis of the SLIV/U2B″/U2A′ cocrystal structure,8 we calculate there is burial of 629 Å2 of polar surface area and 1184 Å2 of apolar surface at the U2B″/U2A′ interface. Applying estimates of binding enthalpy from surface burial14 yields a predicted binding enthalpy (ΔH) of −15 kcal/mol at 22 °C. The measured apparent heat capacity and enthalpy of binding for SNF/U2A′ far exceed this estimate, so unless the binding of SNF to U2A′ is very different from the binding of U2B″ RRM1 to U2A′, there must be other contributions.
Contributions to the observed enthalpy could come from coupling of protonation or ion binding and/or release to complex formation. Cacodylate was used as the buffer in most of the calorimetric titrations in part because the ionization enthalpy of cacodylate is very small (−0.72 kcal/mol).15 To estimate the effect of linked protonation equilibria, experiments were repeated in ACES buffer, which has a much higher ionization enthalpy (7.17 kcal/mol) (both experiments conducted at pH 7.0). This analysis showed a net release of approximately eight protons from the solvent on binding. The source of the large linkage between binding and protonation needs to be investigated to improve our understanding of the binding mechanism.
Conformational changes coupled to binding are a common source of an apparent heat capacity.16,17 We used CD to assess changes in the secondary structure of the proteins upon binding (Figure 5c). CD spectra of SNF, U2A′, and a 1:1 mixture of the proteins show that the spectra are not entirely additive, suggesting some degree of change to the secondary structure upon binding. However, the difference spectrum is small compared to that of other protein–protein interactions with large values of ΔCp. For the SNF/U2A′ interaction, while the changes in overall secondary structure appear to be minor, it is possible that there are significant changes in the tertiary structure of one or both components that are coupled to binding and contribute to the large apparent ΔH and ΔCp.
Protein/RNA Interactions
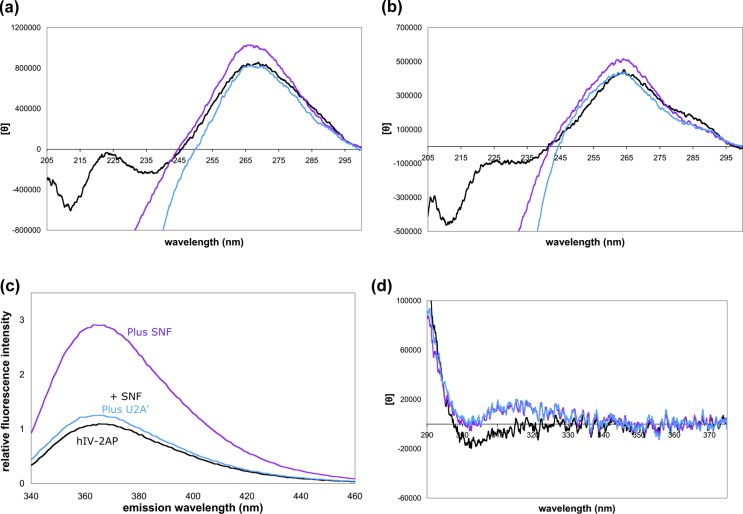
Cocrystals7,8 first suggested that RNA binding to RRMs results in significant distortion of the loops of U1 SLII and U2 SLIV. Most significantly, the RNA loop must open up upon protein binding, which allows formation of the specific contacts between the protein and RNA. To probe conformational changes to the RNA upon protein binding, we measured CD spectra of SLII and SLIV in the presence and absence of SNF and U2A′ (Figure 6a,b). Between 240 and 300 nm, the contribution of the protein to the CD signal is negligible compared to that of the RNA. Changes in the CD spectrum can therefore be attributed to changes in the RNA structure upon binding.
Figure 6.
RNA conformations in binary and ternary complexes. (a) CD spectra of SLII as free RNA (black), RNA with SNF (purple), and RNA with SNF and U2A′ (blue). Stacking of nucleobases increases the ellipticity at 260 nm, so the RNA bases appear to be changing their relative orientations. (b) CD spectra of SLIV as free RNA, RNA with SNF, and RNA with SNF and U2A′. (c) Fluorescence emission spectra of SLIV with 2-aminopurine in the loop show large changes upon protein binding. (d) Low-energy CD spectra of 2-aminopurine in the SLIV loop, with (blue and purple) and without (black) proteins [2 μM RNA or 2 μM SNF or SNF/U2A′ in 50 mM KCl and 10 mM sodium cacodylate (pH 7)].
SNF binding results in an overall increase in the magnitude of the CD signal of the RNA band centered at ∼265 nm, consistent with an increased level of base stacking. Further addition of U2A′ (and formation of the ternary complex), however, results in a significant decrease in the intensity of the CD bands, suggesting unstacking of the loop nucleobases.
The 3′-UCC of the SLII loop does not make contact with U1A;18 neither does the 3′-ACC of SLIV make contact with U2B″ or U2A′ in the cocrystal.8 We previously replaced the 3′-loop adenine of SLIV with 2-aminopurine (2AP) and showed that it does not affect the RNA binding affinity of SLIV for SNF;19 this nucleotide is stacked with its neighboring bases in the free RNA but becomes flipped out of the stack upon binding to SNF.19 Unexpectedly, when U2A′ is added to the preformed SNF/SLIV complex, the 2AP fluorescence intensity is quenched (Figure 6c). The signal can be recovered by addition of a large excess of SNF (data not shown), which presumably increases the relative population of the bimolecular SNF/RNA complex.
At wavelengths greater than 300 nm, 2-aminopurine can show an induced CD band that is sensitive to the environment of the nucleobase.20 Comparing the low-energy CD spectra of free RNA and bound RNA in either the bimolecular or ternary complex shows a substantial increase in the magnitude of the induced CD signal at 315 nm, suggesting a change in the electronic environment of 2AP (Figure 6d). Fluorescence quenching upon U2A′ binding may be due to the increased flexibility of neighboring bases that transiently stack with the 2AP, while 2AP remains in an environment that retains an induced CD. Currently, we do not have a molecular explanation for the 2AP signal changes when proteins are bound, but the data indicate that the RNA undergoes conformational changes in both complexes.
Discussion
The biological implications of the cooperativity that produces the SLIV/SNF/U2A′ ternary complex are complex. The most obvious is the localization of the U2A′ protein to the U2 snRNP and its exclusion from the U1 snRNP. The function of the U2A′ protein in the U2 snRNP is not clear, although there are some experimental results that suggest it is a crucial element of U2 snRNP stability and spliceosome assembly.21,22 Second, as noted in earlier studies, U2A′ does enhance the affinity of SNF for U2 snRNA SLIV. We conclude that the RNA sequence modulates the cooperativity and so determines the localization of U2A′.
The molecular origin of the cooperativity (α) is the predominant unknown that arises from these results. Because the degree of cooperativity determines the form of the SNF complex in vivo, the physical basis of the thermodynamic signature is important to understand. More specifically, we want to understand the origin of α = 350-fold enhancement (positive heterotropic linkage) of binding of SNF to either SLIV or U2A′ (upon binding by the other) with a corresponding free energy (Δg°) of −3.5 kcal/mol. The cooperativity is dependent on the RNA, because linkage effects between binding of SNF to U2A′ and SLII are slightly positive but weak (α = 2.2; Δg° = −0.5 kcal/mol). The RNA dependence of the linkage effects is sufficient to explain why U2A′ is effectively partitioned to the U2 snRNP and excluded from the U1 snRNP.
Conformational changes of proteins and RNA coupled to binding are known contributors to observed large and negative apparent binding enthalpies as well as a large apparent ΔCp. Formation of the SLIV/SNF/U2A′ complex certainly requires conformational changes of the RNA, as shown by our spectroscopic data that monitor the SLIV hairpin loop. SNF likely undergoes conformational changes upon RNA binding, as well, much as U1A RRM1 undergoes a conformational change when bound to SLII (loop 3 protrudes through the RNA loop). SNF RRM1 itself is sampling conformational space, as determined by its NMR spectra: the entire RRM1 undergoes conformational averaging on the chemical shift time scale, suggesting that it is best described as an ensemble of structures.23 In addition, the free and bound conformation of the U2A′ LRR domain could be significantly different, or its conformational sampling could be altered. Coupled conformational changes are likely to be a major contributor to the observed cooperativity, the heat capacity, and the large apparent enthalpy of binding.
Conformational changes coupled to binding might imply that a macromolecule alters its conformation only when a ligand is bound, and such is the premise of the concept of induced fit.24,25 However, the free states of SNF and the RNAs are best described as ensembles of structures. Their binding is best described by conformational selection26,27 in which the structural ensemble is thought to include conformations that are competent to bind ligand. A recent example of this process for RNA/protein binding is seen at the single-molecule level, looking at the conformational ensemble of an RNA five-way junction before and after a protein binds (S4 protein binding to a rRNA five-way junction).28 Combining the mechanisms of induced fit and conformational selection29,30 with the thermodynamics that couple conformational changes to binding will be a challenge in the SLIV/SNF/U2A′ system.
What Is α? Implications for Allostery in SNF Interactions
In 1961, Monod and Jacob introduced the term “allosteric”,31 and the first model to explain the allosteric effect was proposed in 1965.32 The model postulated that the protein existed in an equilibrium between at least two states. Since then, additional models for allostery have emerged (most notably the KNF or sequential model33). However, the term “allostery” has been used to encompass a much broader range of phenomena; almost any “action at a distance” has been described as allostery. The feature common to most of what is described as allostery is the presence of an allosteric binding site. This is a site that is distant from the functional (orthosteric) site; the allosteric site can be a catalytic site or a binding site for a second molecule. When the allosteric site is occupied, the activity of the molecule at the orthosteric site is altered.
If the SLIV/U2B″/U2A′ cocrystal structure is representative of the SLIV/SNF/U2A′ ternary complex, then binding sites for the two ligands (the RNA and U2A′) are distinct. Our data show that binding of RNA to the SNF RRM affects binding of U2A′, and vice versa. Thus, the system meets the two criteria for allostery.
The system is unusual in terms of descriptions of allostery because the ligands (and the ligand binding surfaces) are quite large. Using U2B″ as a model, we calculated that 40% of the SNF RRM1 surface is part of an intermolecular interface. More important than the size of the ligands, however, is the fact that at least one ligand (the RNA) clearly experiences its own conformational heterogeneity, which is modulated by binding. Allostery in larger macromolecular complexes34,35 will need to account for conformational heterogeneity of “ligands” as well as conformational changes of the “macromolecule”.
If in a considerable simplification of the system, we consider the RNA/SNF/U2A′ complex in terms of a two-state SNF equilibrium ensemble, α is given by (see the Supporting Information)
| 6 |
where KC is the equilibrium constant between the two states of SNF and β and γ are the ratios of the binding constants of each state of SNF for each ligand. As a consequence, α is limited by KC, and regardless of β and γ, the maximal value of α is ∼1/KC. For unbound SNF, this means that the free energy difference between the low- and high-affinity states must be at least 3.5 kcal/mol to account for the experimental data [Δg = −RT ln(α) = −3.5 kcal/mol], but this difference is equal to the SNF RRM1 folding free energy [ΔG°(folding) = −3.5 ± 0.3 kcal/mol].4 The observed linkage (α) between U2A′ and RNA binding to SNF could occur if the major conformation of free SNF has a weak affinity for the two ligands but a minor conformation has a high affinity for the ligands. This scenario would require that both the RNA and U2A′ binding surfaces of SNF are substantially different in the two conformations.
Assuming two-state exchange is the basis of the allosteric effect, the difference in linkage between SLII and SLIV binding and U2A′ binding could be explained by substantially different affinities of the RNAs for the two states (in eq 6, βSLIV ≫ βSLII). However, we know that at the very least, the conformational landscape of the RNAs is best described as an ensemble of states, so we must consider whether the internal equilibria of the ligands can substantially alter the measured linkage parameter and/or allosteric response. If we introduce two-state exchange phenomena in one ligand, we obtain the following dependence of α:
| 7 |
where KL is the equilibrium constant for the ligand exchange process and β and μ are the ratios of the binding constants to the two states of the macromolecule for the two states of the ligand (Figure 2 of the Supporting Information). This model requires an allosteric response of the macromolecule (if KC = 0, then α = 1). Given identical ligand exchange-independent parameters, α(exchange) can be greater than or less than α(no ligand exchange). The analysis can be extended to include internal equilibrium of both ligands, with similar results.
The ensemble allosteric model (EAM) is a more general model of allostery36 that includes both MWC and KNF models as special cases. In the EAM, the two ligand binding sites can be treated formally as separate “domains” that can interact. Each domain can sample distinct conformations. Assuming two-state exchange, the equilibria between states of both domains (in the absence of interactions between them) are given by K1 and K2. This is modified by a factor when the two domains interact. If simple two-state ligand internal equilibria are introduced into the EAM, modulation of the linkage parameter α is also seen. Like the simpler model, an allosteric response (α ≠ 1) requires that the macromolecule undergo exchange. If K1 or K2 is zero or if there is no interaction between the domains, then α = 1. While ligand internal equilibria can modify the degree of the allosteric response, this model predicts that allostery requires an energetic change in both domains. It also predicts that the two domains thermodynamically interact when the two binding sites are occupied.
While it is possible that the two RNAs have very different ΔΔG values for the states of SNF (which could account for the difference in the linkage effect), it is also possible that differences in the conformational landscapes of the RNAs (and how they bind protein) are important in the difference between αSLII and αSLIV. Ligand internal equilibria can have a dramatic impact on the observed allostery of the system, but determining the thermodynamic origins of allostery in this system and in other systems will be challenging. Attention has recently focused on allosteric effects that are mediated by changes in protein dynamics, as well as changes in protein structure.37−39 We suggest that such effects are probably ubiquitous and important in the assembly and function of larger macromolecular complexes. This is particularly likely in RNA–protein complexes, where both macromolecules are flexible.
RNA Recognition by Proteins
Protein recognition of RNAs is a complex process; while many structural studies have provided insight into the binding of discrete protein domains to particular tracts of RNA, most RNA binding domains are found in the context of larger proteins, which often contribute to RNA binding. Careful studies of multidomain protein recognition of RNA targets have been undertaken;40 these studies highlight the heterogeneity of mechanisms used to achieve RNA binding specificity.
Large changes in the free energy of binding have been reported for protein/RNA/protein complexes, in which binding by one protein is coupled to a large conformational change in the RNA, which results in a large apparent increase in the affinity for the second protein. One example occurs in 16S rRNA where S15 protein binding to the rRNA was found to increase the free energy of binding of the S6/S18 heterodimer to the 16S rRNA by at least 6.5 kcal/mol.41 Substantial work has shown that protein/protein interactions, coupled with protein/RNA interactions, very significantly impact the catalytic activity of archaeal RNase P,42,43 although the thermodynamics and kinetics have not been completely resolved.
Our results show that a protein/RNA interaction can have a very large (350-fold) impact on protein/protein binding; this is an RNA-specific effect, as a highly similar RNA sequence elicits very little change in the protein/protein interaction. The effect has biological consequences, as it is sufficient to explain the protein partitioning behavior of the system and localize U2A′ exclusively to the U2 snRNP. We consider it likely that such phenomena of coupled binding are important in localizing many proteins within RNPs.
Acknowledgments
We thank W. Tom Stump for lab management.
Glossary
Abbreviations
- RRM
RNA recognition motif
- LRR
leucine-rich repeat
- ITC
isothermal titration calorimetry
- snRNP
small nuclear ribonucleoprotein
- EMSA
electrophoretic mobility shift assay.
Supporting Information Available
Simulations of protein distributions on snRNAs and the dependence of α on systems where both components undergo exchange, as well as the formalism for the description of α. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This work was supported by National Institutes of Health Grants 5R01 GM096444-01 to K.B.H. and F31-GM089576-01 to S.G.W.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Will C. L.; Lührmann R. (2011) Spliceosome structure and function. Cold Spring Harbor Perspect. Biol. 3, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polycarpou-Schwarz M.; Gunderson S. I.; Kandels-Lewis S.; Seraphin B.; Mattaj I. W. (1996) Drosophila SNF/D25 combines the functions of the two snRNP proteins U1A and U2B′ that are encoded separately in human, potato, and yeast. RNA 2, 11–23. [PMC free article] [PubMed] [Google Scholar]
- Harper D. S.; Fresco L. D.; Keene J. D. (1992) RNA binding specificity of a Drosophila snRNP protein that shares sequence homology with mammalian U1-A and U2-B″ proteins. Nucleic Acids Res. 20, 3645–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. G.; Hall K. B. (2010) Coevolution of Drosophila snf protein and its snRNA targets. Biochemistry 49, 4571–4582. [DOI] [PubMed] [Google Scholar]
- Salz H. K. (1992) The genetic analysis of snf: A Drosophila sex determination gene required for activation of Sex-lethal in both the germline and the soma. Genetics 130, 547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J.; Cui G.; Li C.; Liu C.; Shang E.; Lai L.; Jin C.; Wang J.; Xia B. (2009) Structure and novel functional mechanism of Drosophila SNF in sex-lethal splicing. PLoS One 4, e6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oubridge C.; Ito N.; Evans P. R.; Teo C. H.; Nagai K. (1994) Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372, 432–438. [DOI] [PubMed] [Google Scholar]
- Price S. R.; Evans P. R.; Nagai K. (1998) Crystal structure of the spliceosomal U2B″-U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature 394, 645–650. [DOI] [PubMed] [Google Scholar]
- Dosztányi Z.; Csizmok V.; Tompa P.; Simon I. (2005) IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434. [DOI] [PubMed] [Google Scholar]
- Williams S. G.; Hall K. B. (2011) Human U2B″ protein binding to snRNA stemloops. Biophys. Chem. 159, 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall K. B. (1994) Interaction of RNA hairpins with the human U1A N-terminal RNA binding domain. Biochemistry 33, 10076–10088. [DOI] [PubMed] [Google Scholar]
- Tycowski K. T., Kolev N. G., Conrad N. K., Fok V., and Steitz J. A. (2006) The evergrowing world of small nuclear ribonucleoproteins. In RNA World, 3rd ed., pp 327–368, Cold Spring Harbor Monograph Archive, Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- LiCata V. J.; Liu C.-C. (2011) Analysis of free energy versus temperature curves in protein folding and macromolecular interactions. Methods Enzymol. 488, 219–238. [DOI] [PubMed] [Google Scholar]
- Baker B. M.; Murphy K. P. (1998) Prediction of binding energetics from structure using empirical parameterization. Methods Enzymol. 295, 294–315. [DOI] [PubMed] [Google Scholar]
- Goldberg R. N.; Kishore N.; Lennen R. M. (2002) Thermodynamic quantities for the ionization reactions of buffers. J. Phys. Chem. Ref. Data 31, 231–369. [Google Scholar]
- Spolar R. S.; Record M. T. (1994) Coupling of local folding to site-specific binding of proteins to DNA. Science 263, 777–784. [DOI] [PubMed] [Google Scholar]
- Doyle M., and Hensley P. (2005) Thermochemistry of binary and ternary protein interactions measured by titration calorimetry: Complex formation of CD4, HIV gp120, and anti-gp120. In Proteomics and Protein-Protein Interactions Protein Reviews (Waksman G., Ed.) pp 147–163, Springer, New York. [Google Scholar]
- Williams D. J.; Hall K. B. (1996) RNA hairpins with non-nucleotide spacers bind efficiently to the human U1A protein. J. Mol. Biol. 257, 265–275. [DOI] [PubMed] [Google Scholar]
- Rau M.; Stump W. T.; Hall K. B. (2012) Intrinsic flexibility of snRNA hairpin loops facilitates protein binding. RNA 18, 1984–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N. P.; Baase W. A.; von Hippel P. H. (2005) Low energy CD of RNA hairpin unveils a loop conformation required for λN antitermination activity. J. Biol. Chem. 280, 32177–32183. [DOI] [PubMed] [Google Scholar]
- Pan Z. Q.; Prives C. (1989) U2 snRNA sequences that bind U2-specific proteins are dispensable for the function of U2 snRNP in splicing. Genes Dev. 3, 1887–1898. [DOI] [PubMed] [Google Scholar]
- Hamm J.; Dathan N. A.; Mattaj I. W. (1989) Functional analysis of mutant Xenopus U2 snRNAs. Cell 59, 159–169. [DOI] [PubMed] [Google Scholar]
- Williams S. G.; Harms M. J.; Hall K. B. (2013) Resurrection of an Urbilaterian U1A/U2B″/SNF protein. J. Mol. Biol. 425, 3846–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson J. R. (2000) Induced fit in RNA-protein recognition. Nat. Struct. Biol. 7, 834–837. [DOI] [PubMed] [Google Scholar]
- Koshland D. E. (1958) Application of a Theory of Enzyme Specificity to Protein Synthesis. Proc. Natl. Acad. Sci. U.S.A. 44, 98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller A.; Rieder U.; Aigner M.; Blanchard S. C.; Micura R. (2011) Conformational capture of the SAM-II riboswitch. Nat. Chem. Biol. 7, 393–400. [DOI] [PubMed] [Google Scholar]
- Leulliot N.; Varani G. (2001) Current topics in RNA-protein recognition: Control of specificity and biological function through induced fit and conformational capture. Biochemistry 40, 7947–7956. [DOI] [PubMed] [Google Scholar]
- Kim H.; Abeysirigunawarden S. C.; Chen K.; Mayerle M.; Ragunathan K.; Luthey-Schulten Z.; Ha T.; Woodson S. A. (2014) Protein-guided RNA dynamics during early ribosome assembly. Nature 506, 334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P.; Palotai R.; Nussinov R. (2010) Induced fit, conformational selection and independent dynamic segments: An extended view of binding events. Trends Biochem. Sci. 35, 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes G. G.; Chang Y.-C.; Oas T. G. (2009) Conformational selection or induced fit: A flux description of reaction mechanism. Proc. Natl. Acad. Sci. U.S.A. 106, 13737–13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J.; Jacob F. (1961) Teleonomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harbor Symp. Quant. Biol. 26, 389–401. [DOI] [PubMed] [Google Scholar]
- Monod J.; Wyman J.; Changuex J. P. (1965) On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 12, 88–118. [DOI] [PubMed] [Google Scholar]
- Koshland D. E.; Némethy G.; Filmer D. (1966) Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5, 365–385. [DOI] [PubMed] [Google Scholar]
- Safaee N.; Kozlov G.; Noronha A. M.; Xie J.; Wilds C. J.; Gehring K. (2012) Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol. Cell 48, 375–386. [DOI] [PubMed] [Google Scholar]
- Duke T. A.; Le Novère N.; Bray D. (2001) Conformational spread in a ring of proteins: A stochastic approach to allostery. J. Mol. Biol. 308, 541–553. [DOI] [PubMed] [Google Scholar]
- Hilser V. J.; García-Moreno E. B.; Oas T. G.; Kapp G.; Whitten S. T. (2006) A statistical thermodynamic model of the protein ensemble. Chem. Rev. 106, 1545–1558. [DOI] [PubMed] [Google Scholar]
- Boehr D. D.; Nussinov R.; Wright P. E. (2009) The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.-J.; del Sol A.; Nussinov R. (2008) Allostery: Absence of a change in shape does not imply that allostery is not at play. J. Mol. Biol. 378, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q.; Karplus M. (2008) Allostery and cooperativity revisited. Protein Sci. 17, 1295–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackereth C. D.; Sattler M. (2012) Dynamics in multi-domain protein recognition of RNA. Curr. Opin. Struct. Biol. 22, 287–296. [DOI] [PubMed] [Google Scholar]
- Recht M. I.; Williamson J. R. (2001) Central domain assembly: Thermodynamics and kinetics of S6 and S18 binding to an S15-RNA complex. J. Mol. Biol. 313, 35–48. [DOI] [PubMed] [Google Scholar]
- Crowe B. L.; Bohlen C. J.; Wilson R. C.; Gopalan V.; Foster M. P. (2011) Assembly of the complex between archaeal RNase P proteins RPP30 and Pop5. Archaea 2011, 891531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.; Oruganti S. V.; Gopalan V.; Foster M. P. (2012) Thermodynamics of coupled folding in the interaction of archaeal RNase P proteins RPP21 and RPP29. Biochemistry 51, 926–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avis J. M.; Allain F. H.; Howe P. W.; Varani G.; Nagai K.; Neuhaus D. (1996) Solution structure of the N-terminal RNP domain of U1A protein: The role of C-terminal residues in structure stability and RNA binding. J. Mol. Biol. 257, 398–411. [DOI] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. (1996) VMD: Visual Molecular Dynamics. J. Mol. Graphics 14.1, 33–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.