Abstract
Purpose
Proteases have been implicated in cancer progression and invasiveness. We have investigated the activities, as opposed to simple protein levels, of selected aminopeptidases in urine specimens to serve as potential novel biomarkers for urothelial cancer.
Experimental design
The unique urinary proteomes of males and females were profiled to establish the presence of a gender-independent set of aminopeptidases. Samples were also collected from patients with urothelial cancer and matched controls. A SOP for urine processing was developed taking into account hydration variation. Five specific aminopeptidase activity assays, using fluorophoric substrates, were optimized for evaluation of marker potential.
Results
Nineteen exopeptidases and 21 other proteases were identified in urine and the top-5 most abundant aminopeptidases, identical in both genders, selected for functional studies. Depending on the enzyme, activities were consistently lower (P ≤ 0.05), higher or unchanged in the cancer samples as compared to controls. Two selected aminopeptidase activities used as a binary classifier resulted in a ROC curve with an AUC = 0.898.
Conclusion and clinical relevance
We have developed functional assays that characterize aminopeptidase activities in urine specimens with adequate technical and intra-individual reproducibility. With further testing, it could yield a reliable biomarker test for bladder cancer detection or prognostication.
Keywords: activity biomarkers, aminopeptidases, bladder cancer, fluorescence, urine
1 Introduction
Urothelial cancer of the bladder is estimated to affect over 700,000 new individuals and be responsible for nearly 15,000 deaths in 2013 in the United States [1]. It is the fourth most common solid tumor in men and eleventh most common in women in the U.S. and is a major source of healthcare expenditures [2]. The overall 5-year survival rate is above 75%. Annual expenditures for care exceeded $1 billion in 2000 [3], with a significant proportion of cost arising from surveillance in survivors. Non-muscle-invasive bladder cancer has a high rate of recurrence – up to 80% of patients – necessitating frequent invasive investigations to monitor for disease. A simple non-invasive marker would help offset the cost and burden of the surveillance schedule. On the other hand, patients with muscle-invasive disease treated with radical surgery have a 5-year risk of cancer-specific death between 33–74% [4], and earlier detection would theoretically translate into improved survival. The identification of useful biomarkers may allow for earlier detection of cancer and prevention of death, as well as reduction of the economic burden of the disease overall.
An ideal biomarker for bladder cancer diagnosis or surveillance is one that is accurate, noninvasive, and reproducible, with high sensitivity and specificity. Urine is particularly well-suited for biomarker discovery 5], given its proximity to the primary malignancy and ease of collection. Many urinary biomarkers have been investigated and described 6], with urine cytology remaining the gold standard. Several others are FDA-approved and applied regularly in clinical practice, including NMP-22 [7], a nuclear matrix protein found at elevated levels in urine from individuals with bladder cancer, and UroVysion FISH [8]. Urine cytology claims the best specificity, as high as 95%, while the other markers have values ranging from 50–80%. A newly developed biomarker used independently is unlikely to surpass the accuracy of the gold standards for diagnosis, but a goal of discovery would be to integrate a new marker in the process of clinical decision making to improve upon the diagnostic or prognostic ability of already available tools.
The classic path to cancer biomarker discovery and verification has been by measuring differential levels of proteins in tissues, plasma, serum or urine, using immunohistochemical- or mass spectrometric-based screens. Overall, there have been few assays translated into clinical practice [9, 10] and this disconnect pleads for conceptually novel biomarker discovery and validation schemes. An example of an alternate approach is by interrogating the activity of proteins, in particular enzyme families that are relevant with respect to the disease of interest. In the case of cancer, proteases are one such class as several of its members have been found implicated in promoting both tumor progression and suppression [11–14]. Examples of proteolytic activities have been observed in past onco-peptidomic studies [15–17], whereby subsets of serum peptides provided class discrimination between patients with different types of solid tumors and control individuals without cancer [16]. Nearly all relevant peptides sorted into a dozen or so nested sets of sequences, each the result of exopeptidase activities that confer cancer-specific differences superimposed on the proteolytic events of the ex vivo coagulation and complement degradation pathways [16]. In addition, proteomic screens of cultured cancer cells indicated sizable panels of secreted proteases and protease inhibitors [18–20].
This current investigation sought to determine if aminopeptidases were present in urine in sufficient amounts to have utility in detection of urothelial cancer. Our aims were to (i) identify candidate aminopeptidases present at detectable levels in urine of both males and females, and to (ii) explore the functional activity of these enzymes as a potential indicator for presence of bladder cancer. We therefore carried out gender-specific proteomic screens and developed robust, quantitative assays to selectively measure activities of five individual aminopeptidases in urine without the need for any sample pre-fractionation or pre-treatment. Our new approach and related tests are uniquely suited to probe a potentially altered balance of aminopeptidases and/or their modulators in urine of cancer patients and to prospectively use that information for diagnostic or prognostic purposes.
2 Materials and methods
2.1 Sample collection
Urine samples from healthy volunteers with no known malignancies and from patients diagnosed with urothelial cancer of the bladder were all collected at Memorial Sloan-Kettering Cancer Center (MSKCC) following a standard clinical protocol. Details on patient age and pathologic diagnosis are given in Supplementary table 4 and in section 3.4. All specimens were collected under an Institutional Review and Privacy Board approved research protocol at the same location and according to the exact same standard operating procedure. Samples (30–50mL) were collected and either frozen or immediately centrifuged at 2000 rpm (Beckman GS-6KR) for 15 minutes at 4°C and the supernatant concentrated and desalted by ultrafiltration through a Millipore Ultracel 10kD centrifugal filter by centrifugation at 2000 rpm (Beckman GS-6KR) for 2×30 minutes at 4°C, which concentrates the sample approximately 30-fold. The protein concentration of the solution was measured using the Bradford BioRad assay with a bovine serum albumin standard on a Beckman DU640 spectrophotometer at 595 nm, and taken to normalize the samples for the enzymatic activity assay. Measurements of creatinine and albumin were made in some cases with a Siemens DCA Vantage benchtop automated assay. Aliquots of the concentrated urine are then stored at −80°C.
2.2 Proteomic screen
Gender-specific pools of concentrated urinary protein (2.5-μg per sample) were partially size-fractionated by SDS-PAGE (~1cm total separation distance past the stacking gel). In addition, pooled samples from each gender were depleted of the 14-most abundant blood plasma proteins using a Seppro IgY14 spin column (Sigma Aldrich), protein eluates quantitated and, again, separate aliquots of 1-μg each loaded for gel fractionation. The four lanes of the lightly stained gel were each sliced into five 2-mm pieces for in situ tryptic digestion and capillary LC-MS/MS analyses. Details of mass spectrometric and data analysis are given under Supplementary methods.
2.3 Aminopeptidase activity assays
Amino acid and dipeptide derivatives of 7-Amino-4-methylcoumarin (AMC) were used as substrates for measurements of aminopeptidase activities in urine samples. Ala-AMC, Glu-AMC, Gly-Pro-AMC, Lys-Pro-AMC, and Pro-Arg-AMC substrates were purchased from MP Biomedicals, Inc., or Bachem Corp., and stored at −20°C as 100-mM stock solutions in DMSO. Free AMC was purchased from AnaSpec, Inc., amastatin and bestatin from MP Biomedicals, and sitagliptin from Merck (under the name JANUVIA®). Each activity was analyzed under the respective optimal conditions as shown in Table 2. The conditions were initially taken from the literature [21–25] and then further optimized through systematic studies in our laboratory, evaluating recombinant aminopeptidases, in combination with a large number of substrates and inhibitors, in an effort to create conditions that favor interrogating activities of individual enzymes (Yaneva and Tempst, in preparation).
Table 2.
Direct activity assays of urinary aminopeptidases
| Target Aminopeptidase | Urinary protein (μg) | Substrate 0.5 mM | pH | NaCl (mM) | CaCl2 (mM) | DTT (mM) | Inhibitors
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PMSF | EDTA | AMA | BEST | SITAG | CYST | |||||||
|
| ||||||||||||
| ANPEP | 1 | Ala-AMC | 7.5 | 150 | + | + | ||||||
| ENPEP | 5 | Glu-AMC | 8.0 | 200 | 50 | + | + | |||||
| DPP4 | 5 | Gly-Pro-AMC | 7.5 | 150 | + | Δ | ||||||
| DPP7 | 5 | Lys-Pro-AMC | 6.0 | + | + | + | + | |||||
| CTSC | 0.5 | Pro-Arg-AMC | 6.0 | 50 | 5 | + | + | + | Δ | |||
ANPEP, alanyl aminopeptidase; ENPEP, glutamyl aminopeptidase; DPP(4, 7), dipeptidyl aminopeptidase (4, 7); CTSC, cathepsin C; AMC, 7-amino-4-methylcoumarin.
Buffers: 50 mM Tris. HCl (pH 7.5 and 8.0); 25 mM MES (pH 6.0); all reaction mixtures contained 1 mg/mL BSA.
Inhibitors: 2 mM PMSF; 2 mM EDTA; 10 μM amastatin (AMA); 10 μM bestatin (BEST); 10 μM sitagliptin(SITAG); 10 μg/mL cystatin (CYST); Δ = difference between −/+ inhibitor
In general, all assays were performed for 2h at room temperature in the presence of excess substrate (0.5 mM) and with selected inhibitors in a 100-μL total volume. The amount of urine protein in each assay was determined empirically to yield a read-out in the linear part of the curve (see table 2). Sample and buffer were first added to each well and the assay was then started by adding substrate (10 μL) using a multichannel pipetor. For all analyses in the presence of inhibitors, samples were pre-incubated with the respective inhibitor for 30 min at RT before adding the substrate. The fluorescence of the released AMC, as a measure of the enzyme activity, was monitored in a Gemini EM microplate fluorescence reader (Molecular Devices, Inc.), using opaque 96-well microtiter plates and operated in kinetics mode with excitation and emission wavelengths of 380 and 460 nm, respectively. The fluorescent signal was calibrated using known concentrations of free AMC. All assays were performed in duplicate using different plate readers, with results presented as the averages and after adjustment for background measurements of the sample alone and the substrate alone in blank wells.
2.4 Statistical analysis
Analysis of variation (ANOVA) was done using a Wilcoxon rank-sum test (Mann-Whitney) before and after adjusting for age, with Benjamini-Hochberg multiple testing correction as needed. Odds ratios were calculated for each aminopeptidase activity and the 90% confidence interval (CI) computed using resampling methods. We then evaluated the success of candidate binary classifiers as indicators for the presence of bladder cancer, using the best predictive models that utilize either a single activity assay or the best combination of 2 assays, by establishing the area under the receiver-operator characteristic curve (AUC) before and after multiple randomizations of the class labels.
3 Results and discussion
3.1 Gender specificity of the urinary proteome
Several exopeptidases have been detected in urine in past proteomic screens [26, 27] but little is known about their activities. Furthermore, those analyses have invariably been done using mixed samples collected from both genders. It is therefore not known whether any gender-specific or -uneven distribution exists for any of these enzymes. Obviously for a disease like bladder cancer that affects both men and women, we didn’t want to select a functional biomarker that, for example, derives solely from the prostate. We therefore collected urine samples from 5 male and 5 female healthy volunteers to generate two gender-specific pools that were then processed separately as described under materials and methods. MS data derived from all slices of each individual lane were merged and used to search the UniProt protein database, all as described under supplementary materials and methods. The full search results are listed in the supplementary proteomics data section and have been deposited in the PRIDE open access database.
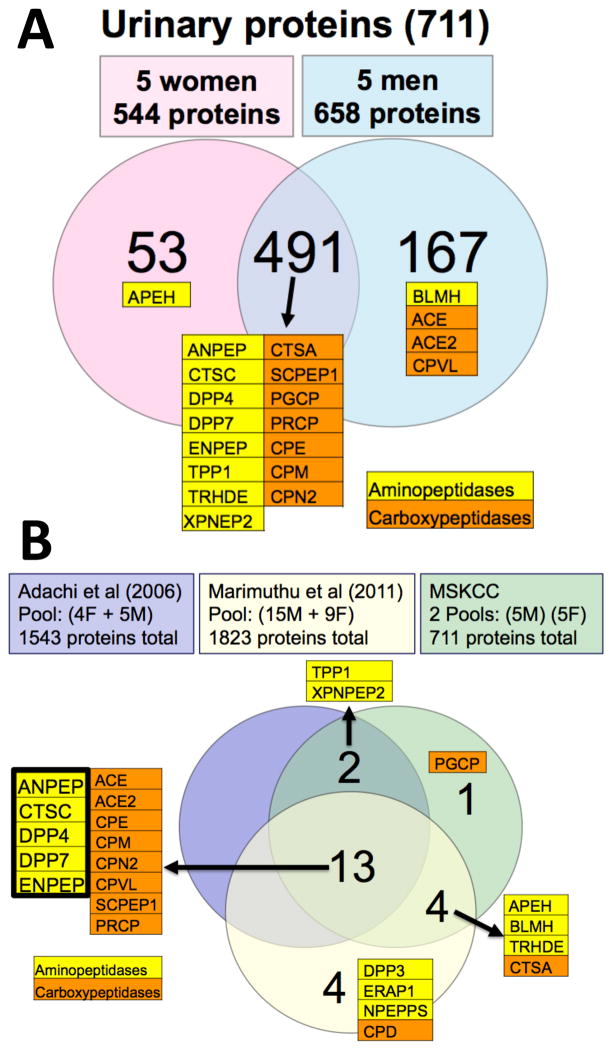
Ten aminopeptidases, 10 carboxypeptidases and 21 additional proteases were positively identified; judging from the spectral counts, 15 of those 20 exopeptidases were found in urine of both genders in approximately the same amounts (Figure 1A; Suppl. Table 1). Overall, we detected 20 exopeptidases (~25% of the estimated ~80 total number human exopeptidases in the MEROPS [28] database) and 41 proteases (~7.3% of the estimated ~560 total number human proteases in MEROPS) as part of a total 711 confidently identified urinary proteins (representing ~3.6% of the estimated total number of human genes). When comparing our results with two previous studies [26, 27], we have now evidence for at least 24 human urinary exopeptidases (~30% of all estimated human exopeptidases); of those, 5 aminopeptidases and 9 carboxypeptidases are shared between the three studies (Figure 1B).
Figure 1. Aminopeptidases in the human urinary proteome.
A. Gender-specific proteomic analysis (this work). Pooled female and male urine samples were separately analyzed by LC-MS/MS of tryptic peptides, before and after immuno-depletion of 14 abundant plasma proteins (see section 2.2, supplementary methods and materials, and supplementary proteomics data). The number of proteins uniquely detected (2-peptide minimum) in each gender, as well as those in common, are shown in the Venn diagram. Identified exopeptidases are indicated.
B. Comparison of three urinary proteomic analyses. Exopeptidases positively identified by Adachi et al [26], Marimuthu et al [27] and in this study are shown in the Venn diagram. Note that the published urinary proteomes are of mixed gender composition. The five aminopeptidases detected in all three studies were taken for further functional assays.
While not the focus of our limited proteomic analysis, we did observe some significant gender-specific differences in the urinary proteome (Table 1). Among the male-specific ones were, not surprisingly, PSA (KLK3), prostate stem cell antigen (PSCA) and lactotransferrin (LTF) [29–31], thus validating several others including the androgen-regulated and/or -binding proteins submaxillary gland androgen-regulated protein 3B (SMR3B), protein-glutamine gamma-glutamyltransferase 4 (TGM4) and mammoglobulin-B/secretoglobin 2A1 (SCGB2A1) [32, 33]. On the female side, we found the unique presence of several serpins and, intriguingly, the EGF-receptor (EGFR/HER1). Lastly, several members of the proline-rich protein family are represented in the urinary proteome in a gender-specific, mutually exclusive manner.
Table 1. Human urinary proteins with a high degree of gender-specificity.
Proteins identified in the immuno-depleted samples (see section 2.2) are listed, together with the UniProt accession numbers, estimated molecular mass, and total spectral counts. Known prostate-derived proteins are in bold [29–33]. The complete protein identification set is listed under Supplementary Proteomics Data.
| Human Urinary Proteome: | Uniprot Ac.# | ~Mr | Female Depleted | Male Depleted |
|---|---|---|---|---|
|
|
||||
| Identified Proteins: | Assigned Spectra | |||
|
| ||||
| Prostate-specific antigen GN=KLK3 | KLK3_HUMAN | 29 kDa | 0 | 178 |
| Lactotransferrin GN=LTF | TRFL_HUMAN | 77 kDa | 0 | 131 |
| Protein-glutamine gamma-glutamyltransferase 4 GN=TGM4 | TGM4_HUMAN | 77 kDa | 0 | 111 |
| Submaxillary gland androgen-regulated protein 3B GN=SMR3B | SMR3B_HUMAN | 8 kDa | 0 | 53 |
| Salivary acidic proline-rich phosphoprotein 1/2 GN=PRH1 | PRPC_HUMAN | 17 kDa | 0 | 45 |
| Programmed cell death 6-interacting protein GN=PDCD6IP | PDC6I_HUMAN | 97 kDa | 0 | 41 |
| Neogenin GN=NEO1 | NEO1_HUMAN | 160 kDa | 0 | 32 |
| Moesin GN=MSN | MOES_HUMAN | 68 kDa | 0 | 32 |
| Desmoplakin GN=DSP | DESP_HUMAN | 332 kDa | 0 | 31 |
| Extracellular glycoprotein lacritin GN=LACRT | LACRT_HUMAN | 14 kDa | 0 | 27 |
| C-type lectin domain family 14 member A GN=CLEC14A | CLC14_HUMAN | 52 kDa | 0 | 25 |
| Bone marrow proteoglycan GN=PRG2 | PRG2_HUMAN | 25 kDa | 0 | 20 |
| Prostate stem cell antigen GN=PSCA | PSCA_HUMAN | 13 kDa | 0 | 20 |
| Proline-rich protein 4 GN=PRR4 | PROL4_HUMAN | 15 kDa | 0 | 19 |
| Limbic system-associated membrane protein GN=LSAMP | LSAMP_HUMAN | 37 kDa | 0 | 18 |
| Folate receptor alpha GN=FOLR1 | FOLR1_HUMAN | 30 kDa | 0 | 16 |
| Mammaglobin-B GN=SCGB2A1 | SG2A1_HUMAN | 11 kDa | 0 | 14 |
| Desmoglein-2 GN=DSG2 | DSG2_HUMAN | 122 kDa | 0 | 14 |
| Galectin-7 GN=LGALS7 | LEG7_HUMAN | 15 kDa | 0 | 14 |
|
| ||||
| Small proline-rich protein 3 GN=SPRR3 | SPRR3_HUMAN | 18 kDa | 154 | 2 |
| Serpin B3 GN=SERPINB3 | SPB3_HUMAN | 45 kDa | 147 | 5 |
| Leukocyte elastase inhibitor GN=SERPINB1 | ILEU_HUMAN | 43 kDa | 47 | 0 |
| Cornulin GN=CRNN | CRNN_HUMAN | 54 kDa | 47 | 4 |
| small proline-rich protein 1A | IPI00914840 | 10 kDa | 30 | 0 |
| Epidermal growth factor receptor GN=EGFR | EGFR_HUMAN | 134 kDa | 30 | 0 |
| Small proline-rich protein 2E GN=SPRR2E | SPR2E_HUMAN | 8 kDa | 23 | 0 |
| Fatty acid-binding protein, epidermal GN=FABP5 | FABP5_HUMAN | 15 kDa | 18 | 1 |
| Involucrin GN=IVL | INVO_HUMAN | 68 kDa | 18 | 0 |
| Trefoil factor 3 GN=TFF3 | TFF3_HUMAN | 9 kDa | 16 | 0 |
| Glutathione S-transferase A2 GN=GSTA2 | GSTA2_HUMAN | 26 kDa | 15 | 0 |
3.2 Targeted urinary aminopeptidase activity assays
The five aminopeptidases (ANPEP, alanyl aminopeptidase; ENPEP, glutamyl aminopeptidase; CTSC (a.k.a. DPP1), cathepsin C; DPP4, dipeptidylpeptidase 4; DPP7 dipeptidylpeptidase 7) that were identified in common in all three urinary proteomic analyses were selected for further activity profiling for several reasons. First, they are all robustly represented in urine of both genders in approximately the same relative amounts (Figure 1B; Suppl. Table 1), thus satisfying the major criterion of our search. Second, judging from the known substrate specificities [28, 34] and based on past efforts by our group to develop activity assays unique for several individual aminopeptidases (Table 1; Yaneva and Tempst, unpublished), it was a reasonable hypothesis to be able to discriminate among the activities of these five proteases as well as to distinguish them from the other, lower-abundant urinary aminopeptidases (Suppl. Table 1), from all carboxypeptidases (Suppl. Table 1), and from all other known urinary proteases [27]. Finally, they have not been previously recognized or evaluated as urinary disease markers of any sort [35], thus directing our efforts towards bona fide discovery.
Fluorescence-based protease assay substrates, such as amino acids or dipeptides N-terminally coupled to 7-amino-4-methylcoumarin (AMC) have been used to monitor aminopeptidase activities in serum [36–38]. Our group has characterized 25 recombinant aminopeptidases for amino acid (P1 position) or sequence (P2 and P1; P2′ and P1′) specificities using 30 different substrates, as well as studied the effects of assay medium (buffer, pH, ionic strength, divalent metals, etc.…) and of various inhibitors: (i) broad-protease class (e.g., EDTA, PMSF, cystatin), (ii) broad aminopeptidase-specific (e.g., bestatin, amastatin), and (iii) single enzyme-specific inhibitors (e.g., sitagliptin inhibits DPP4 but not DPP7). This allowed us to develop 10 unique, substrate-and-conditions-dependent assays specific for either single enzymes or small sets (e.g., 2–3) of enzymes using small volumes of undepleted serum or plasma (Yaneva and Tempst, unpublished). We call these ‘direct assays’ as no prior manipulation (e.g., prefractionation, target immuno-capture) of the samples is required. Five of these assays measure activities of the aminopeptidases we have selected for analysis in the current study. The conditions are listed in Table 2 and have only been slightly modified from serum analysis in that 0.5–5μg of total urinary protein is used (for reasons set forth in section 3.3), instead of 2–20μL of serum, depending on which enzyme activity is measured.
3.3 Optimization of assay conditions
Normal individuals without renal dysfunction excrete, on average, 50–150 mg protein per 24 hours [39]. Albumin accounts for about one third of this amount and Tamm-Horsfall protein for another third. The remainder is comprised of well over a thousand lower abundance proteins (see 3.1). In a collected specimen of urine, the total protein concentration will vary, depending on hydration, diet, and renal function. Indeed, total protein concentration of fresh samples from healthy individuals ranged from 12 to 60 μg/mL as measured by a Bradford assay. Aminopeptidase assays are performed in 96-well flat-bottom microtiter plates. Total assay volume is 100μL with a practical sample volume limit of 50μL. Given this limitation, we found that use of fresh urine was not feasible as an adequate amount of protein could not be attained in the well to perform an assay with activity in the linear range (200–15,000 RFU; see below). For instance, when attempting an activity assay of ANPEP (the most abundant aminopeptidase in urine – Suppl. Table 1), addition of up to 50 μL of fresh sample achieved readings of less than 500 RFU at 2 hours. Therefore, we tested various methods to concentrate protein from fresh samples, including precipitation with acetone and membrane ultrafiltration with a 10kDa cutoff (data not shown). Centrifugal filtration (see materials and methods) achieved good protein yields and effectively concentrated the specimen approximately 30-fold, typically from 15 mL to ~0.5mL, with final protein concentrations of 0.3–2 mg/mL. In this way, 2–10μL of processed sample contained enough protein to observe robust signals for each of the five enzyme activity assays (see below).
Unlike serum that exhibits fairly uniform protein concentration, the levels in urine vary for the reasons stated above. Activity assay results can therefore not be expressed on a per volume basis. Instead, enzymatic measurements have been normalized using the total amount of secreted albumin or creatinine over a 24-h period [39]. We wanted to establish a relatively simple and accurate normalization method without having to collect patient or control urine samples around the clock. To this end we measured the amounts of creatinine, albumin and total protein in 8 urine samples, 2 regular daytime specimens each from 4 individuals (Suppl. Table 2). Two-hour end-points of ANPEP activity assays (in duplicate) were also determined for all 8 samples using the equivalent of 1-μL of concentrated sample per assay. Activity readings (RFU) were then expressed per μg total protein (TP), or RFUs per μg albumin (ALB), or RFUs per 0.1μg creatinine (CR), and the mean, standard deviation and coefficient of variation calculated for each: 17.4% (RFU/TP), 29.1% (RFU/ALB) and 36.8% (RFU/CR). As a result, we adopted total protein-based normalization for all further studies. To further test the robustness of our normalization method we conducted an experiment whereby urine specimens were collected from one female and one male volunteer over 4 days and under highly varying conditions: (1) standard daytime collection; (2) hyperhydrated (by extra fluid intake); (3) dehydrated (restricted fluid intake); (4) first morning void. Duplicate aliquots (low-μL) containing 1 μg of concentrated total protein were taken for ANPEP activity analysis. The coefficient of variation for the normalized measurements over the 4 days varied from 12.4 to 18.6%, depending on the individual (Suppl. Table 3). We also investigated the effect of repeated freeze/thaw cycles and it was found to adversely affect aminopeptidase activities (data not shown), indicating content degradation or loss of protein and/or activity. Our final SOP thus calls for freezing and storage at −80°C of fresh urine specimens, followed at a later time by thawing, processing and aliquoting, followed again by immediate freezing and storage for future one-time use. So at the time of activity measurement, each sample had been frozen and thawed twice.
To optimize each aminopeptidase assay for activity (RFU) read-outs in the linear portion of the curve, we first established the fluorescence dose-response by mixing free AMC at various concentrations in assay buffer containing 10 μL of urinary protein concentrate. The plots shown in Suppl. Figure 1 indicate a linear curve from ~200 to 15,000 RFU, with R2 values ≥0.997. Based on this information, we titrated the total protein amounts needed to yield a linear response using an assay for the activities of urinary DPP4 + DPP7 combined (i.e., Gly-Pro-AMC as substrate without the presence of sitagliptin). This was achieved when ~0.2 up to 10μg of total protein was used (R2 ≥0.992) (Suppl. Figure 2). This process was repeated for each of the other proteases we intended to evaluate. Once optimal procedures had been established, we verified technical reproducibility under slightly suboptimal conditions (e.g., 500 ng total urinary protein for an ANPEP assay, as shown in Suppl. Figure 3) and obtained coefficients of variation well below 10%.
3.4 Altered urinary aminopeptidase activities in bladder cancer patients
To explore the use of the aminopeptidase activity tests for functional biomarker discovery, we analyzed five different proteolytic activities in urine samples from 16 male bladder cancer patients and from 16 male, comparatively age-matched healthy controls (Suppl. Table 4), using the optimized conditions as described under section 3.3 and in materials and methods. The patients had a predominantly urothelial histology. Additional histology identified included squamous differentiation in 2, glandular in 2, and plasmacytoid in 2. Specimens were collected prior to or at the time of surgery, with transurethral resection (TUR) performed in 6 and radical cystoprostatectomy (RC) in 10. Pathologic stage from TUR or RC was T2 in 9 patients, T3 in 6 and T4 in 1; concomitant carcinoma in situ (CIS) was identified in 5 patients. Three patients received neoadjuvant chemotherapy prior to the time of specimen collection, and three patients had received prior intravesical therapy remote to the time of specimen collection. Of the specimens collected and banked during the time coinciding with this study, there were only two women that had tumor stage > T2 (muscle-invasive), which did not represent a cohort with enough variability to report a range or any valuable assay data. We therefore performed this exploratory investigation in samples from men with a definite intention to expand our investigation to female patients when enough samples were collected.
The results of duplicate analyses (Suppl. Table 4) indicated that, first, the inter-individual variability is more pronounced among cancer patients (c.v. ≥ 70%) than in the control group (c.v. <50%, except for DPP7 activities), which may reflect the heterogeneity of disease state and therapeutic history. It is conceivable that prior therapy may have altered the activity signal, and with a larger sample cohort, this may be possible to stratify or study. At this point, however, we cannot say if there was a difference in signal based on therapies even though we feel one would not expect such a difference given that the urine samples were collected at the time of the reported stage, meaning there was still residual tumor in the bladder.
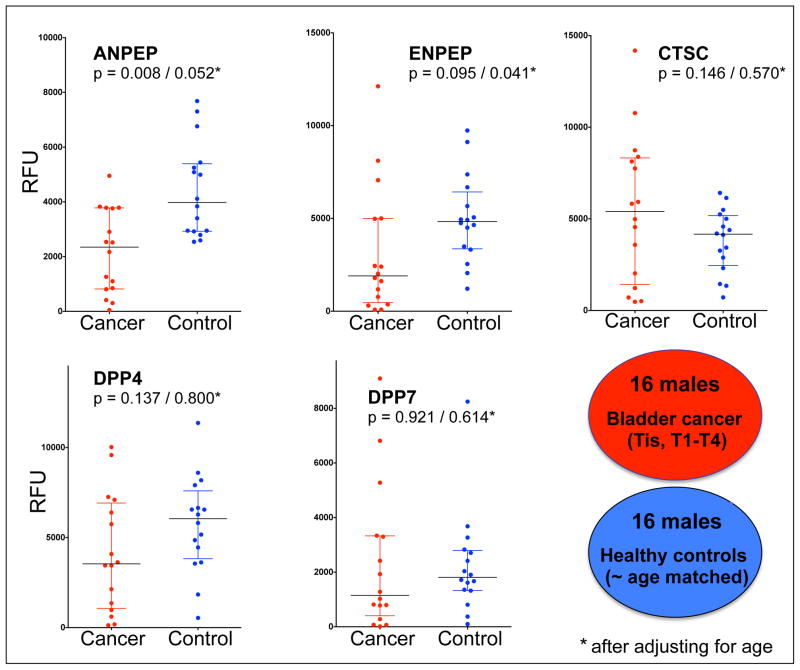
When presented as dot plots (Figure 2), the results furthermore illustrate that the median aminopeptidase activities were generally lower in the cancer group, except for CTSC activity that was slightly higher. Analysis of variation (ANOVA) indicated that ANPEP and ENPEP activity assays could distinguish between the cancer and control groups with P values ≤0.05 after adjusting for age (Figure 2). To further eliminate the probability that age could have been a confounding factor in the analysis, we divided both the cancer (CA) and control (NL) groups in two subgroups each of the 8 youngest and 8 oldest individuals for further comparison (Suppl. Table 4). Whereas the ‘younger’ CA group (CA-1) and ‘older’ NL group (NL-2) are near perfectly matched (the mean age of CA-1 is 61.4 ± 5.1 and that of NL-2 is 61.3 ± 5.6), measured ENPEP and especially ANPEP activities could readily distinguish between the two groups with P values of, respectively, 0.065 and 0.038 (Suppl. Figure 4).
Figure 2. Specific activities of five aminopeptidases in urine of bladder cancer patients and healthy controls.
Activities were measured using fluorescence-based assays as described in section 2.3 and table 2. Data points are the average of duplicate analyses performed on two different plate readers and are expressed as relative fluorescence units; median and interquartile ranges are indicated on the dot plots. Analysis of variation (Wilcoxon rank sum test) was carried out before and after (*) adjusting for age. Details on patients and healthy volunteers are provided in supplementary table 4.
3.5 Aminopeptidase activities as prospective urinary biomarkers for cancer
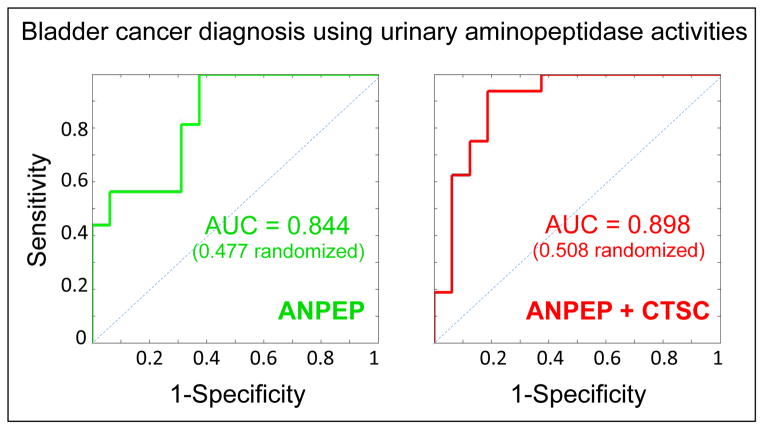
Going back to the complete cancer and control groups, we calculated that urinary ANPEP and ENPEP activities were reasonable predictors of bladder cancer (ANPEP: odds ratio 0.369; 95% confidence interval 0.079, 0.913; p = 0.095; ENPEP: odds ratio 0.496; 95% confidence interval 0.195, 0.90; p = 0.060; unit value is 1,000 RFU), whereby lower values are associated with disease as the dot plots in Figure 2 already seemed to imply. The large 95% confidence intervals are the result of the relatively small sample sets and it is imperative that our analyses are repeated with larger numbers of patients and control individuals enrolled in the study to confirm the current findings. On the basis of the available numbers, the performance of ANPEP activity as a classifier between bladder cancer and control was further illustrated by an area under the ROC curve (AUC) of 0.844 (0.477 after multiple randomizations of the 32 class labels), by far the highest among all aminopeptidase activities tested for this purpose (Figure 3). Addition of a second variable didn’t improve the AUC value except for CTSC activity; the ANPEP+CTSC pair allowed class prediction with AUC = 0.898 (0.508 after multiple randomizations of the class labels). Therefore, even though the measured CTSC activity did not statistically differ between the cancer and control groups in its own right (P value = 0.57 after adjusting for age), it appears to augment the capacity of the ANPEP activity measurements to do so.
Figure 3. Urinary aminopeptidase activities as candidate classifiers of bladder cancer.
Receiver operator characteristic (ROC) curves are shown and the areas under the curve (AUC) calculated using ANPEP activity or ANPEP+CTSC paired activities (see Figure 2 and supplementary table 4) as the variable(s). AUC calculations were then repeated after multiple randomizations of the class labels.
Interestingly, it appeared that the activities of urinary ANPEP are lower in the bladder cancer than in the control group. This seems in contrast with earlier reports stating that ANPEP is highly expressed in some carcinomas [40] and that the concentration of its soluble form is elevated in several types of cancer [41]. Increased ANPEP expression may also be functionally correlated with metastasis of cancer cells by promotion of angiogenesis [42]. Of course, the comparison is between urinary activities on the one hand and elevated amounts in tissue or blood plasma on the other, and so there could be several explanations for this discrepancy. First, we don’t know whether the relative ANPEP activities in urine reflect the relative concentrations since the latter have never been established for bladder cancer patients. Even if ANPEP levels were similar or perhaps higher in cancer patients, activities could be modulated by denaturation, allosteric changes or the presence of inhibitors. We and others have noted a large number of protease inhibitors in urine such as serpins, cytostatins, TIMPs, ITIH4 and others (Supplementary proteomic dataset; [27]). But whereas these same inhibitors are also abundantly present in blood plasma/serum [43], they don’t appear to interfere appreciably with exopeptidase activities [16, 17]. Thus, any putative inhibitor would likely be of a different class and may emanate from the tumor; however, there is no direct experimental evidence to support such a model at this time. It should also be noted that the majority of bladder cancer patients with muscle invasive disease suffer from hematuria, resulting in higher urinary protein concentration [44]. However, this is unlikely to affect normalized aminopeptidase activities (i.e., relative to protein concentration) as ANPEP and the other four aminopeptidases we have analyzed are all present in plasma [43]. Thus, as plasma proteins leak into the urine the increase of the proteases and their concomitant activities would be proportional to the increase of total protein. The normalization of urinary enzymatic activities should therefore remain unaffected and not introduce experimental error in the ANPEP or other activity measurements in patient urine. To further refute the notion that normalized aminopeptidase activities (ANPEP activity/μg protein) may in fact be correlated with total urinary protein concentrations (μg/mL) we calculated the coefficient of determination and found low R2 values for both the patient (R2 = 0.117) and healthy control groups (R2 = 0.014), indicating such correlation is weak to nonexistent. Lastly, the statistically relevant reduction of ANPEP activity in urine of patients appears unique among the aminopeptidases that were analyzed. The activities of DPP4 and 7 are only marginally lower and the activity of CTSC is in fact higher, implying that decreased ANPEP activity is not merely the result of disease related hematuria as that should affect changes in activity of all five proteases similarly.
In summary, we have described a new approach and related tests uniquely suited to probe a potentially altered balance of aminopeptidases and/or their modulators in urine of bladder cancer patients. Fluorescent assays as employed here have advantages over most conventional protein measurements, including low cost, parallel high-throughput analysis, ease of use, portability, and being fully transparent to the entire urine proteome. These tests offer the option of a targeted, enzymatic readout that may be either a supplement or a practical alternative for the classical biomarker discovery and verification techniques. We anticipate that as we scale up these efforts using the same general methodology, we will expand the number of protease activity tests for prediction of cancer, including urinary endoproteases that may be assayed using FRET-based fluorescent read-outs. The assays may also have diagnostic value, either alone or in combination with existing tests, for identifying cancer subtype and stage or may mark a given clinical outcome of interest. Thus, the current study provides a framework for screening of additional samples, across a wider range of disease stage and grade, toward the goal of validating our findings and developing a reliable predictive measurement for detection of malignancy.
4 Concluding remarks
Urothelial cancer of the bladder is responsible for numerous deaths and a major source of healthcare expenditures. Identification of useful, non-invasive, novel biomarkers may allow improving upon direct diagnostic or prognostic ability of already available tools. However, the classic path to cancer protein biomarker discovery has yielded disappointingly few assays translated into clinical practice so far. In this work, we have taken an alternate approach by interrogating the activity of proteins, more specifically of urinary aminopeptidases, as a means to biomarker discovery. The results of a proof-of-principle study suggest that ANPEP activity, either by itself or in combination with CTSC activity, may be a potential candidate as urinary indicator for the presence of bladder cancer. We expect that future uses and implementation of aminopeptidase or other enzymatic activity tests will go well beyond those provided in the current account. If those future developments are successful and, as a result, effective functional cancer biomarker panels emerge, it would have a major clinical impact.
Supplementary Material
Clinical Relevance.
Urothelial cancer of the bladder is responsible for many deaths and a major source of healthcare expenditures. Identification of useful, novel biomarkers may allow improving upon direct diagnostic or prognostic ability of already available tools. In this work, we have taken an alternate approach by interrogating the activity of one class of enzymes – aminopeptidases – in urine of patients, wanting to establish a distinction based on enzymatic function as opposed to simple protein levels as a means to biomarker discovery. Our limited study presented here suggests that a panel of two selected urinary aminopeptidase activities may emerge as a candidate binary classifier and as an indicator for the presence of bladder cancer. If future developments and large-scale screens are successful and, as a result, effective functional cancer biomarker panels materialize, it would have a major clinical impact for non-invasive cancer detection and prognostication.
Acknowledgments
This work was supported by US National Institutes of Health U24 CA126485 grant (to PT), which was part of the NCI Clinical Proteomic Technology Assessment for Cancer (CPTAC) Network; by the Weiner Fund (to BHB); and by a NIH-NRSA T32 CA82088 training grant, an American Urological Association Foundation Research Scholar Award and a Society of Women in Urology Elisabeth Pickett Award (to JMT).
Abbreviations used
- AMC
7-amino-4-methylcoumarin
- ANPEP
alanyl aminopeptidase
- CTSC
cathepsin C
- DPP4
dipeptidylpeptidase 4
- DPP7
dipeptidylpeptidase 7
- ENPEP
glutamyl aminopeptidase
- FDA
United States food and drug administration
- FRET
fluorescence resonance energy transfer
- PSA
prostate-specific antigen
- RFU
relative fluorescence units
- SOP
standard operating procedure
Footnotes
The authors have declared no conflict of interest.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Botteman MF, Pashos CL, Redaelli A, Laskin B, et al. The health economics of bladder cancer: a comprehensive review of the published literature. Pharmacoeconomics. 2003;21:1315–1330. doi: 10.1007/BF03262330. [DOI] [PubMed] [Google Scholar]
- 3.Konety BR, Joyce GF, Wise M. Bladder and upper tract urothelial cancer. J Urol. 2007;177:1636–1645. doi: 10.1016/j.juro.2007.01.055. [DOI] [PubMed] [Google Scholar]
- 4.Herr HW, Dotan Z, Donat SM, Bajorin DF. Defining optimal therapy for muscle invasive bladder cancer. J Urol. 2007;177:437–443. doi: 10.1016/j.juro.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 5.Pisitkun T, Johnstone R, Knepper MA. Discovery of urinary biomarkers. Mol Cell Proteomics. 2006;5:1760–1771. doi: 10.1074/mcp.R600004-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Vrooman OP, Witjes JA. Urinary markers in bladder cancer. Eur Urol. 2008;53:909–916. doi: 10.1016/j.eururo.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Grossman HB, Soloway M, Messing E, Katz G, et al. Surveillance for recurrent bladder cancer using a point-of-care proteomic assay. JAMA. 2006;295:299–305. doi: 10.1001/jama.295.3.299. [DOI] [PubMed] [Google Scholar]
- 8.Hajdinjak T. UroVysion FISH test for detecting urothelial cancers: meta-analysis of diagnostic accuracy and comparison with urinary cytology testing. Urol Oncol. 2008;26:646–651. doi: 10.1016/j.urolonc.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 10.Ransohoff DF. How to improve reliability and efficiency of research about molecular markers: roles of phases, guidelines, and study design. J Clin Epidemiol. 2007;60:1205–1219. doi: 10.1016/j.jclinepi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 11.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 12.Matrisian LM, Sledge GW, Jr, Mohla S. Extracellular proteolysis and cancer: meeting summary and futuredirections. Cancer Res. 2003;63:6105–6109. [PubMed] [Google Scholar]
- 13.Lopez-Otin C, Matrisian LM. Emerging roles of proteases in tumour suppression. Nat Rev Cancer. 2007;7:800–808. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- 14.Palermo C, Joyce JA. Cysteine cathepsin proteases as pharmacological targets in cancer. Trends Pharmacol Sci. 2008;29:22–28. doi: 10.1016/j.tips.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 15.Villanueva J, Martorella AJ, Lawlor K, Philip J, et al. Serum peptidome patterns that distinguish metastatic thyroid carcinoma from cancer-free controls are unbiased by gender and age. Mol Cell Proteomics. 2006;5:1840–1852. doi: 10.1074/mcp.M600229-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Villanueva J, Shaffer DR, Philip J, Chaparro CA, et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest. 2006;116:271–284. doi: 10.1172/JCI26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villanueva J, Nazarian A, Lawlor K, Yi SS, et al. A sequence-specific exopeptidase activity test for functional biomarker discovery. Mol Cell Proteomics. 2008;7:509–518. doi: 10.1074/mcp.M700397-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Gronborg M, Kristiansen TZ, Iwahori A, Chang R, et al. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics. 2006;5:157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Kulasingam V, Diamandis EP. Proteomics analysis of conditioned media from three breast cancer cell lines. Mol Cell Proteomics. 2007;6:1997–2011. doi: 10.1074/mcp.M600465-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Lawlor K, Nazarian A, Lacomis L, Tempst P, et al. Pathway-based biomarker search by high-throughput proteomics profiling of secretomes. J Proteome Res. 2009;8:1489–1503. doi: 10.1021/pr8008572. [DOI] [PubMed] [Google Scholar]
- 21.Turner AJ. Membrane alanyl aminopeptidase. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. 2. Academic Press; San Diego: 2004. pp. 289–294. [Google Scholar]
- 22.Wang J, Cooper MD. Aminopeptidase A. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. 2. Academic Press; San Diego: 2004. pp. 289–294. [Google Scholar]
- 23.Ikehara Y, Ogata S, Misumi Y. Dipeptidyl-peptidase IV from rat liver. Methods Enzymol. 1994;244:215–227. doi: 10.1016/0076-6879(94)44018-2. [DOI] [PubMed] [Google Scholar]
- 24.Underwood R, Chiravuri M, Lee H, Schmitz T, et al. Sequence, purification, and cloning of an intracellular serine protease, quiescent cell proline dipeptidase. J Biol Chem. 1999;274:34053–34058. doi: 10.1074/jbc.274.48.34053. [DOI] [PubMed] [Google Scholar]
- 25.Rao NV, Rao GV, Hoidal JR. Human dipeptidyl-peptidase I. Gene characterization, localization, and expression. J Biol Chem. 1997;272:10260–10265. doi: 10.1074/jbc.272.15.10260. [DOI] [PubMed] [Google Scholar]
- 26.Adachi J, Kumar C, Zhang Y, Olsen JV, et al. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marimuthu A, O’Meally RN, Chaerkady R, Subbannayya Y, et al. A comprehensive map of the human urinary proteome. J Proteome Res. 2011;10:2734–2743. doi: 10.1021/pr2003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nat Rev Cancer. 2008;8:268–278. doi: 10.1038/nrc2351. [DOI] [PubMed] [Google Scholar]
- 30.Reiter RE, Gu Z, Watabe T, Thomas G, et al. Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci USA. 1998;95:1735–1740. doi: 10.1073/pnas.95.4.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buckett WM, Luckas MJM, Gazvani MR, Aird IA, et al. Seminal plasma lactoferrin concnetrations in normal and abnormal semen samples. J Andrology. 1997;18:302–304. [PubMed] [Google Scholar]
- 32.Dubbink HJ, Verkaik NS, Faber PW, Trapman J, et al. Tissue-specific and androgen-regulated expression of human prostate-specific transglutaminase. Biochem J. 1996;315:901–908. doi: 10.1042/bj3150901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao F, Mirwald A, Papaioannou M, Baniahmad A, et al. Secretoglobin 2A1 is under androgen control mediated by a peculiar binding site for Sp family transcription factors. Mol Endocrinol. 2005;19:2964–2978. doi: 10.1210/me.2004-0408. [DOI] [PubMed] [Google Scholar]
- 34.Barrett AJ, Rawlings ND, Woessner JF. Handbook of Proteolytic Enzymes. 2. Academic Press; San Diego: 2004. [Google Scholar]
- 35.Shao C, Li M, Li X, Wei L, et al. A tool for biomarker discovery in the urinary proteome: a manually curated human urine protein biomarker set. Mol Cell Proteomics. 2011;10:M111.010975. doi: 10.1074/mcp.M111.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kojima K, Mihara R, Sakai T, Togari A, et al. Serum activities of dipeptidyl-aminopeptidase II and dipeptidyl-aminopeptidase IV in tumor-bearing animals and in cancer patients. Biochem Med Metab Biol. 1987;37:35–41. doi: 10.1016/0885-4505(87)90007-7. [DOI] [PubMed] [Google Scholar]
- 37.Durinx C, Neels H, Van der Auwera JC, Naelaerts K, et al. Reference values for plasma dipeptidyl-peptidase IV activity and their association with other laboratory parameters. Clin Chem Lab Med. 2001;39:155–159. doi: 10.1515/CCLM.2001.026. [DOI] [PubMed] [Google Scholar]
- 38.Carrera MP, Ramirez-Exposito MJ, Valenzuela MT, Garcia MJ, et al. Glutamyl- but not aspartyl-aminopeptidase activity is modified in serum of N-methyl nitrosourea-induced rat mammary tumours. Anticancer Res. 2004;24:801–805. [PubMed] [Google Scholar]
- 39.McPherson RA, Pincus MR. Henry’s Clinical Diagnosis and Management by Laboratory Methods. 22. Elsevier Saunders; Philadelphia: 2011. [Google Scholar]
- 40.Perez I, Varona A, Blanco L, Gil J, et al. Increased APN/CD13 and acid aminopeptidase activities in head and neck squamous cell carcinoma. Head Neck. 2009;31:1335–1340. doi: 10.1002/hed.21099. [DOI] [PubMed] [Google Scholar]
- 41.van Hensbergen Y, Broxterman HJ, Hanemaaier R, Jorna AS, et al. Soluble aminopeptidase N in malignant and nonmalignant effusions and intratumoral fluid. Clin Cancer Res. 2002;8:3747–3754. [PubMed] [Google Scholar]
- 42.Rangel R, Sun Y, Guzman-Rojas L, Ozawa MG, et al. Impaired angiogenesis in aminopeptidase N-null mice. Proc Natl Acad Sci USA. 2007;104:4588–4593. doi: 10.1073/pnas.0611653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farrah T, Deutsch EW, Omenn GS, Campbell DS, et al. A high-confidence human plasma proteome reference set with estimated concentrations in Peptide Atlas. Mol Cell Proteomics. 2011;10:M110.006353. doi: 10.1074/mcp.M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Budman LI, Kassouf W, Steinberg JR. Biomarkers for detection and surveillance of bladder cancer. Can Urol Assoc J. 2008;2:212–221. doi: 10.5489/cuaj.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkler SG, Lacomis L, Philip J, Erdjument-Bromage H, et al. Isolation and mass spectrometry of transcription factor complexes. Methods. 2002;26:260–269. doi: 10.1016/S1046-2023(02)00030-0. [DOI] [PubMed] [Google Scholar]
- 46.Erdjument-Bromage H, Lui M, Lacomis L, Grewal A, et al. Micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J Chromatogr A. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.