Abstract
We re-examine the pH dependence of partial processes of QH2 turnover in Glu-295 mutants in Rhodobacter sphaeroides to clarify the mechanistic role. In more crippled mutants, the bell-shaped pH profile of wildtype was replaced by dependence on a single pK at ~8.5 favoring electron transfer. Loss of the pK at 6.5 reflects a change in the rate-limiting step from the first to the second electron transfer. Over the range of pH 6–8, no major pH dependence of formation of the initial reaction complex was seen, and the rates of bypass reactions were similar to wildtype. Occupancy of the Qo-site by semiquinone (SQ) was similar in wildtype and the Glu→Trp mutant. Since heme bL is initially oxidized in the latter, the bifurcated reaction can still occur, allowing estimation of an empirical rate constant <103 s−1 for reduction of heme bL by SQ from the domain distal from heme bL, a value 1000-fold smaller than that expected from distance. If the pK ~8.5 in mutant strains is due to deprotonation of the neutral semiquinone, with Q.- as electron donor to heme bL, then in wildtype this low value would preclude mechanisms for normal flux in which semiquinone is constrained to this domain. A kinetic model in which Glu-295 catalyzes H+ transfer from QH., and delivery of the H+ to exit channel(s) by rotational displacement, and facilitates rapid electron transfer from SQ to heme bL by allowing Q.- to move closer to the heme, accounts well for the observations.
Keywords: bifurcated reaction of Q-cycle, control and gating, semiquinone occupancy, H+ exit pathway, kinetic model
1. Introduction
The cytochrome (cyt) bc1 complex (ubihydroquinone:cyt c oxidoreductse, E.C. 1.10.2.2, or complex III of the mitochondrial respiratory chain ), catalyzes oxidation of ubihydroquinone (quinol, QH2) by cyt c (or cyt c2 in bacteria). The complex in both mitochondria and bacteria is a homodimer, with a catalytic core of three subunits, cyt b, cyt c1, and the Rieske iron-sulfur protein (ISP), in each monomer. In bacterial complexes, no other subunits are structurally defined, but in Rhodobacter sphaeroides, the isolated complex has an additional subunit (SU IV) of uncertain function, which is absent from that of the closely related Rb. capsulatus. Mitochondrial complexes have up to 11 subunits, many of uncertain function [1, 2].
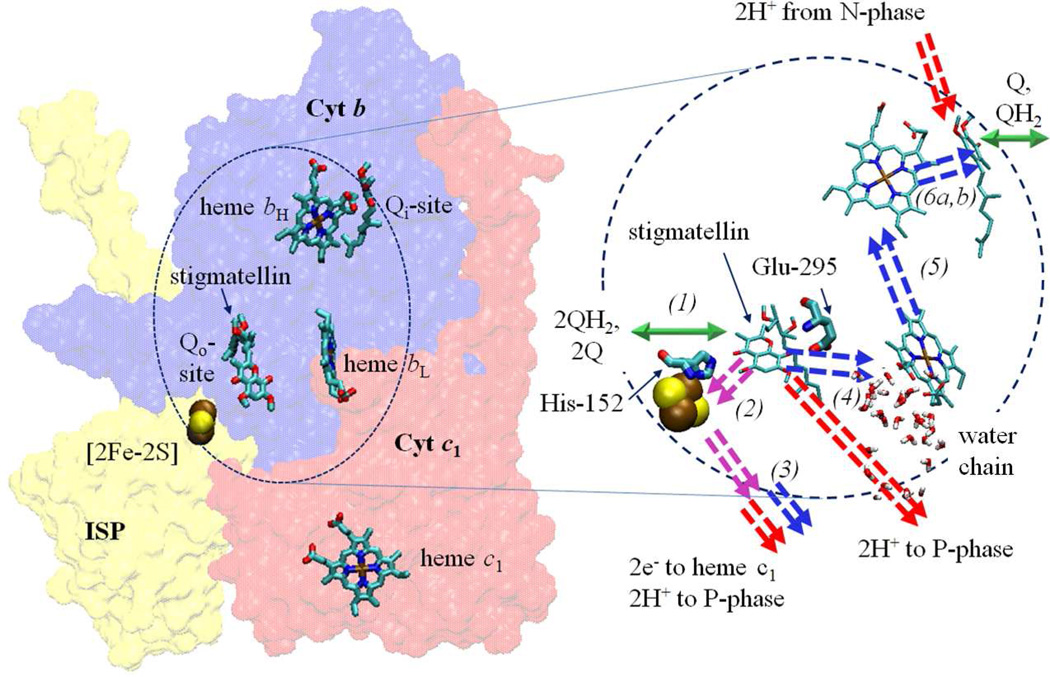
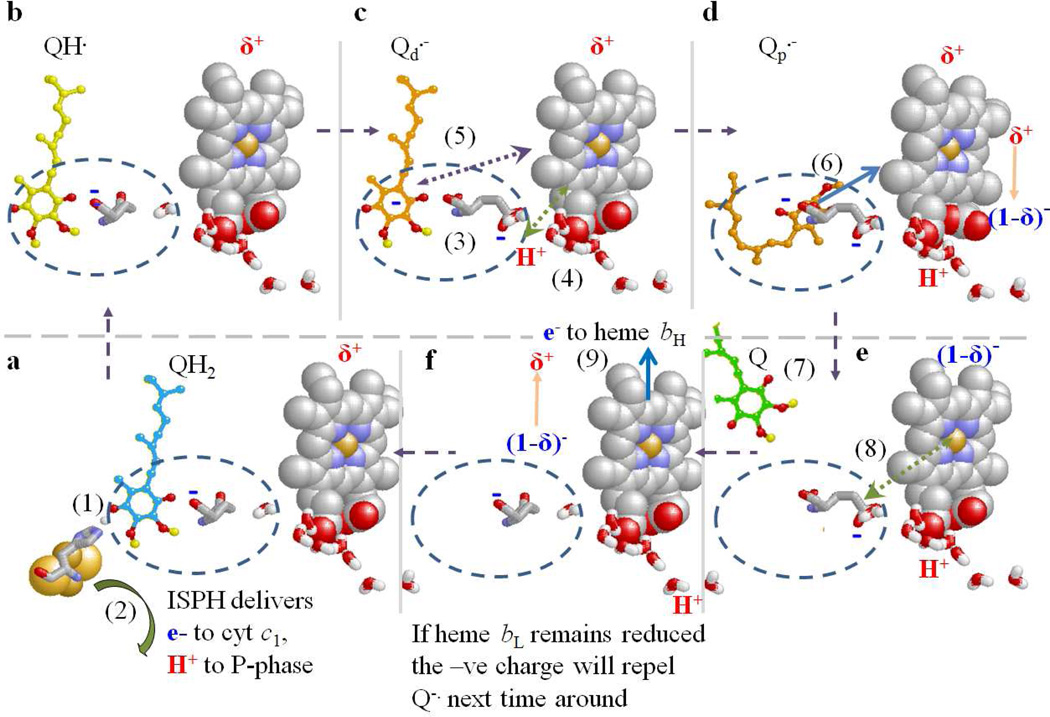
In the Q-cycle mechanism through which the complex operates (Fig. 1), oxidation of QH2 at the Qo-site occurs through a bifurcated reaction delivering the electrons to two different acceptor chains [3–6]. The first electron reduces the high potential chain (ISP and cyt c1), and generates an intermediate semiquinone (SQ). The electron from SQ reduces the low potential chain, consisting of hemes bL and bH of cyt b, which deliver the electron across the membrane to reduce ubiquinone (Q) or SQ at the Qi-site in an electrogenic process that contributes to the electrical component of the proton gradient used to drive ATP synthesis.
Figure 1. Q-cycle mechanism.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6a, 6b) |
In the first electron transfer, both an electron and a proton are transferred on reduction of ISPox, leaving as a product the neutral semiquinone, QH.. The proton-coupled electron transfer of this step accounts for the high activation energy of the overall reaction [7], and mechanistic parameters have been established from detailed studies of the dependence of rate on physicochemical properties of ISP [8–13]. The mechanism of the second electron transfer is more controversial. The Q-cycle operates with high efficiency so that in the absence of backpressure from the proton gradient [14], virtually all electrons go through the bifurcated process. If electron exit from the low potential chain is blocked, bypass reactions occur, which under aerobic conditions lead to reduction of O2 to superoxide, and a cascade of secondary reactive oxygen species (ROS) and related radicals that lead to cellular damage, aging, etc. [15–17] A question of mechanistic and anthropocentric interest is how evolution has honed the mechanism to minimize these damaging short-circuits.
1.1 Structural considerations
Structures at atomic resolution are available from mitochondrial and bacterial sources [18–23], but some aspects of the structure-function interface can only be addresses through models because the enzyme-substrate (ES-) complexes for both Qo-site electron transfers are meta-stable. Speculation about the mechanism of the initial step depends on the nature of its ES-complex (ES1) at the Qo-site (Fig. 1, eq. 1). The two substrates, QH2 and ISPox, cannot coexist in equilibrium at any Eh; the driving force (ΔE′) is generated in vivo by metabolism, or by activation of linked photochemical processes, and ES1 has so far been inaccessible to crystallographic exploration. Models for the reaction complex have been based on the binding of inhibitors, well-defined in structures. In particular, that for stigmatellin has been used as a guide in discussion of both ES1 and enzyme-product (EP-) complexes [24–26]. The protein side-chains liganding the inhibitor (His-152 of ISP and Glu-295 of cyt b, using Rb. sphaeroides numbering), have attracted particular attention. The ES1-complex has been modeled by replacing stigmatellin with QH2, which fits well within the electron density of the inhibitor [25]. However, the polarity of the H-bond between ISP and the occupant is different; His-152 of the reduced ISP (ISPH) is the H-bond donor to a carbonyl group of stigmatellin, while in ES1 a phenolic -OH of QH2 would donate an H-bond to ISPox, with His-152 in the imidazolate form [8, 13, 27]. The difference in polarity largely reflects the change in pK of His-152 on reduction to give ISPH, which is fully protonated in the neutral range. NMR studies in Thermus thermophilus bc1 complex [28] have now unambiguously associated the values for pKox1 and pKox2 with histidines equivalent to His-152 and His-131 of ISP, with values ~7.6 and ~9.6 in Rb. sphaeroides, while the pKred values of both in ISPH are in the range ~12–13 [29].
1.2 The first electron transfer
At saturating substrate concentrations, the first electron transfer is a rate-limiting proton-coupled electron transfer, the reaction is endergonic [7, 8, 30–33], and the products are ISPH and the neutral semiquinone, QH.. The reaction is proton-first-then-electron; the rate depends on the contribution of pKox1 of ISP to the Brønsted barrier determining distribution of the proton along the H-bond [34], and on the contribution of Em,ISP to the electron transfer driving force and activation barrier. The behavior is well-described according to a Marcus-Brønsted relationship [13, 27], and varies on modification of these values in ISP mutants. When substrates QH2 or ISPox were limiting, the rate also varied as expected from the controlling role for substrate concentration in formation of the ES1-complex (eqs. 1, 7, 8), such that with one substrate held constant at saturating concentration, variation of the other gave Michaelis-Menten behavior [7] [35], allowing calculation of relative binding coefficients of both substrates:
| (7) |
| (8) |
The pH dependence of quinol oxidation shows an increase over the range pH 6 to 7, described by an apparent pK (pKapp) of ~6.5 [7]. We suggested that the difference between pKapp and pKox1 ~7.6 (in wildtype) of the isolated ISP must reflect KISPox (eq. 8), the binding constant for formation of the ES1-complex. With ISP mutant strains, pKapp varied in parallel with change in pKox1 [7, 13], demonstrating a direct correlation.
1.3 The second electron transfer
The ES-complex (ES2) for the second step (eq. 4–6) is the bound QH. product of the first (eq. 2), and is also inaccessible to crystallography. In normal forward chemistry, the second electron transfer is fast enough to rapidly remove SQ so that none can be detected [36–38]. Different mechanisms have been proposed for this step, including variants of concerted mechanisms [31, 39, 40], double-gated mechanisms [41], double-occupancy mechanisms [42–44], SQ stabilized in a complex with ISPH [45, 46], and low occupancy by SQ with rate enhanced by mobility [7, 24, 26]. This choice of mechanisms is representative of the controversy as to the state from which electron transfer to heme bL occurs. We had suggested that an anionic species, Q.-, might migrate close to the heme so as to react rapidly at low occupancy [24, 26, 47, 48]. As recently demonstrated by EPR [30, 32, 37], under conditions in which it can accumulate, SQ is retained in the Qo-site volume as the anionic species Q.- [30], likely with Glu-295 carboxylate as H+ acceptor. The properties are in line with the mechanism proposed [24], but discrepancies between labs as to occupancy translate to uncertainties in interpretation of kinetic data.
The focus of the present study is Glu-295 of cyt b (E295), highly conserved in α-proteobacteria and mitochondria. The -PEWY- span containing this residue forms one side of the Qo-site. Our previous studies had shown that mutagenesis resulted in strongly inhibited rates of QH2 oxidation, and as structures became available, an obvious explanation became apparent [24, 49]. In stigmatellin-containing structures, the carboxylate group of E295 is H-bond acceptor from a hydroxyl group of the inhibitor [24, 26]. In the native structure, and those occupied by myxothiazol (or several other similar MOA-type inhibitors with an E-β-methoxy-acrylamide (or -acrylate) active group), the side chain is rotated by some 160° [26, 50] so that the carboxyl group connects to a water chain leading to the P-phase aqueous interface [21, 24, 26, 51]. This difference in configuration suggested a functional role for E295 in removal of the second proton from quinol oxidation [21, 24, 51]. The E295 would accept a proton from the neutral QH. (formed in the first electron transfer) via the ring –OH, and deliver it by rotation of the (now) carboxylic sidechain to the proton exit channel, leaving the anionic Q.- in the site. Clearly, mutation of E295 would be expected to lead to inhibition of this step. Dissociation and H+ transfer down the water chain would then provide a pathway for exit to the P-phase. When stigmatellin occupied the site, structures show that the E295 side-chain and two water molecules filled the volume between the inhibitor and heme bL [21]. However, in the structures containing myxothiazol or similar MOA-type inhibitors, this volume was occupied by the pharmacophoric MOA group [26, 50]. Assuming that QH. formed after the first electron transfer was initially in the same volume as in ES1 (the site distal from heme bL), H+ transfer and rotation of the E295 side-chain would open up the volume proximal to heme bL when no inhibitor was present. This suggested an additional indirect functional role; by opening up that volume, rotation of E295 might allow movement of Q.- closer to heme bL, to facilitate transfer of its electron [24, 26]. The ~5 Å change in distance would increase the rate constant by >1000-fold, allowing efficient operation at much lower SQ occupancy. The sidechain rotation exposes the backbone >NH of E295 as an H-bond partner for an O-atom of the MOA-group of the inhibitor; in the absence of inhibitor, this would be a potential ligand for SQ in the proximal volume.
Although several publications have raised objections to specific features of the mechanism proposed [31, 38, 53–56], some support for the proton exit scenario was provided by studies of the role of E295 in Zn-binding and inhibition [52]. In this paper, we have mutated the - PEWY-glutamate to D, G, Q, W, K, and L, and explored the effects on the bifurcated reaction and bypass processes, and their pH dependence compared to wildtype. We have measured the occupancy of SQ in wildtype and in mutant E295W, and used these values to estimate the rate constant for oxidation of SQ at the distal domain of the Qo-site by ferriheme bL. We address the points criticized, and introduce a kinetic model that illustrates the feasibility of the mobile SQ hypothesis, but leaves unresolved details of how gating to minimize bypass reactions is implemented. We discuss possible mechanisms in terms of a ballet of molecular dancers, with choreography directed by coupled coulombic forces between electron, proton, Q.-, heme bL, and carboxylate groups, all of which have charges in play at different times during the second electron transfer reaction.
2. Materials and Methods
2.1 Materials
Biochemical reagents were obtained from commercial sources. Molecular engineering supplies and protocols followed standard conventions [10, 57, 58]. Horse heart cytochrome c, antimycin-A, and decyl-ubiquinone, were purchased from Sigma and used without further purification. POPC was obtained from Avanti-biolipids. Ascochlorin was a kind gift from Dr. Nobuko Minagawa, Department of Biochemistry, Niigata University of Pharmacy and Applied Life Sciences, Niigata 956-8603, Japan.
2.2 Experimental protocols
2.2.1 Pre-steady state kinetics of quinol oxidation at the Qo-site
Chromatophores are formed by a pinching off vesicular invaginations of the bacterial cell membrane. Formation of sealed vesicles occurs on cell breakage by mechanical disruption. This traps a periplasmic aqueous volume, including cyt c2 at a stoichiometry similar to that of bc1 complex, which is in the membrane. Photochemical reaction centers (RC) are included in the cell membrane, with a stoichiometry about twice that of bc1 complex, but this can vary with growth conditions. On excitation by actinic light, the RCs generate the substrates (ferricyt c2 diffusing in the trapped aqueous phase, and QH2 diffusing in the membrane) for the bc1 complexes, which turn over to regenerate the substrates for the RCs (ferrocyt c2 and Q). In coupled membranes, turnover generates a proton gradient, with the major component contributed by membrane potential, Δψ. Kinetics of turnover of the photosynthetic chain were measured as previously described, following absorbance changes of the photochemical RC and cytochromes, after actinic flash (~5 µs at half-height) illumination of a dark-adapted suspension of chromatophores under controlled conditions of redox poise, temperature, and pH [3, 59–61]. Kinetic traces were recorded at 542, 551, 561, 566, 569 and 575 nm, and specific components convoluted from absorbance changes at the following wavelength differences: RC oxidized donor (P+), 542 nm; cyt c1 plus c2 (cyt ct), 551–542 nm; heme bH, 561–569 nm; heme bL, (566–575 nm) – 0.5(heme bH), with further small corrections for contributions from cyt c2 and RC dependent on relative stoichiometry. When appropriate, the electrochromic absorbance change of carotenoid pigments that monitor Δψ, were measured at 503 nm. In most experiments reported, electrochromic changes were minimized by addition of valinomycin and nigericin to collapse the proton gradient. In experiments to measure the kinetics of Δψ formation, the ionophores were omitted, and the changes at 503 nm were monitored separately.
Activation of RC generates the substrates for the bc1 complex; when the Q-pool is partially reduced, the time scale for activation of turnover is determined by diffusion of ferricyt c2 and its oxidation of the high potential chain (cyt c1 and ISP) with t½ ~150 µs, allowing exploration of kinetics in the sub ms range. Because equilibration in both high and low potential chains is more rapid than the rate-limiting first electron transfer, when electron transfer out of the b-heme chain is inhibited by antimycin, the rate of reduction of heme bH provides a measure of the rate of oxidation of ubiquinol during a single turnover of the Qo-site. By control of redox poise and pH, and with knowledge of the relative stoichiometries of the components, kinetic parameters as a function of substrate concentration, pH, or any other controllable reaction condition, can be readily extracted from the data [3, 7, 62]. Details of reaction media, redox mediators, pH buffers, temperature control, and redox poising are given in the Figure legends. For kinetic modeling, we used the program Dynafit (Biokin Ltd., Pullman WA) [63].
2.2.2 Molecular engineering, and sample preparation
Rb. sphaeroides strains expressing wildtype and mutant bc1 complexes were constructed essentially as described in [57]. The expression of the expected mutation was checked by PCR amplification of plasmid DNA from cells used for each chromatophore preparation used experimentally. Chromatophores were harvested from Rb. sphaeroides essentially as in [64]. Isolated bc1 complex was purified in a single step through a His6-tag affinity column as described in [57], followed by extensive dialysis. Concentrations of bc1 complex were quantified through absorption spectrophotometry by using the dithionite reduced – ferricyanide oxidized difference spectrum at 561–569 nm for heme bH, using ε=20 mM−1cm−1. The stoichiometry of subunits was check by semi-quantitative PAGE analysis.
2.2.3 Preparation of freeze-quenched EPR samples
Isolated bc1 complex at 20 µM from wild type or E295W chromatophores was incubated with 200 µM oxidized equine cytochrome c, 20 µM ferricyanide, and 100 µM antimycin A (to inhibit electron transfer from heme bH, and to prevent formation of SQ at the Qi-site), with and without the Qo-site inhibitor ascochlorin at 80 µM. Absorption spectra confirmed that the bc1 complex and equine cytochrome c were oxidized prior to mixing (data not shown). To initiate the reaction, the bc1 complex was mixed with 100 µM decyl-QH2 in a 1:1 volume ratio, to final concentrations of 10 and 50 µM respectively. Both the bc1 complex and decyl-QH2 were prepared in a buffer of 50 mM MOPS at pH 8.5, 100 mM KCl, 20% glycerol, and 25 µg/ml 1-palmitoyl-2-oleoyl- glycero-3-phosphocholine (POPC). Decyl-QH2 was prepared from decyl-Q using borohydride reduction, and diluted into degassed buffer immediately prior to use. Samples were mixed using a Biologic MPS-20 freeze-quench apparatus, housed in an anaerobic box equipped with gloves to allow manipulations prior to mixing. Before each experiment, the flow-lines were cleared with 250 µl of sample, which was removed by suction. All buffers and samples were degassed under argon for a minimum of 20 minutes before use in the anaerobic chamber. The reaction was started by mixing 250 µl of each solution, and quenched after 50 ms. Myoglobin-sodium azide control was conducted as in [30], yielding an average freezing time of 35ms. This, combined with mixing time and calculated time of flight, gave a total dead time of 50ms. Samples were injected directly into liquid pentane at −125° C, and then sealed and stored in liquid nitrogen. Following collection and freezing, the vials containing the frozen samples were thawed in a bath of liquid hexane brought to −60° C through immersion in liquid nitrogen in a secondary container. Frozen samples were packed into ~8 mm OD EPR tubes using stainless steel rods through a Teflon funnel immersed in hexane at −60° C. The sample volume in all cases allowed a filling of the EPR tube to a height > 2 cm, the maximum height of sample detection. Spin concentration of the SQ was determined using an internal standard (~2 µM Mn2+ in final sample), and quantified by double-integration of the EPR spectrum and comparison against a ladder of external Mn2+ standards, to compensate for variations in packing density, and fluctuations in detector sensitivity over time. Spectra were measured at 1.0 mW power [30], 5.0 gauss modulation amplitude, 100 kHz modulation frequency, centered at 3320 gauss with a sweep of 160 gauss, effective amplifier gain of 104, using microwave frequency ~9.30 GHz, with the value recorded for each measurement to allow calculation of g-values. Traces are the average of 75 scans of 25 s each.
2.2.4 Electron Transfer rates for forward and bypass reactions
Protocols were developed for measurement of forward electron transfer and bypass reactions in the pseudo-steady-state, achieved by actinic illumination by a series of closely spaced flashes. Pseudo-steady-state experiments were performed under conditions similar to those for measurement of the single turnover kinetics but with some modifications. For most experiments, reactions were measured under conditions, in which the quinone pool was initially half-reduced (for example, at Eh~ 90 mV at pH 7.0). The following rates were measured, as illustrated in Figs. 2, 3:
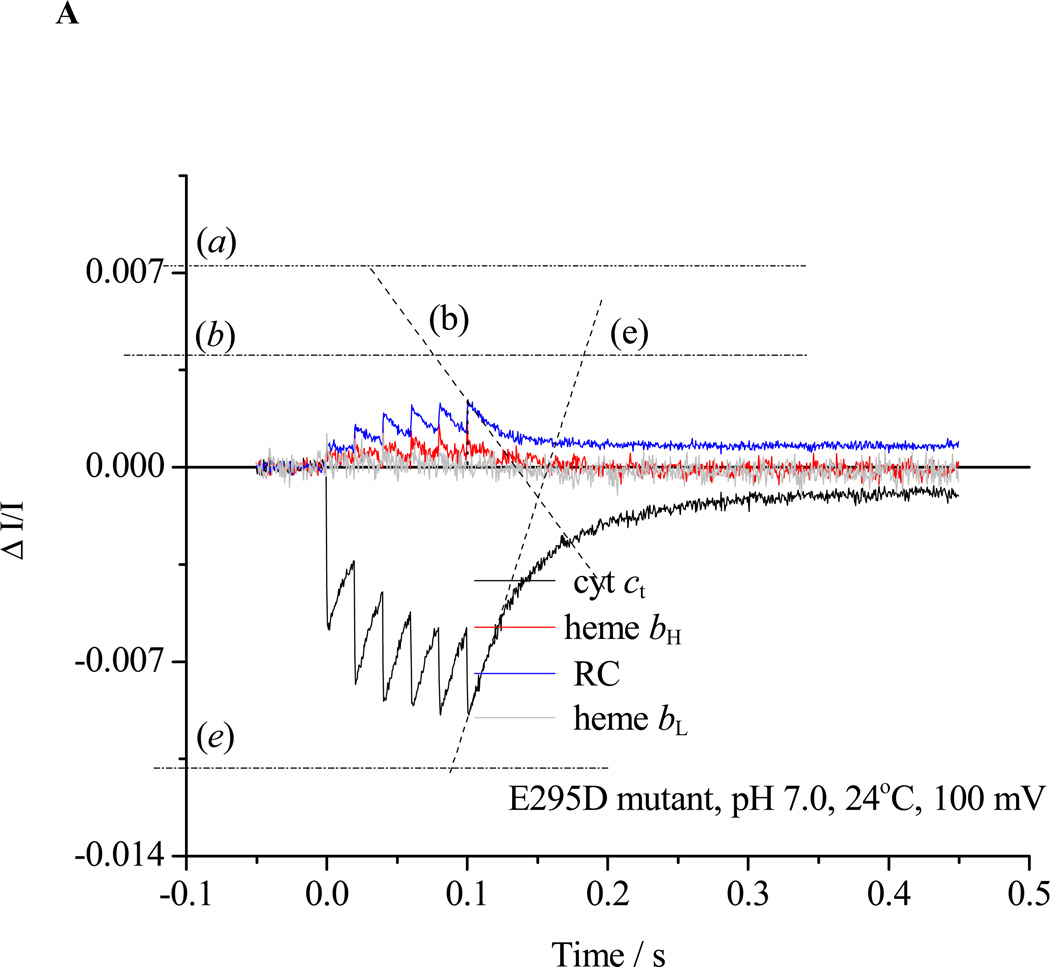
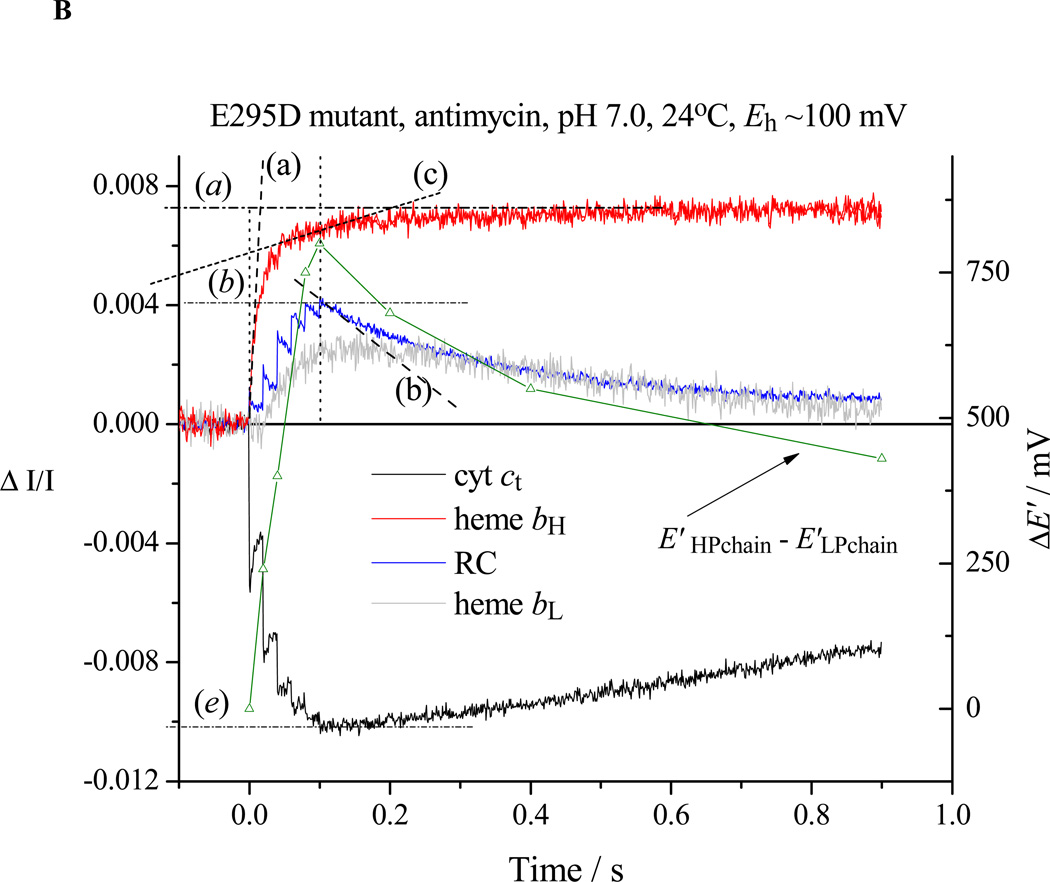
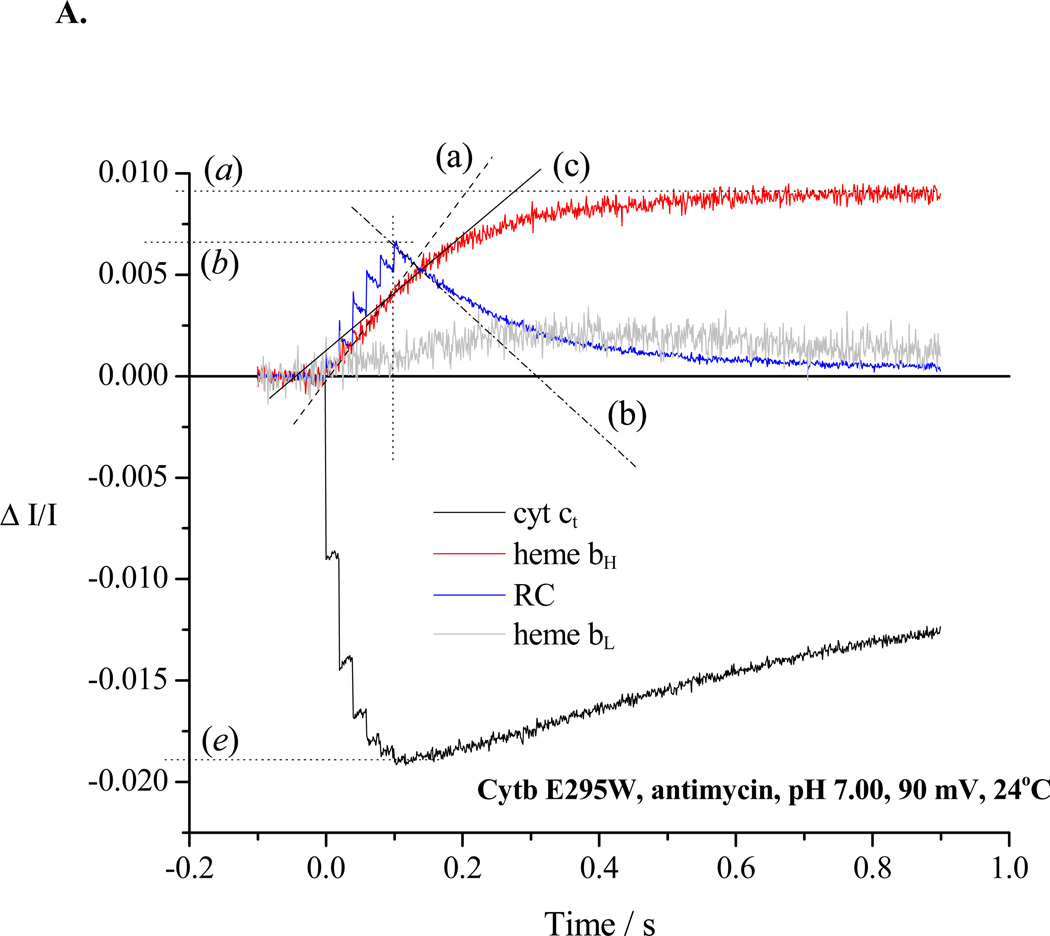
Re-reduction of P+ after a train of 6 saturating flashes spaced at 20 ms (slope (b)). In the absence of inhibitor in wildtype (not shown) or in partially inhibited strains (cf. E295D in Fig. 2A), the flux through the electron transfer chain in the pseudo-steady-state includes the bifurcated reaction at the Qo-site of the bc1 complex, bypass reactions, and back reactions of the RC, and is given by the rate of re-reduction of RC (slope b) plus that for cyt ct (slope (e)). In the absence of inhibitors in severely crippled mutant strains, or in the presence of antimycin, this flash regime was sufficient to fully oxidize RC, cyt c2 and cyt c1, and the rate of rereduction of RC then measured total flux (Fig. 2B, trace (b)).
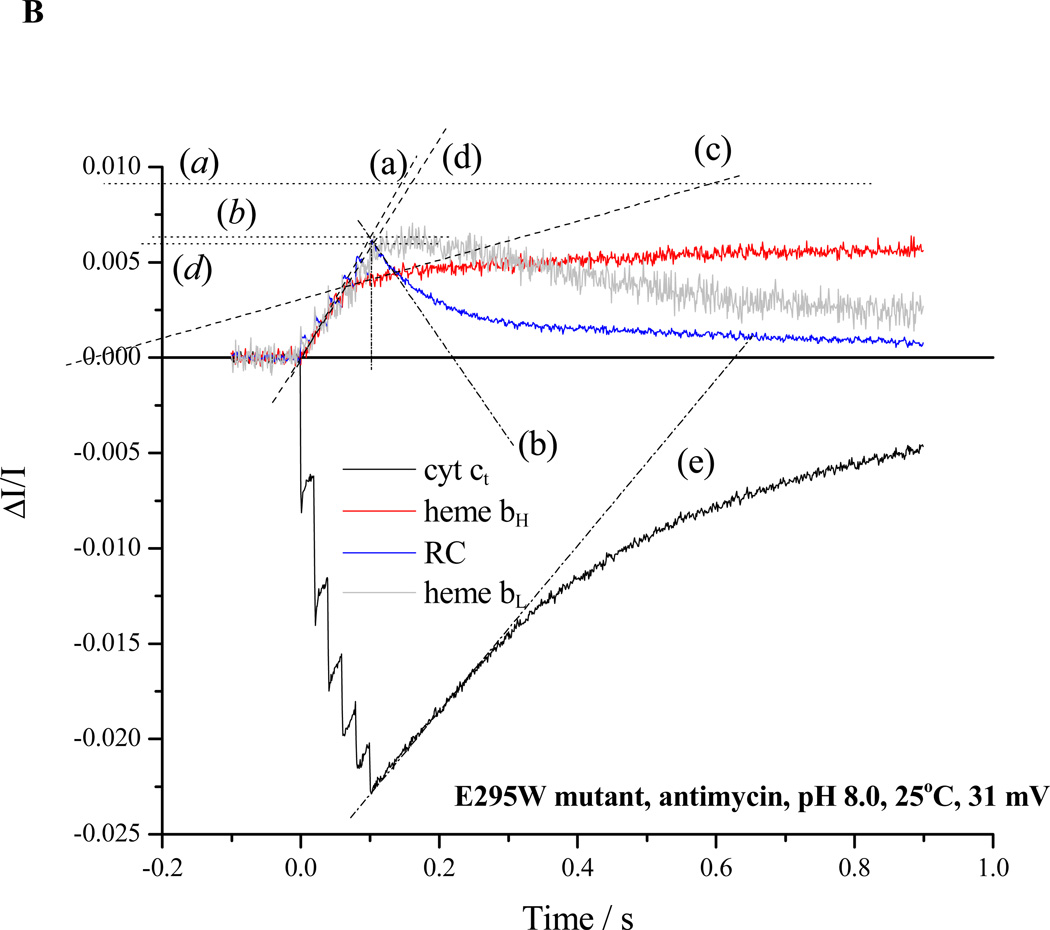
Rates for the bifurcated reaction were determined from the rate of heme bH reduction following the first flash in the presence of antimycin (Fig. 2B, trace (a)). If heme bH was initially oxidized, no reduction of heme bL was seen after the first flash [3, 59], but was delayed until heme bH was substantially reduced, as expected from the relative Em values [59, and see section 4.3]. For severely crippled strains, reduction of heme bH was only partial during the 20 ms before the next flash, and the initial slope measured during the train was taken (Fig. 3A, trace (a)). When heme bH was initially partly or completely reduced, the initial rate of reduction of heme bL could be similarly assayed, starting after the first flash (Fig. 3B, trace (d)), with the total flux through the bifurcated reaction obtained from the sum of initial rates into the two b-hemes.
In the presence of antimycin (Figs. 2B, 3), the turnover of the Qo-site becomes inhibited as electrons back-up in the b-heme chain, and the residual electron transfer measured after a train of flashes (trace (b)) is mainly from bypass reactions. In severely crippled mutants, the b-heme chain was not completely reduced after the 6th flash (Fig. 3A, trace (a)), and some residual bifurcated reaction still continued (slope (c)). The rate could be subtracted from the RC re-reduction rate to give the fraction contributed by bypass and back reactions.
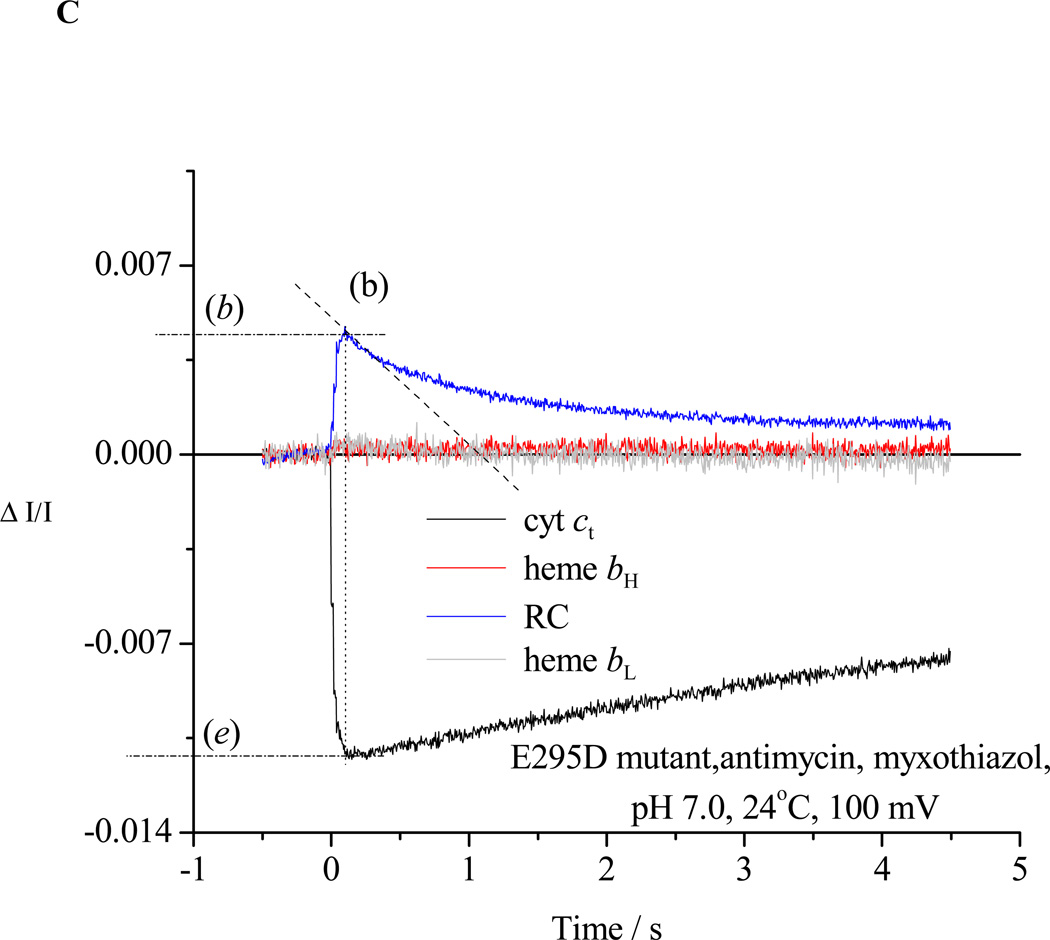
In the presence of antimycin and myxothiazol, the bifurcated reaction is completely inhibited in all strains, and measurement of RC re-reduction rates gives residual bypass reactions and back reactions (Fig. 2C).
In the pseudo-steady-state after six flashes, the total flux is given by rate of re-reduction of cyt ct plus that of RC. In wildtype and partially crippled stains like E295D, the relaxation of the chain between flashes is sufficient to allow regeneration of a significant population of reduced cyt c before the flash, and the oxidation of this fraction gives rise to a re-reduction of RC faster than detectable at the time scale and time constant used. This component was ignored, and the rates measured from a point 500 µs after the last flash were considered as representative.
Figure 2. Kinetic traces to illustrate protocols used to assay different partial processes.
Chromatophores from E295D mutant were suspended in a medium containing 100 mM KCl, 50 mM MOPS buffer at pH 7.0, with mediators to catalyze equilibration of redox centers with the ambient redox potential, Eh, poised at 100 mV. Under these conditions, the ubiquinone pool was ~30% reduced, and the ES1-complex close to saturated. The b-hemes of the bc1 complex, and the quinone acceptors of the reaction center (RC) were oxidized, and the high potential chain, and the RC donor, were reduced. The reaction mixture in an anaerobic cuvette was stirred, except during data acquisition. Turnover was initiated by a group of six saturating flashes (~5 µs at half-height), spaced 20 ms apart. Each flash activated >90% of open RCs to generate the substrates of the bc1 complex. A. Kinetics in the absence of inhibitors; B. Kinetics in the presence of antimycin; C. Kinetics in the presence of antimycin and myxothiazol. Traces are colored as follows: red, heme bH reduction (ΔA561–569); black, heme c1 + c2 (cyt ct) (ΔA551–542); blue, RC P+ (ΔA542); gray, heme bL (ΔA566–575 – 0.5(ΔA561–569) with small additional corrections for P+ and cyt ct depending on relative stoichiometry). The rates of different partial processes were determined from the slopes and amplitudes labeled as follows: (a) Initial rate of reduction of heme bH. In the presence of antimycin, this assays the rate of QH2 oxidation through the bifurcated reaction. (a) Maximal amplitude of heme bH absorbance change in the presence of antimycin (1 heme / bc1 monomer), allowing normalization of rate. (b) Rate of reduction of P+, the oxidized RC donor, immediately after the 6th flash, which assays the the pseudo-steady state flux from processes leading to relaxation of the system. (b) Maximal amplitude of P+ absorbance change to assay total RC, allowing normalization of rate. Since myxothiazol blocks the bifurcated reaction, traces in the presence of myxothiazol assay myxothiazol-insensitive bypass reactions, back-reactions of RC, etc. (c) Rate of the heme bH reduction (bifurcated reaction) immediately after the 6th flash (to allow correction of bypass rates. This assays the rate in bc1 complexes in which reduction of heme bH is incomplete. Subtraction of this rate from that measured in (b) allows assay of bypass reactions. (d) Rate of reduction of heme bL. At the Eh and pH of this experiment, reduction of heme bL was delayed until heme bH was substantially reduced. If heme bH is initially reduced, reduction of heme bL is observed after the first flash (see Fig. 3B). (d) Maximal amplitude of heme bL absorbance change. (e) Rate of reduction of cyt ct immediately after the 6th flash. This flux must be added to that determined in (b) to obtain the total flux through the high potential chain. (e) Maximal amplitude of cyt ct absorbance change to allow normalization. See text for further details.
Figure 3. Partial processes in turnover of the Qo-site in E295W mutant.
A. Kinetics of cytochromes and reaction center at pH 7.0; reaction conditions and labeling of traces as in Fig. 2. B. Kinetics at pH 8.0; reaction conditions as in Fig. 2, but with 50 mM MOPS replaced by 50 mM EPPS.
Rates were normalized to the maximal amplitude of the component, measured on activation by multiple flashes in the presence of antimycin (levels indicated by (a), (b), (d) and (e) correspond to amplitudes for heme bH, RC, heme bL and cyt ct, respectively). This could be achieved by poising the redox potential at Eh,7 ~120 mV, where the high potential chain was initially fully reduced, the low potential chain oxidized, and the Q-pool 10% reduced. For the bc1 complex, it was assumed that a monomer contained 1 equiv. each of hemes bH, bL, c1, and ISP. The stoichiometric ratio of RC to bc1 complex was taken using an extinction coefficient for P+ measured at 542 nm as 10 mM−1cm−1, and for heme bH measured at 551–569 nm as 20 mM−1cm−1.
2.2.5 Estimation of turnover from the electrogenic flux
The electrogenic flux associated with turnover of the photosynthetic apparatus was determined by measurement of absorbance changes due to the electrochromic response of carotenoids measured at 503 nm, when induced by a single saturating flash [3, 65–73]. The flux of charge across the membrane associated with electron transfer between the Qo- and Qi-sites of the bc1 complex and coupled release and uptake of H+, was determined by subtraction of the trace measured in the presence of myxothiazol and antimycin (reflecting the RC-linked electrogenic processes) from the change in the absence of inhibitors (reflecting both RC and bc1 turnover), and normalized to give units e−/(bc1 complex)/s, using the stoichiometric ratio of RC to bc1 complex in each strain. The flux assayed at Eh,7 ~100 mV matches the rate of heme bH reduction when the latter is measured in the presence of antimycin [72], and this relationship was assumed at other pH and Eh values.
2.2.6 Estimation of enzyme kinetic coefficients
Values for Vmax and Km for QH2 as substrate for the Qo-site reaction were obtained from linear fits to standard Eadie-Scatchard plots (v/S against v). Redox poise of the ubiquinone pool was varied by changing the ambient redox potential, Eh, and [QH2] was calculated using the standard Nernst-Peters equation, and estimated values for total Q + QH2, and QH2 in the membrane (which varied with redox poise), including QH2 generated in the flash [74]. When the redox poise was measured in situ by reduction of the Qpool, the concentration of both Q and QH2 changed, and values for Km measured reflect in part the differential binding properties of Q and QH2, as discussed at length in [8, 62]. The same set of data could be used to determine apparent affinity of QH2 in formation of the ES-complex, with similar caveats [8]. The pH dependence of turnover in the range ≤ 8.0 was measured from rates of reduction of heme bH (or bH plus bL) in the presence of antimycin. Kinetics of quinol oxidation at pH ≥8.5 were measured from rates of charge transfer across the membrane, assayed via the electrochromic carotenoid band-shift determined from the absorbance change at 503 nm [67, 72]. The trace in the presence of myxothiazol (RC uninhibited, no turnover of Qo-site) was subtracted from that in the absence of inhibitors (both RC and bc1 complex turning) over) to show the fraction of the change due to bc1 complex turnover.
In strains with the first electron transfer as the limiting step, the apparent pK controlling formation of the ES-complex could be determined from the pH dependence of Em for ISP and the dependence of rate on pH at saturating concentration of QH2 [7, 8, 13, 29, 75, 76] (see Introduction and eq. 8).
3. Results
3.1 General features of kinetics in E295 mutant strains
In the present work, we have extended our earlier investigations [9] to anaerobic conditions at controlled redox poise, pH, etc., in order to further explore the reaction parameters. The mutant strains studied (E295D, L, W, Q, K, G) all showed substantially inhibited rates for the bifurcated reaction when compared to wildtype. The behavior when assayed under anaerobic conditions was similar to that under aerobic conditions reviewed earlier [9]. The degree of inhibition of the bifurcated reaction was substantially higher than reported in experiments using isolated mitochondrial bc1 complexes [55], although the antimycin-inhibited rates were similar.
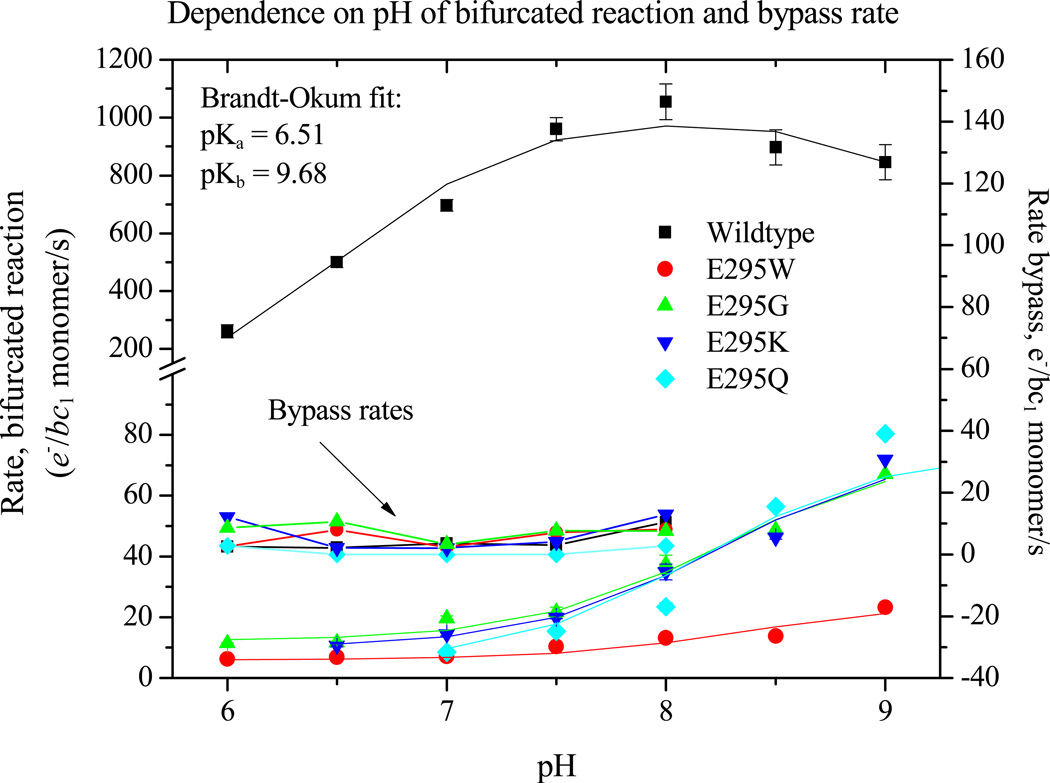
Turnover of the Qo-site in wildtype and mutant strains, and rates for bypass reactions, were assayed after a single flash, or in the pseudo-steady state established after 6 consecutive flashes at 20 ms apart as described under Experimental Protocols (section 2.2.4). Rates were measured from the kinetics of absorbance changes of the bc1 complex and photochemical reaction center (Fig. 3A, B), or from associated electrogenic processes (Fig. 4). In the more severely crippled strains, reduction of heme bH was still incomplete after the sixth flash (at 100 ms), and continued after illumination because the driving force established by oxidation of the high potential chain continued to sustain the forward reaction. In contrast, the level of reduction of heme bL leveled off almost immediately, and then declined; this behavior reflects the difference in Em values of the acceptor hemes (see section 4.2). At higher pH, heme bH was partly reduced in equilibrium with the quinone pool at Eh close to Em [6, 59], and in that fraction of centers, reduction of heme bL started after the first flash (Fig. 3B). Fig. 5 summarizes results from measurement of initial rates of the bifurcated reaction and myxothiazol-sensitive bypass rates over the range pH 6.0 to 9.0 for wildtype and mutant strains. All rates measured through reduction of the b-hemes were corrected for fluxes not associated with the bc1 complex by subtraction of the residual rate of re-reduction of P+ after inhibition of the bifurcated reaction by myxothiazol. These residual fluxes came mainly from back-reactions of the reaction center, and minor contributions from mediators, etc., which could not be distinguished from myxothiazol-insensitive short-circuit reactions of the bc1 complex. The total myxothiazol-insensitive flux represented ~1 e−/bc1/s (~0.1 % of the steady-state flux in wildtype, or ≤10% of the flux in the presence of antimycin). Note that this myxothiazol insensitive flux is much less significant than reported in mitochondrial complexes [77, and references therein].
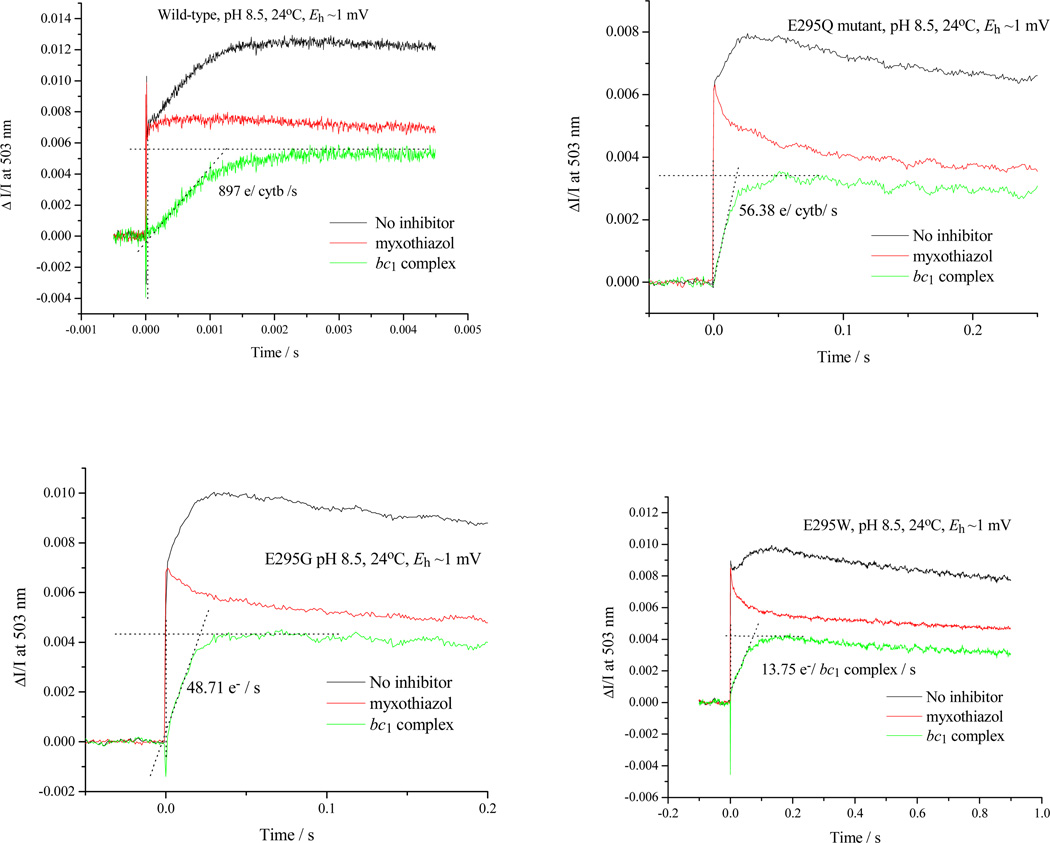
Figure 4. Qo-site turnover at high pH assayed through electrogenic reaction of the bc1 complex.
The absorbance change at 503 nm was measured in the absence and presence of myxothiazol, and the difference kinetics used to determine electrogenic processes associated with Qo-site turnover. The change at 503 nm is the due to electrochromic response of carotenoids to the electric field generated across the membrane by electron transfer from the Qo- to the Qi-site, and accompanying H+ displacements. The change due to reaction center, seen as the only contribution in the presence of myxothiazol, was subtracted.
Figure 5. The dependence of partial processes on pH.
Partial processes were assayed as shown in Figs. 2 and 3, under the conditions of Fig. 2, except for the substitution for 50 mM MOPS of the following buffers: pH 6.0 and 6.5, 50 mM MES; pH 7.5, 50 mM HEPES; pH 8.0 or 8.5, 50 mM EPPS; and pH 9.0 or 9.5, 50 mM CHES.
Under aerobic conditions, bypass rates assayed in chromatophores in the presence of antimycin in the pseudo-steady state at pH 7.0 were, for all the E295 mutants, similar to those in the wildtype [9], and were comparable to those previously seen in mitochondrial systems under aerobic conditions [77, 78]. When oxygen is present, reduction to superoxide occurs, likely with the Qo-site SQ as donor [77]. When isolated bc1 complex was used in these earlier experiments [77], similar rates were seen under anaerobic conditions as long as oxidized cyt c was present as an alternative acceptor. In chromatophores, cyt c2 is trapped within the enclosed vesicle, so that it can act as electron acceptor, and no addition of c-type cytochrome was needed to observe anaerobic bypass rates using the above protocol. When oxidation of SQ is blocked either by mutation, or by backup of the low potential chain in the presence of antimycin, the pseudo-steady-state flux determined from the rate of reduction of the terminal acceptor, P+, under anaerobic conditions, includes all bypass reactions involving SQ. Since the rates are similar to those under aerobic conditions, cyt c2, which is reduced by SO, likely acted as acceptor under both conditions.
A notable feature in all strains was the pattern of equilibria among redox centers following a train of flashes. The strongly inhibited rates for reduction of the low potential chain in mutant strains, associated with the slowed SQ oxidation, allowed us to observe kinetics, and the change in poise of the components of high and low potential chains, and the equilibria of the bifurcated reaction as the driving force increased on successive flashes (Figs. 2B, 3). Constraints arising from thermodynamic limitations in the Q-cycle [3, 5, 44, 59, 72, and see section 4.2] were obvious, and provide a basis for calculation of parameters need for estimation of thermodynamic poise of the partial reactions involving SQ (see section 4.4).
3.2 The pH dependence of the rate of the bifurcated reaction
The kinetic analysis was extended to measurement of the pH dependence of rates of partial processes so as to dissect out the contributions from dissociable species (Figs. 3–5). The pH dependence was tested with Eh poised to keep [QH2] in the quinone pool close to saturating (50% reduced). In the strain with wildtype bc1 complex, this approach yields a bell-shaped curve for the rate of the bifurcated reaction, fit by two [79] or three [55, 80] pK values, the highest of which (pK~9.5) gave the falling edge at high pH. We have interpreted the curve as reflecting contributions of the pK values of ISP to stability of the ES1-complex [7, 8, 62]; the rising edge of the bell shape is related to pKox1 (but displaced by the affinity of QH2 for ISPox), and the falling edge to pKox2, of the histidine ligands to the [2Fe-2S] cluster in the ISPox state [62, 80, 81]. This behavior was replaced in the E295 mutant strains tested (E295G, Q, K, W) by a simpler behavior. There was little pH dependency in the range below pH 7.5, but a strong increase in rate developed over the range above pH 8.0, well described by a single pK ~8.5 (Fig. 5). The degree of inhibition compared to wildtype varied over the range tested. In the lower pH range, where the degree of inhibition was highest, it varied with the rise of the bell-shaped curve, but over the higher range where the bell-shaped curve was falling and the curve for the mutants was rising, the degree of inhibition was smaller. A similar modified pH dependence was previously noted in Rb. capsulatus [56] and in mitochondrial complexes [55, 82]. The disappearance of the behavior associated with the pK values of ISPox, and the identity of the group conferring the simpler dependence on a single pK, are discussed extensively below (section 4.6).
3.3 Effects of mutation of Glu-295 on the binding affinity of quinol at the Qo-site
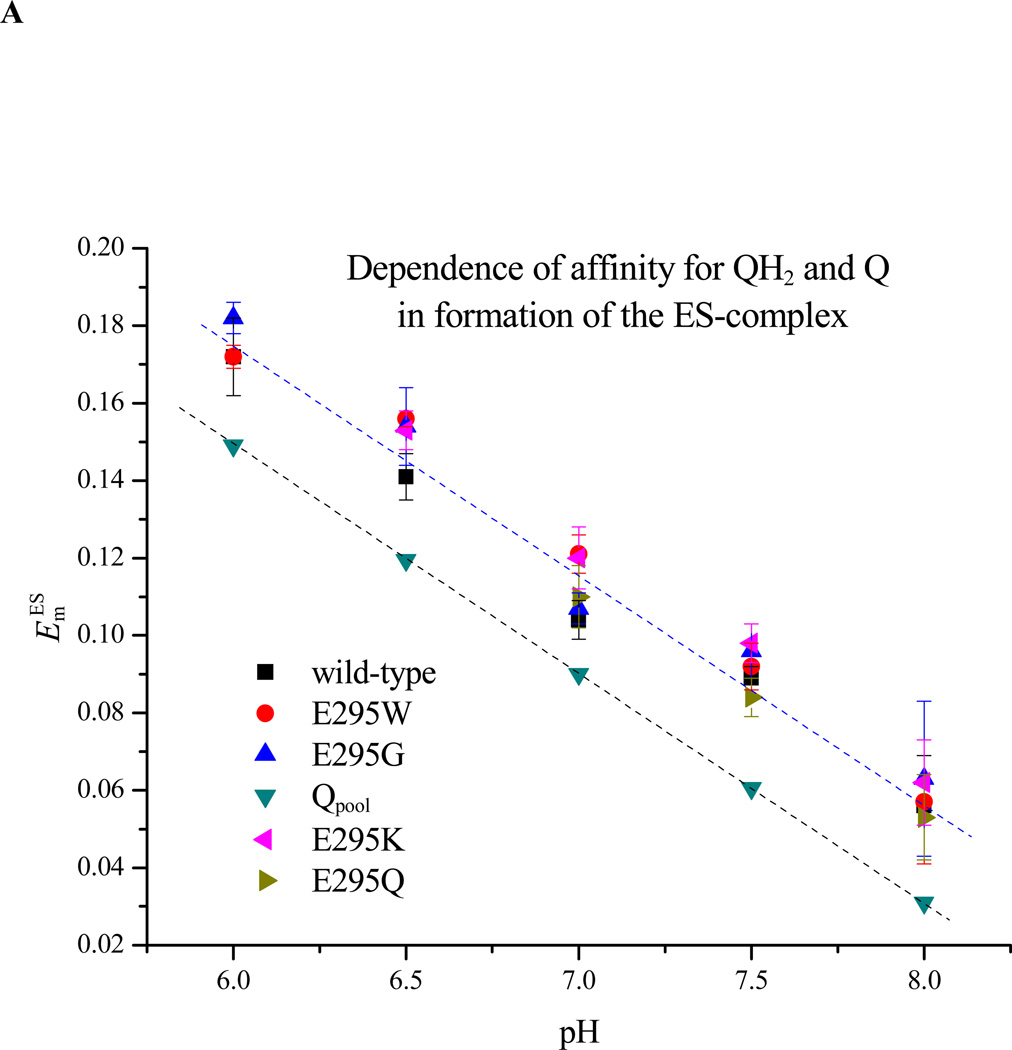
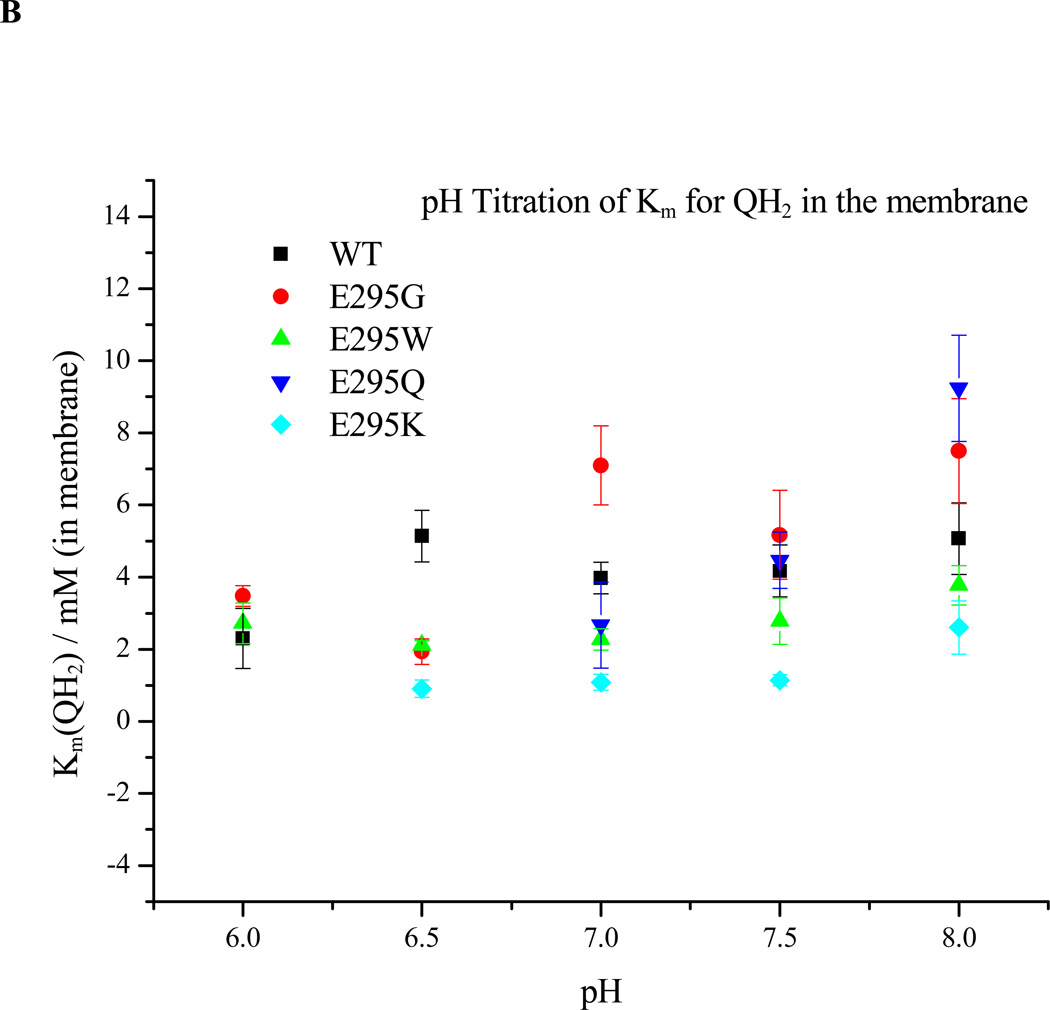
Using the above protocols, we have measured the fluxes associated with different partial processes as a function of [QH2], varied by changing the ambient redox potential, Eh, and of pH. All mutants tested showed the acceleration in rate of the bifurcated reaction as [QH2] increased on reduction of the pool seen in wildtype, as expected from Michaelis-Menten considerations, and followed the same pattern as in wildtype. The change in rate with [QH2] was analyzed either using conventional approaches to give apparent Km and Vmax values [35], or in terms of relative affinity of Q and QH2 for the Qo-site ([8], and see section 4.6), and both parameters could then be determined as a function of pH to look for any dependency of affinities in formation of the ES1-complex, which might reflect a role for dissociation of essential residues (Figs. 6A, B). Because bypass fluxes are not readily determined from measurement of electrogenic processes, studies of pH dependence were restricted to the range ≤pH 8.
Figure 6. Dependence on pH of ES1-complex formation. A. Apparent Em for formation of the ES1-complex assayed from the dependence of rate on Eh.
The rate of QH2 oxidation, assayed through the rate of heme bH reduction in the presence of antimycin, was measured as the quinone pool was reduced by lowering the ambient redox potential, Eh. Since rate is proportional to [ES], the mid-point of the titration give Em ES. The Em values for wildtype and all mutants were displaced from that of the pool by ~30 mV, but all showed the same pH dependence. B. Dependence on pH of Km for QH2. The same data could be used to calculate Km values for QH2 for each strain tested, plotted here as a function of pH (see Materials and Methods section).
The dependence of rate on degree of reduction of the quinone pool reflects the saturation of the ES1-complex by QH2, which in this system is maximal a few hundred µs after generation of the substrates on flash activation. When normalized to [bc1 complex], the rate gives the fraction of centers in the ES1 form. The curve of maximal rate as a function of Eh is then a titration of maximal occupancy of the ES1-complex, [ES1]. The mid-point potential for formation of ES1, , is shown as a function of pH for wildtype and E295 mutants in Fig. 6A. The mid-point potentials (taken from the midpoint of the plot of rate of QH2 oxidation, estimated from heme bH reduction, as a function of Eh) were estimated by fitting the titration curve to the Nernst equation for a redox couple with n=2, appropriate to the Q/QH2 couple. The apparent association constant, KQH2, for quinol binding on formation of the ES-complex can then be calculated from using eq. 7 [8]. As seen in Fig. 6A, the ES1-complex mid-point potentials for wildtype and E295G, K, W, and Q mutants were not very different over the range pH 6.0 – 8.0, and followed the slope for titration of quinone free in the pool, displaced by the difference in value between . From this, the mutations did not have any large effect on the apparent KQH2 of the ES-complex. For the E295K and Q mutants, mid-point potentials for the ES-complex were, respectively, slightly higher and lower than the wildtype over this pH range, and complemented the trend seen for Km. However, within the error of the measurement, the difference from the wildtype was marginal.
Previous kinetic data on E295 mutants showed an apparent Km for QH2 up to 2.5-fold higher in E295D and E295G than in wildtype [24, 55, 82]. The increase was interpreted as showing a weak contribution from an H-bond between E295 and QH2 to the binding on formation of ES1. However, the structures show variation in the configuration of the glutamate with different distal domain inhibitors [1, 23, 83, 84]. In the present work (Fig. 6B), changes in apparent Km for E295G, W, K, and Q mutants compared to wildtype were also small, but in E295W and K, were in the opposite direction to that previously reported for E295G. Only minor variations were observed over the pH range assayed. In view of the approximations involved in deriving a value for membrane concentration of ubiquinol, and the variation between chromatophore preparations in the stoichiometric parameters involved, we consider the weak variation in values for Km as a function of pH to be of less significance than the lack of any major pH dependence of this parameter over the range shown.
3.4 SQ formed at the Qo-site on addition of QH2 to the oxidized complex
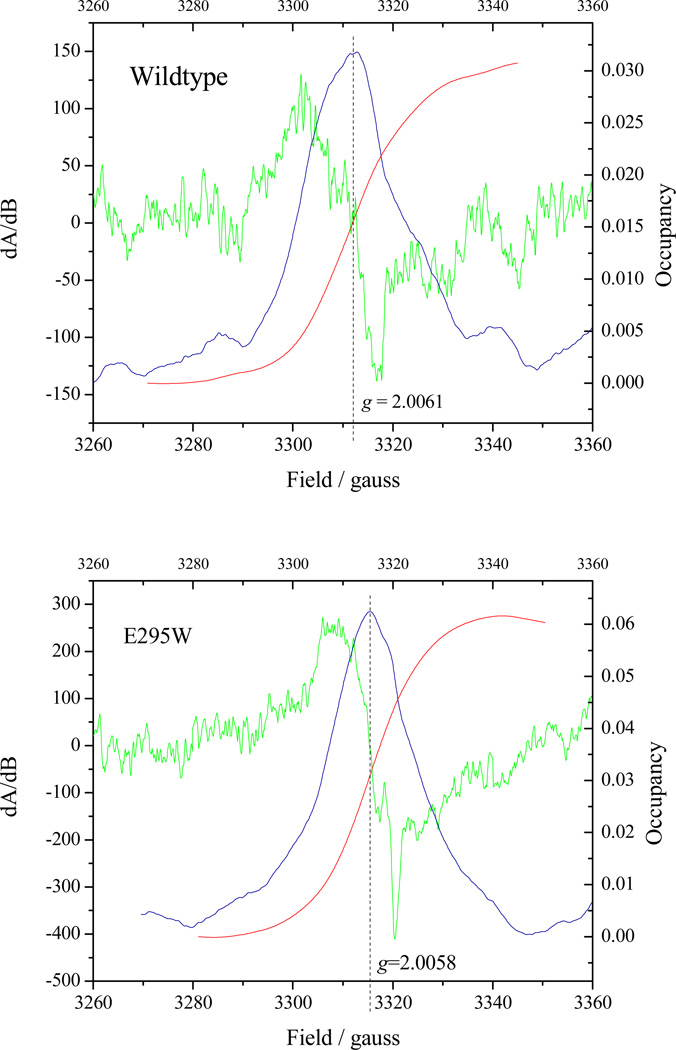
Published values for SQ occupancy of the Qo-site are conflicting, and no data for E295 mutants has been reported. We have used protocols similar to those reported in [30, 32]. Our results are consistent with those of Cape et al. [30]; Fig. 7 shows the SQ signals after correction for the ascochlorin insensitive component (ascochlorin occupies the distal domain of the Qo-site [84], and is expected to displace all other occupants). The yields of SQ were similar in wildtype and E295W (though higher in the latter), and at the high end of the range previously reported. Although the subtracted spectra are somewhat noisy, the g-values (2.0061 in wildtype and 2.0058 in E295W) were, within the accuracy of the noise, similar to those determined in [30], where chemically generate Q.- had g=2.0056. Our line-widths, however, were both wider (24 in wildtype and 18 in E295W), perhaps because of some contaminating background signal that was not subtracted. Our attempts to measure SQ on illumination of chromatophores [32] were inconclusive. When the delay before measurement was short enough to leave a substantial driving force in the high-potential chain, the EPR signal due to residual P+ swamped the SQ signal, as might be expected from the kinetics of decay of the optical signal for P+ at 542 nm in Figs. 2–3, making accurate determination of occupancy difficult.
Figure 7. SQ occupancy at the Qo-site in wildtype and E295W mutant.
The green curve shows the CW-EPR derivative spectrum of the Qo-site SQ, obtained by subtraction of the signal when ascochlorin was added to the reaction mixture so as to remove components not associated with Qo-site turnover. The blue curve shows the result of a first integration (to generate the absorbance spectrum), and the red curve a second integration to determine the occupancy, shown by the right-hand scale. Antimycin was present in all experiments to eliminate contributions from the Qi-site SQ. See Materials and Methods section for details.
3.5 Redox properties of the hemes in wildtype and E295G mutant
The E295G mutant was chosen as an example to test if mutation caused any large changes in the thermodynamic or spectral properties of the b-hemes. The most probable effects would come from loss of the charge of the glutamate side chain, and from changes in packing at the Qo-site, and this mutant might be expected to show both effects. Thermodynamic characteristics were determined by full-spectrum redox titration over the wavelength range 520–580 nm. Redox potentials of cyt bL, bH, and c1 hemes of the E295G mutant at pH 7.0 (Em.7 (heme bL) = − 90 mV; Em.7 (heme bH) = 40 mV; Em.7 (heme c1) = 270 mV; Em.7 (cyt b150) = 150 mV (due to SQ:bH heme interactions (2)), and Em.7 (cyt c2) = 340 mV) were not significantly different from those of wildtype strain, and the spectra were essentially the same. A more complete study of a different set of E295 mutants in Rb. capsulatus (E295V, K) [56] found no significant change with respect to wildtype in thermodynamic properties determined over a wider pH range (pH 5–10).
4. Discussion
The hypothesis tested was first proposed in a simple and explicit form 13 years ago (24). It had suggested direct roles for E295 in H+ exit and generation of the Q.- as substrate for the electron transfer step, and an indirect role in facilitating SQ migration in the site to enhance the rate. All were essential, so inhibition of the overall process could have been attributed to inhibition of any step. Critical points supported by our new data can be summarized as follows. E295 certainly plays an important role in oxidation of SQ. The SQ is formed, and its oxidation is inhibited in E295 mutants. At high pH in the antimycin-inhibited wildtype, the SQ was in the dissociated form (30). From hypothesis, this is the form from which electron transfer occurs. The SQ detected in wildtype is generated only after heme bL has become reduced, and therefore unable to act as acceptor of the electron from SQ. As a consequence, no additional information about the electron transfer step could be obtained. However, in E295W, the SQ is formed with heme bL mainly in the oxidized form. In this case, the SQ is likely constrained to the distal domain, and the terms needed to derive directly a value for the rate constant for oxidation of the semiquinone from this domain, are the rate of electron transfer to the b-heme chain and the occupancy measured. The value determined, 103 s−1, is ~1000-fold lower than that calculated on the basis of distance. Our data on binding of Q and QH2 show essentially the same values in the E295 mutants as in wildtype over the pH range 6–8, suggesting that one feature of the original hypothesis, - weak involvement of E295 in stabilization of the ES1 complex, - must be revised.
We address the question of dissociation state of SQ in E295W, which requires speculation based on incomplete information. The data show unambiguously that in the more crippled strains the rate of electron transfer increased with an apparent pK~8.5. Since the SQ spectrum is similar in E295W and wildtype in our experiments, the most economical hypothesis is that the increase reflects an increase in [SQ] due stabilization of the Q.- form as the pH is raised above the pK for this dissociation. If this is so, then the inhibition observed in the more crippled E295 mutants must reflect a stronger effect on the electron transfer than on the proton exit. In the following sections we discuss these points in greater detail.
4.1 Involvement of E295 in different partial processes
Earlier work [24, 26] showed that mutation of E295 in Rb. sphaeroides to D, Q, and G caused substantial inhibition of the rate of quinol oxidation at the Qo-site, suggesting a mechanistic involvement in the reaction; resistance to stigmatellin inhibition, and a slight increase in the Km for QH2 suggested a role in substrate binding. However, a normal gx = 1.80 EPR signal associated with EP-complex between ubiquinone, Q, and ISPH was seen for all the mutants, indicating that binding of Q at the distal site was not perturbed [26, 56]. These results supported the suggestion that different domains of the Qo-site might be of importance in catalysis for the two partial reactions delivering electrons to the high and low potential chains, with the proximal domain and the -PEWY- glutamate involved in the second electron transfer. More recent work [9, 36] on these and other E295 mutants has shown that bypass rates in such strains were similar to those seen in wildtype assayed under antimycin inhibition [9], suggesting that SQ occupancy in the mutants was also similar under these conditions. In the more crippled mutants (Qo-site turnover ~1% of wildtype rate), the bypass rates were in the same range as the residual bifurcated reaction assayed through heme bH reduction, when measured in the neutral pH range. Where tested, these results are confirmed under more rigorous conditions in the present work, but the role in substrate binding is shown to be unimportant.
4.2 The equilibria of the bifurcated reaction
In wildtype, the Qo-site reaction is controlled by the thermodynamic parameters of the modified Q-cycle, as seen in the equilibria measured for the reactants involved [3, 6]. These reflect three features of the reaction: (i) the rate-limiting first electron transfer; (ii) rapid equilibration in both high and low-potential chains, reflecting the much more rapid rates for other partial processes; and (iii) the equilibrium constant for the bifurcated reaction. Despite the substantial inhibition in the low potential chain, the equilibria between hemes appeared to be controlled by the same values in the E295 mutants. For the oxidation of QH2, the overall poise is determined by , where subscripts L and H indicate acceptors in the low and high potential chains [59]. For the first QH2 oxidized, the acceptors are heme bH (Eo′ ~40 mV at pH 7) and (predominantly) ISPox (Eo′ ~300 mV), and with Q/QH2 poised at Eo′ ~90 mV, Keq1 has a value of ~ 500. For the second QH2, since the two more favorable acceptors have been consumed, the acceptors are heme bL (Eo′ −90 mV) and (predominantly) heme c1 (Eo′ 270 mV), and Keq2 then has a value ~1.
In chromatophores isolated from strains with wildtype photosynthetic chains grown under standard conditions, there are approximately 2 RC/bc1 complex [3]. When QH2 is available in the pool and the antimycin-inhibited reaction is initiated by oxidation of the high potential chain through flash activation of the RC, the kinetics of QH2 oxidation in the first turnover lead to the reduction of 1 heme bH per monomer. Little if any reduction of heme bL is seen because the poise of reactants for the oxidation of the second QH2 comes up against the low value for Keq2. On subsequent flashes, heme bL goes reduced as ferroheme bH accumulates, because the driving force is pumped up by reoxidation of the high potential chain. In the high-potential chain, the oxidizing equivalents from 2P+ migrate initially to heme c1 and ISP, which then get reduced by QH2 via the bifurcated reaction. The observed rate of reduction of heme c1 is slower than the rate of heme bH reduction because the electron from the first turnover re-reduces preferentially the invisible ISPox [72]. For the same reason, the amplitude of re-reduction of heme c1 after one turnover of the Qo-site (assayed ~10 ms after the first flash) is partial, because the oxidizing equivalent left is shared between ISP and heme c1. These properties were of critical importance in allowing a distinction between the modified Q-cycle [3, 6, 85] and other variants current at the time, including Mitchell’s original version [4, 5].
A kinetic pattern similar to that on the first flash, - full reduction of heme bH, and partial reduction (~50%, depending on pH) of heme c1, - is seen on reduction of isolated oxidized complex by decyl-UQ in the presence of antimycin [31, 53, 86–88], and is well accounted for by the modified Q-cycle through consideration of the constraints discussed above. However, in experiments with the mitochondrial complex, the behavior has recently been interpreted differently. An ingenious mechanism proposed by Trumpower and colleagues [31, 53, 86–88] borrows the bifurcated reaction from the Q-cycle, but introduces control by an unspecified mechanism that operates within the dimer to limit Qo-site activity to only one of the two monomers, giving a half-of-sites functionality. To explain the poise of reactants discussed above, it is then necessary to postulate that the active Qo-site can access its own high potential chain, but both low potential chains through inter-monomer electron transfer at the level of heme bL. To account for the monotonic reduction of heme bH observed, the rate of inter-monomer electron transfer would necessarily be more rapid than the rate-limiting step (the first electron transfer), since successive turnovers of the active Qo-site would reduce first one, then the other heme bH, and a slower rate for the inter-monomer transfer would lead to separation of the kinetics into distinguishable phases. Furthermore, since both electrons go to heme bH centers, the equilibrium constants determining poise from this mechanism would differ from those of the Q-cycle; successive turnovers of the active Qo-site would have heme bH acceptors with similar Em, and reactions with similar high equilibrium constants.
The half-of-sites mechanism has received strong support from three studies in which molecular engineering in bacteria had been used to construct systems allowing expression of heterodimeric bc1 complexes in which differential mutations in the two copies of cyt b enforced inter-monomer electron transfer [53, 54, 89]. These showed the rapid kinetics seen in wildtype. In our attempts to reproduce such a heterodimeric expression system, we could measure the rapid kinetics previously interpreted as showing inter-monomer electron transfer, but showed that these were accounted for by cross-over recombination at the DNA level to reconstruct the wildtype homodimer from the heterodimeric construct [90]. We demonstrated that strains in which the heterodimeric constructs enforced an inter-monomer path could not compete with monomeric function. Although this problem had not been discussed in the earlier papers, in subsequent papers [91, 92] two groups acknowledged that recombination occurred with high frequency in their systems, and addressed the question in greater detail. In a more recent paper [93], the rates reported in isolated heterodimeric complexes constructed so as to enforce the inter-monomer path were <10% those measured in similar constructs with wildtype sequence, - much too slow to be commensurate with the original claims, or with a half-of-sites mechanism. The results previously claimed seem better explained by the same reconstruction of a wildtype operon by the crossover recombination we demonstrated, and by operation of the wildtype monomeric mechanism. Although some slow flux between monomers may well occur, the major flux is clearly carried by a monomeric functionality, and the remaining discussion will be framed in these terms.
4.3 Parameters determining the degree of inhibition at the Qo-site
In turnover of the Qo-site, the rate measured in situ in chromatophores (~760 QH2/bc1/s at pH 7.0 at 25°C, with maximal rate ~103 s−1 at pH 8.0) is that of the limiting reaction, - the first electron transfer from QH2 to ISPox. In E295 mutant strains tested, the overall rate decreased by between 13 and 160-fold compared to wildtype (maximal rate for E295D mutant was ~59 s−1 at pH 9.0; minimal for E295W mutant was ~ 4.79 s−1 in the range pH 6–7). Although this is a substantial inhibition, representation of the degree of inhibition with respect to turnover in wildtype can be misleading because the intrinsic rate of the second step in wildtype is necessarily faster than the limiting rate, and a true measure of inhibition would take account of this. To get a better understanding of the true degree of inhibition, it is necessary to refer the inhibited rates to the operational for the 2nd electron transfer under conditions of maximal flux. In principle, can be calculated from the observed rate and the occupancy of SQ, using , but requires a value for the SQ occupancy. As previously noted [10, 72], useful constraints for wildtype can be established directly from measurements of lag-phases [10, 38, 72, 94, 95]; the rate of formation of intermediate states of the second step would be limited by the rate of the first step, and any significant population of such a state would give rise to a delay in electron delivery to the terminal acceptor (heme bH in antimycin-inhibited chromatophores). Observed lags are in the range 20–30 µs [38, 72, 95] and give a maximal value for combined occupancy of intermediate states (formation of SQ and ferroheme bL) as ~0.02 SQ/Qo-site. In a technical tour-de-force, Zhu et al. [38] using rapid-mix freeze-quench and EPR to assay the reactants, failed to detect any SQ attributable to the bifurcated reaction under conditions in which acceptors were available in both chains. Although the authors concluded that no SQ was involved, a more realistic interpretation [10] is that the detection limit was not sufficient to see either SQ, or differences in the ISPH and heme bL kinetics, in the range expected (see kinetic model below). Cape et al. [30] detected no SQ under aerobic conditions, and since the Zhu et al. [38] work was under such conditions, the failure to detect SQ could alternatively be explained on that basis. However, since their time resolution was sub-millisecond, it seems likely that they would have seen the SQ if it had been formed at measureable occupancy, so their experiment provides a useful constraint. Estimates of occupancy under conditions in which SQ might be expected to accumulate also show some discrepancy. Cape et al. [30] reported values in the range 0.01 – 0.1 (measured at pH 8 under anaerobic conditions), and Zhang et al. [32] reported values ≤ 0.01, measured at pH ~9, where the SQ stability is expected to be maximal.
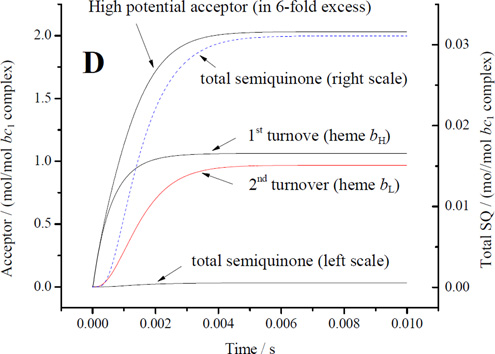
The driving force available in the Cape et al. [30] experiments, in which decyl-QH2 was added to the oxidized complex, should have been limited by thermodynamic constraints intrinsic to the Q-cycle [3] discussed above (section 4.2). On addition of QH2 to the oxidized complex, the Qo-site has to undergo at least one turnover, in a reaction with Keq1 ~500 (see section 4.2 for derivation of Keq values), leading to the consumption of the two favorable acceptors, before any SQ could be expected to accumulate. The second turnover would then reach equilibrium with heme bL and heme c1 in a reaction with Keq2 ~1, and in the absence of additional oxidant would reach a poise leaving heme bL still largely oxidized [32, 53]. The detection of SQ in [30] was therefore somewhat surprising. It seems likely that some additional oxidant was available to pull the reaction further over. Our own data (Fig. 7) show occupancies at the high end of their range under conditions in which an excess of oxidized cyt c was provided, supporting this speculation, and providing firmer data for calculation of values for kinetic parameters.
In the experiments of Zhang et al. [32, 37], the bc1 complex was mutated to remove heme bH, so that SQ might be expected to form as the second QH2 was oxidized. The poise of the Qo-site reaction, measured in chromatophores, was pushed towards products by pumping up the driving force using a group of flashes to re-oxidize the high potential chain while the Q pool was still substantially reduced. The occupancy under these forcing conditions would be expected to be much higher than that in [30], but the measured value was lower. Zhang et al. [32, 37] framed their discussion in terms of the stability constant, KS, for the SQ formation, and estimated a value for KS in the range 10−14 to 10−15. The rationale was to find the difference in poise for the two SQ couples appropriate to the difference in redox potential between the high and low potentials chains under the conditions of experiment, estimated to be ~800 mV from the poise of reactants. The poise can be expressed in terms of ΔE′ values (see below), but they used the relation between the Em values and stability constant to calculate KS. Since at maximal yield from disproportionation (the peak of the bell-shaped curve of a redox titration), , one might expect an occupancy in the range ~4 × 10−7 under these conditions, much lower than the value they reported, or that from the Cape et al. [30] experiment. Use of KS in the context of the Qo-site reaction is confusing because the process in which the SQ is formed and consumed physiologically is not a disproportionation, but is given by eqs. 2–6 (see Fig. 1 legend), and it is these equations that should be used. At the low flux of the inhibited systems, the two SQ couples would likely be poised close to equilibrium with the high (highPC) and low (lowPC) potential chains, with , but through eqs. 2–6, so that
| (eq. 9) |
might be thought of as the driving force for SQ formation. The ΔE′ values after a group of flashes can be obtained directly from spectrophotometric data like those of Figs. 2 and 3. From simple mass-action considerations, the [SQ] in eq. 2 would increase as increased, and [SQ] in eq. 3 would increase as decreased, leading to high occupancy at high driving force. (Note however that in the more crippled strains, the low potential chain had not yet reached equilibrium after six flashes (Fig. 3), and the occupancy of SQ would have been higher immediately after the flash than when equilibrium with heme bL was eventually achieved.) The parameter of interest is the displacement from equilibrium represented by eq. 9; in effect has the same form as , but here represents for eq. 9. In deriving values, the driving force ~800 mV (Fig. 2B) immediately after a group of flashes, when most RCs are in the P+ form, would be appropriate for calculation of occupancy. However, the EPR spectra in [32] showed no significant P+ in the sample assayed 1–3 s after illumination, so the driving force must have decayed substantially by the time of measurement. From Figs. 2B and 3B (under similar conditions), at 1 s after illumination, P+ had decayed to <10%, the c-hemes were >70% reduced, and heme bL >70% oxidized, so the value of ~800 mV is clearly wrong for discussion of their results; a value ~400 mV would seem more appropriate (cf. ΔE’ curve in Fig. 2B). The SQ occupancy expected from eqs. 2 and 3, would then be much lower than immediately after the last flash, which is in accord with the relatively low occupancy they found. In our own hands, all attempts to measure SQ occupancy of the Qo-site in chromatophores under conditions of high driving force (samples frozen <50 ms after illumination by a group of flashes) led to EPR spectra in which the g~2.005 region was dominated by P+, precluding an accurate estimation of SQ. Despite differences in power saturation, we were not able to resolve the two signals satisfactorily (not shown).
4.3.1. The importance of E295 in mechanism
The Qo-site reactions in Glu-295 mutants were also studied by Osyczka et al. [56] in chromatophores from Rb. capsulatus. They measured rates similar to those reported in our earlier study [24] and here. However, they chose to interpret their results in terms of the resilience of the Qo-site, with glutamate as a nonessential and replaceable component of the reaction. They based their conclusion on the fact that, when expressed in terms of observed rates, the degree of inhibition even in the slowest mutants was never complete, and, in their E295H strain at high pH, approached ~50% of the wildtype rates. Their interpretation of an unimportant role was biased by use of the rate of the limiting first electron transfer as the reference state in determination of the degree of inhibition. As discussed above (section 4.3), the inhibition is much more dramatic when compared to the rate constant for the second electron transfer, so their choice led them to underestimated the importance of E295.
Wentz et al. [55, 96] and Seddiki et al. [82] explored mutations at this site (E272) in the yeast mitochondrial complex. A feature of their studies was an even more modest degree of inhibition in glutamate mutants compared to wildtype. The difference between their observations and ours comes not from the rates observed in the mutant strains, which were similar, but from comparison of these with different rates for the wildtype. In the present work, we measured Qo-site turnover (~103 s−1 at pH 8) (Fig. 5) with the complex in its native membrane and the native Q-10 reacting from the lipid phase. In the studies with the yeast complex, the activity in wildtype was ~10-fold slower (75 s−1 [82], 52 s−1 [55], or 120 s−1 [95]) when assayed in the isolated protein through steady-state cyt c reductase activity using decyl-QH2 as substrate. Even slower steady-state rates have been reported in bacterial complexes when determined using the detergent solubilized enzyme in a similar assay (cf. [54, 97, 98]). However, when a single turnover of the bacterial enzyme was measured in similar preparations through photoactivation of the high potential chain, rates of heme bH reduction and heme c1 re-reduction were comparable to those seen in situ [95], showing that the pathways for Qo-site catalysis within the enzyme were not impaired by detergent isolation. Likely these rapid rates reflected the single turnover using bound (Q-2)H2, initially present at concentration saturating occupancy of the ES1-complex. Possibly, the steady-state flux was limited by the rate of exchange of decyl-Q (or Q-2) couples, partitioned between separate micellar phases in the detergent solubilized mix.
As discussed at length above, any observable inhibition likely represents a loss of activity in the second step, which then becomes the limiting process, reflecting minimally a 100-fold inhibition with respect to the effective . The inhibition is always substantial and reflects an important role of Glu-295 in catalysis. The fact that inhibition is never complete is intrinsic to the mechanism proposed, and reflects bypass reactions and the residual rate of the bifurcated reaction on accumulation of SQ.
4.4 Semiquinone occupancy at the Qo-site under forward and under forcing conditions
In our experiments under conditions similar to those of Cape et al. [30], the occupancy was at the higher end of the range they reported. The value expected under normal forward chemistry (with acceptors available in both chains) would, of course, be much lower than under these forcing conditions and likely too low for direct measurement [cf. 32]. In the following discussion we will assume initially an occupancy of 0.01SQ/Qo-site, consistent with the limits from kinetic data, and then discuss plausible ranges under uninhibited and under forcing conditions. We will then use the SQ occupancy we measured to estimate an empirical value for the rate constant for oxidation of SQ in the distal domain by heme bL, and from this derive other thermodynamic and kinetic parameters.
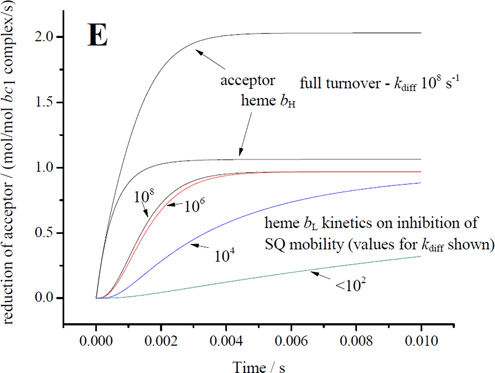
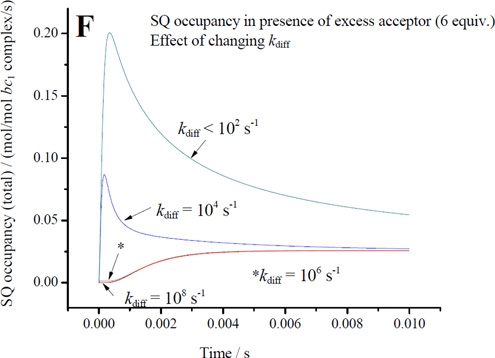
For electron transfer using parameters λ = 0.7 V, and ΔGo′ = −0.1 V, assuming a Marcus treatment of the Arrhenius term [10, 99], and the dependence of intrinsic rate constant on distance [100], the 11.5 Å for an edge-to-edge distance from an occupant of the distal domain to heme bL gives a value for of ~106 s−1, and for the 6.5 Å from the proximal position gives ~109 s−1, values similar to those used in previous discussions [7, 9, 44, 101, 102]. How reasonable are these values? The maximal rate measured in uninhibited chromatophores (~103 e−/bc1/s−1 at pH 8), is determined by the rate constant for the first electron transfer with a saturated ES1-complex (occupancy ~1). To match this rate for the second electron transfer using occupancy of 0.01, a minimal value for would have to be in the range 105 s−1, compatible with the value derived from distance. Given the uncertainty of occupancy values from earlier experiments, a simple mechanism in which the SQ could donate electrons to heme bL from the distal domain could not be excluded [44]. However, three factors make this naïve scenario questionable: (i) the reaction is certainly not a simple electron transfer, (ii) the occupancy under uninhibited flux must be much lower than the value under inhibited conditions, so likely the value used in the example above is an overestimate, and (iii) the intrinsic rate of removal of SQ would have to substantially exceed the rate of generation in order to account for the pattern of dependence on driving force in mutants; the full effect from changes in Em for ISP [62] is as expected from Marcus theory if the first electron transfer is limiting, and the lack of any consistent effect from changes in Em for heme bL on the overall rate (for example, enhanced rate when Em of heme bL is raised) is consistent with a much faster intrinsic rate for the second electron transfer [103]. Assuming a factor of ten for each of these last two points, we would need a value > 107 s−1, raising the question of consistency with mechanisms based on electron transfer from the distal domain using the value suggested by the distance.
When similar considerations are applied to the E295 mutants, the naïve treatment of rate constant becomes more obviously flawed. An important technical point to note is that, because rates in the E295 mutants are so slow, they can be measured at a time, <50 ms, corresponding to that at which SQ is sampled. With t½ ~100 ms, most centers would still have heme bL oxidized, so that an empirical rate constant for the bifurcated reaction could be determined directly. For the bulky sidechains (E295Q, E295W), the SQ would likely be constrained to the distal domain, and SQ occupancy would likely approach the maximal value allowed by the equilibrium constant for the first electron transfer, K1, and by driving force. Measurement of occupancy for SQ in E295W therefore provides, for the first time, an empirical value for the rate constant of the bifurcated reaction. Using values measured at pH ~8.5, and by substitution in v = k2([SQ.bL ox] we get a value for of ~103 s−1, which is 3-orders of magnitude smaller than the value calculated from distance. We use approximate values ([SQ] in the range 0.05 – 0.1/Qo-site, rate ~ 60 e−/s/bc1 seen with E295Q in Fig. 4) because we recognize that occupancies might vary in different strains, and because in E295W the bulk of the tryptophan might lead to artificial displacements in the site leading to lower rates than for more native side chains. As a consequence, the rate constant calculated is higher than if we had used rates derived from E295W data.
One clear conclusion is that the rate constant calculated from distance is inappropriate for the reaction under these conditions. Factors other than distance must come into play. Given the complexity of the second electron transfer step, it is perhaps not surprising that other parameters might modulate the rate. It will also be obvious that if the value for (~103 s−1 from the distal domain) is also appropriate in wildtype, and SQ was restricted to the distal domain, then no occupancy consistent with the measured lag phases could also be consistent with the observed uninhibited rate. Use of the same rate constant from the distal domain in wildtype and E295 mutants is the most economical hypothesis, and is supported by the kinetic model (section 4.9), and by the fact that bypass rates are similar in all strains, but the impact of this conclusion depends on whether or not the reactants are the same, and hence on the dissociation state of SQ, discussed further below.
While the rate constant calculated from distance is clearly inappropriate for transfer from the distal domain, if SQ were to move close to heme bL, the reaction might be a simple electron transfer, and the value expected from distance ( ~109 s−1 for 6.5 Å) would then be appropriate, representing a maximal operational value. Any observable inhibition of the bifurcated reaction (as in the E295 mutants) must indicate a rate less than the limiting first step (~103 s−1), and (since occupancy is low) a substantial inhibition, but a realistic kinetic model and justifiable constraints are needed to estimate a degree of inhibition with respect to an operational .
Knowledge of the occupancy for SQ also allows us to use the equilibria in the two chains to provide an estimate of the potentials of the two couples, and of equilibrium constants for the two steps of the bifurcated reaction. Recognizing that in the inhibited system, after six flashes the reactions in the two chains come close to equilibrium, and using
| (eq. 10) |
for the Qo-site reaction, taking values from the experimental EH and the kinetic traces, and ~ 90 mV (for the Q-pool), we get ~500 mV, and ~ −320 mV. Then, using (with [SQ] ~ 0.06), we get Em SQ/QH2 ~ 570 mV, and an equilibrium constant for the first electron transfer, K1 ~10−4.5 ~0.00003. With the Q-pool as reference, Em Q/SQ is then ~ −390 mV, K2 ~105, and the overall equilibrium constant, Koverall ~ 3.5, - the same value as obtained directly from ΔG′o on substitution of Em values in eq. 10. Formation of ES1 showed a midpoint, , displaced from that of the pool by ~30 mV (Fig. 6A), so discussion with respect to that as starting state (the configuration used in the kinetic model of section 4.9), requires consideration of the ~10-fold displacement in favor of QH2 binding indicated by this ΔEm. These values are used in the kinetic model discussed below (section 4. 9).
4.5 Involvement of Glu-295 in transfer of the second electron and proton
Since, from the discussion above, the rate constant from the distal domain would give a rate >100 times too slow, movement of SQ to allow migration closer to heme bL, facilitated by rotational displacement of the E295 sidechain, becomes an attractive possibility [24]. This opens the question of whether the inhibition observed involves the direct role in proton exit, or the indirect role in facilitation of electron transfer. What conclusions about the behavior can be drawn from particular mutants? Several properties of the glutamate side chain might be expected to be of importance, - the acidic nature, polarity, length, bulk, mobility, etc. The role in proton exit has been supported by studies of the role of E295 in Zn-inhibition, which likely involves participation of groups that otherwise stabilize the water chain [52]. However, exploration of this role, and of any role E295 may play in gating [9, 10], must be approached indirectly, since no such intermediate states have been directly observed. The E295D mutant has an acidic side chain, shorter by 1 −CH2 than the wildtype, and shows the least perturbation of behavior. E295G is next in line, and has no side-chain. All the other mutants are more severely crippled, including E295Q, which shares all characteristics of glutamate except the acidic nature. From the differences, we can infer that the acidic property is most important, that the length of the native side chain is required for maximal rate, and that, absent the carboxylate function, a bulky side chain introduces more severe constraints than a small one. We discuss these roles more fully in the following paragraphs.
4.6. Contribution of Glu-295 to the affinity of QH2 at the Qo-site
Although mutation of Glu-295 had a dramatic effect on the overall rate of QH2 oxidation, it is unlikely that the inhibition had any major direct effect on rate constants for the first electron transfer. We would expect to see an effect on Km if glutamate stabilized the ES1-complex. In stigmatellin-containing structures, the E295 carboxylate forms a H-bond with an –OH group of the inhibitor, and this feature was incorporated into early models of the ES1-complex, leading to an expectation that E295 would contribute to the affinity of the site for QH2. Indeed, in earlier work, a ~2-fold increase in Km was measured, in the expected direction [24]. In the present work, the changes in binding affinity of QH2 in the Glu-295 mutants compared to wildtype were also small, but there was no consistent pattern to the sign of change; - Km increased in some strains but decreased in others. From this it seems clear that, in agreement with [56, 82], the contribution, if any, of the H-bond to QH2 affinity is too small to explain any major fraction of the substantial inhibition observed in mutants.
Since the first electron transfer is endergonic, the inhibition in mutants could be readily explained by a reduced rate of removal of the intermediate SQ. This second step can be dissected into two main components, - electron transfer and proton transfer. In order to participate in proton transfer, the E295 side chain would have to be in the carboxylate form to act as H+ acceptor from QH.. The consistency of this assumed polarity would depend on the pK values of QH. and E295. The values in solution might be similar, but pK values in the protein can vary widely from those measured in an aqueous environment. The lack of any major effect of pH on Km suggests that neither the glutamate nor the quinol has a pK that affects ES1-complex formation in the pH range (6 to 8) tested. Since the dependence on pH of tracked that of Em Q-pool, we can also conclude that there was no marked pH dependence on KQH2, or on relative affinities of Q and QH2 when ISPox is the partner. It was previously noted in studies of E295 mutations in Rb. capsulatus [56] that in formation of the gx=1.80 complex, neither the affinity of Q or QH2 (measured from the Em for the change in EPR line intensity of the gx=1.80 complex on reduction of the Q-pool), nor the apparent Em of ISP, were modified in the mutants. However, the gx=1.80 complex is an EP-complex, in which the participating species are Q and ISPH [26, 47], and is relevant to ES1 only insofar as these states are present before flash-excitation (likely in ~60% of centers at Eh,7 ~100 mV [72]). The gx=1.80 complex is of no relevance to the ES2 complex in which QH. is the substrate; none of these results precludes a role for E295 in liganding the QH. state.
The question might be raised as to why in the mutant strains we observe a dependence of rate on substrate concentration for QH2, but not for ISPox, since both are required for formation of the ES1-complex. The answer lies in how the two values are measured. For each value of , the dependence of formation of ES1 on [QH2] was measured at fixed pH. Since the second electron transfer depends on the product of the first, that dependence is reflected in the rate measured, although the second step is limiting. In contrast, dependence of rate on [ISPox] is assayed by changing pH, and the weak contribution from the first electron transfer in the low pH range is swamped by the much larger pH dependence of the 2nd electron transfer in the high pH range.
4.7 The role of E295 in the bifurcated reaction, and the pH dependence in mutant strains
The preceding discussion was framed without reference to the dissociation state of SQ, the parameters calculated all pertain to the general case, and would be valid for a neutral or anionic SQ. In this section we discuss the dissociation state of the SQ species observed.
4.7.1. Interpretation of the pH dependence of the bifurcated reaction in E295 mutants
As noted earlier (sections 1.2, 3.2), the bell-shaped Brandt-Okun [79] pH profile of the wild-type Qo-site reaction can be well accounted for in the range below pH 8 by the involvement of the imidazolate form of His-152 of ISPox, and the role of the pKox1 of this group in formation of the ES-complex [8, 27, 62, 104]. The pK values determined from redox behavior in the isolated ISP, pKox1 ~7.6, pKox2 ~9.6, can be assigned unambiguously to His-152 and His-131 respectively [28, 105]. The value for the pKapp determined experimentally tracks the value of pKox1 on mutation of ISP, and since His-152 is the residue involved in H-bond interaction with QH2, we can take this as demonstrating a direct relationship between pKapp and pKox1, and assign the difference in value (ΔpK ~1.1) to the free-energy contributed by this interaction to formation of the ES1-complex [8] (see section 1.2 and eq. 8 for derivation of ΔpK).
Wentz et al. [55] studied the pH dependence in E272D and Q mutants in the yeast complex, and observed loss of a component corresponding to a pK ~6.2 from a three pK fit [80] (the slope of the wild-type curve was shifted to lower pH than in other studies). They interpreted loss of the pK at ~6.2 as reflecting replacement of the glutamate. However, the direct association of pKapp with His-152 of ISPox makes this explanation implausible [62]. Osyczka et al. [56] also determined the pH dependence of rate in the Rb. capsulatus complex, and for strains common to the two studies, results were similar to ours. However, the pH dependence in their less inhibited strains showed a less dramatic shift in the range below pH 7.5. No simple interpretation or firm conclusions from their results was offered [56].
In our more severely inhibited E295 mutants, the Brandt-Okun behavior was lost, and the pH dependence was relatively weak over the range pH 6–7.5. Instead, the pH profile showed a steep increase in rate over the range pH 8–9, where the wildtype shows a decrease in activity. The pK for the increased rate is in the range ~8.5 ±0.3 for the four strains tested. Note that the protocol for measurement means that this pK reflects only the bifurcated reaction. Since the pK ~6.5 in wildtype is well explained by the involvement of ISP His-152 in formation of the ES1 complex, the loss of the pK ~6.5 cannot by attributed to loss of E295 [55]. Rather, it is likely due to loss of the controlling role for the first electron transfer, and its replacement by a limitation in the second electron transfer in the E295 mutants. The ionizing group revealed must be from some partial process of the second electron transfer step, but could include either the proton exit or the electron transfer pathway or both. The most obvious candidate for the dissociable species is the substrate semiquinone, undergoing dissociation from the neutral to the anionic form. Cape et al. [30] showed that the SQ detected in wildtype was in the anionic form. This was based on comparison of line width and g-value of the Qo-site SQ to species generated chemically, and on ENDOR measurements of proton couplings. Our own spectra for SQ in E295W showed a similar g-value, but the quality of the signal means that we cannot claim strong support for an anionic species on this basis alone, and we have not yet generated samples appropriate for high resolution studies which would provide a definitive assignment. Other possible candidates are protein side chains, but the structures show E295 (lost in the mutants), and His-152 (the ligand from ISP) as the only dissociable groups within 7 Å of the stigmatellin ring (in PDB 2qjy). E295 mutations with Glu replaced by Gly, Trp, or Gln do not have pKs, and for Lys the pK would likely be higher, but all showed titrations with the same pK ~8.5. Of the residues in the Zn-binding site in Rhodobacter bc1 complexes [52], suggested to be associated with proton exit, only E295 could directly effect SQ by dissociation, since His-276 at 7.98 Å, Asp-278 at 10.11 Å, are too distant to act as ligands, and Asn-279 is not dissociable. Although we cannot rule out indirect effects from this set, the site is not conserved between mitochondrial and bacterial complexes. Since the profile of pH dependence in the more crippled strains was similar between yeast bc1 and the bacterial complex, it seems unlikely that residues peculiar to the bacteria site are responsible for the pK ~8.5.
4.7.2. The pathway inhibited in E295 mutants
In experiments with the isolated complex, a significant driving force would be attained within 50 ms on mixing with QH2. In the kinetic experiments using a 6-flash groups, the full driving force (ΔE′ ~800 mV) is established by 100 ms; equilibration of the high-potential chain with SQ/QH2 couple should occur on this timescale in the E295 mutants. However, the rate of electron flow was still markedly inhibited, despite the high occupancy of SQ. The inhibition is clearly after formation of SQ, and before electron transfer to heme bL. Although electrons and protons likely follow separate pathways in the overall process, both species have to be removed for net forward flux, and the two fluxes are likely closely coupled. In which pathway is the inhibition occurring in the E295 mutants? In the mechanism proposed for the wildtype, Q.- is the immediate electron donor, and the species detected in [30] was unambiguously this anionic form. In both wildtype and E295W, the driving force in our experiments was initially the same, and generated a similar occupancy. The higher value in E295W could be trivially explained, as discussed in section 4.9.1 (see the model kinetics in Fig. 9F), using the same equilibrium constant. In that case, the same thermodynamic parameters would have to pertain to the SQ species (see section 4.8.2 for further discussion). If the species in E295W is also anionic, then the increase in rate at high pH would be simply explained by the increase in occupancy of this form, and the electron transfer rate would not be limited by deprotonation. Inhibition would then have to be attributed to some other process after formation of Q.-. How does this relate to the mechanism in wildtype? The SQ is likely formed in the distal domain and the anionic form is the substrate for electron transfer, so, if SQ was anionic in both cases, the rate constant from the distal domain derived from the inhibited rate in mutants would be appropriate for electron transfer to heme bL from this domain in wildtype. If so, some additional process has then to be invoked to account for the uninhibited flux (at much lower SQ occupancy) in wildtype. Movement of SQ in the site [24] seems the most natural candidate, with inhibition of movement as the consequence of mutation. We favor this as the most parsimonious explanation. However, we recognize that it must remain hypothetical until the pK values and dissociation state of the SQ species in all the E295 mutants is resolved.
Figure 9. Simulated kinetic curves on addition of QH2 to the oxidized bc1 complex.
A. Kinetics in the first turnover to show the effect on turnover of slowing the diffusion of SQ from distal to proximal domains of the Qo-site by changing kdiff.
B. Kinetics on reduction by adding 200-fold excess of QH2 to the oxidized complex in the absence of additional acceptor. The reduction of heme bL reflects a second turnover driven by the excess QH2.
C. Occupancy of different states in which the SQ was significantly populated during the first turnover.
D. Kinetics in the presence of 6-fold excess of acceptor for the high-potential chain. Heme bL becomes reduced only after heme bH, and high occupancy of SQ lags reduction of heme bL. The right-hand scale is appropriate for the dashed trace, amplified to show SQ kinetics.
E. Kinetics to show the dependence of heme bL reduction (second turnover) on SQ mobility.
F. Kinetics of SQ occupancy with different degrees of inhibition of SQ diffusion. As diffusion becomes severely limited, the SQ occupancy builds up during the first turnover, and then relaxes to the equilibrium value as electrons pass through to the b-heme chain. Note that, apart from a slight increase in occupancy during the first two turnovers (*), the SQ accumulating during the third partial turnover has the same kinetics over the range of values for kdiff between 108 and 106s−1 (c.f. frame D).
If the SQ is anionic, this need not imply that deprotonation and proton exit were uninhibited. These processes could be slowed by orders of magnitude, and still be fast enough to populate the Q.- state so long as the rate of electron transfer was slower still. However, catalysis of deprotonation by E295 in the wildtype would be replaced in the mutants by a simpler pH dependence and proton equilibration with the external aqueous phase by less efficient pathways. If SQ movement was only partially inhibited in other E295 mutants, proton exit could be the controlling process. In the hypothesis, E295 has functions in both pathways, inhibition of either would slow both, and the degree to which either path was inhibited could be similar.
4.8 Scenarios for gating
Osyczka at al. [44] had argued persuasively that the Q-cycle would be decoupled unless some mechanism prevented short circuit reactions from occurring, but suggested that “only by eliminating the SQ intermediate in a concerted reaction, or by introducing double gating of a SQ intermediate…” could the mechanism be saved. Since the involvement of SQ seemed well established, we have preferred the second option.
4.8.1. Empirical rate constants for bypass processes
Muller et al. [77] suggested plausible bypass reactions and subsequent discussion [cf. 9, 44] has been framed in this context. Our determination of SQ occupancy allows calculation of empirical values for rate constants for these bypass reactions. The main reactions of interest are: (i) electron transfer from SQ to O2 or the high potential chain; and (ii) the reduction of SQ by heme bL− [77] (reduction of Q by the reversal of the normal forward reaction is not a bypass process). Since in the more crippled E295 mutants, rates for bypass reactions are similar to those for the bifurcated reaction, a value can be derived from the parameters determined above: vHP = kHP [SQ]·[2nd reactant] ~5 s−1, and vLP = kLP[SQ·bL−] ~5 s−1. For (i), under aerobic conditions, since the rate of SO formation saturates at ~10−4 M O2 [77], the value for kHP would be in the range ~107 M−1 s−1. In the absence of O2, the local activity of the second reactant (ISPox in an appropriate configuration), is unknown, so speculation must be limited. For (ii) with heme bL fully reduced, kLP[SQ] would have to be ~102 s−1, with kLP ~103 s−1 at [SQ] of 0.1, much lower than the value from distance assumed in the stochastic models (101,102,121). Reduction of either QH. or Q.- to QH2 would necessitate uptake of one or two H+, so it is clearly not a simple electron transfer, and it is unlikely that the distance dependent value for kLP would be appropriate.
4.8.2. Gating mechanisms
We suggested that bypass reactions of type (ii) could be averted by minimizing electron transfer between SQ and heme bL when the heme is reduced, and we proposed a coulombic mechanism to account for gating [9]. Likely, the electron and proton follow different paths, and Q.-, the E295 and heme propionate carboxylates, and the heme itself, all change their charge at different stages of the reaction, so they are all coulombic ‘dancers’ in this ballet. Given the pH dependence of Em for heme bL, the oxidation of QH. by heme bL would overall be a neutral process. Since charge is conserved, separation of paths for electron and proton transfer present possibilities for electrostatic interaction during oxidation of QH., and offer considerable scope for speculation. Any such mechanism must depend on details of the electrostatic profile around the reactants, and on the changes in charge of species during the overall electron/proton transfer reaction. The ferriheme bL carries a formal positive charge, which if uncompensated would provide an electrostatic attractant for the mobile Q.- postulated in our mechanism, thereby facilitating movement to the proximal domain. In this case, the ferroheme might be neutral, and the coulombic repulsion of the gating mechanism previously suggested [9] would then be inappropriate. On the other hand, the low potential of heme bL suggests a local environment in which the protein provides a compensating negative field. If this were so, the ferriheme state might be neutral, and the mechanism previously discussed [9] would remain attractive. However, although a single electron charge is transferred, it could well be to an oxidized heme of partial charge δ+, resulting in a charge (1-δ)− after transfer of the electron; then perhaps the most attractive scenario would involve a mix of these two mechanisms, and both attractive and repulsive effects would come into play at different times during the evolution of the process. The scheme in Fig. 8 sketches such a scenario (see legend for explanation).
Figure 8. Scheme to show possible mechanistic features of the Qo-site ballet.
a. Before the 1st electron transfer, the ES1-complex forms in the distal volume of the Qo-site, stabilized by a strong H-bond to His-161 of ISPox. E295 may H-bond weakly to QH2. Heme bL is in the oxidized form (with local field of δ+ charge). An electron and H+ are delivered to ISPox to form ISPH (1). Immediately after the 1st electron transfer, the ISPH rotates away (2) to deliver its electron to cyt c1 and release the H+ to the P-phase.
b. This leaves the neutral SQ (QH.), H-bonded to E295 carboxylate in the ES2-complex. Heme bL is still oxidized, and available as acceptor for the electron from QH. (or Q.-), but the low occupancy, and low value for k2d (~103 s−1) means the rate is slow.
c. The H+ is transferred to the E295 carboxylate, to form the SQ anion (Q.-), and E295 (3), now in carboxylic form, rotates to open up the proximal volume. The sidechain makes a H-bond to the water chain leading to the P-side aqueous phase. The δ+ field around heme bL lowers the pK, allowing release of the H+ (4), which transfers down the water chain, leaving E295 in the carboxylate form. The δ+ field also attracts Q.- (5), which migrates closer to heme bL, increasing the rate constant (k2p ~109 s−1).
d. The electron from Q.- transfers rapidly to heme bL (6), and the quinone is free to migrate back to the distal volume, and exit the site (7). Reduction of heme bL changes the charge by 1, and the field changes transiently to (1-δ)−.
e. The field is felt by the carboxylate of E295 (8), and the coulombic force flips it back to its initial position.
f. If heme bH is oxidized, the electron is transferred (9), and the field associated with heme bL returns to its initial δ+ charge. The site is now vacant, and available to accept a QH2 for the next turnover. If heme bL remains reduced (after the second turnover), the (1-δ)− field serves to repel the Q.- formed after the next cycle (reactions (1) – (3) representing a partial third turnover).
4.8.3. Proton exit pathways
For gating mechanisms associated with coulombic interactions between the charged participants, all the dancers have to be considered. In this context, the relative pK values governing the proton pathway between QH., E295, and the aqueous phase must also be considered. If the pK at ~8.5 measured in the E295 mutants does reflect dissociation of QH., then two general cases might be argued:
The pK ~8.5 measured in the more crippled E295 mutants might reflect dissociation of QH. appropriate only to the case in which the carboxylate function is missing. If this were the case, the working pK of QH. in the wildtype might be much lower, with a favorable gradient for H+ transfer along a pathway in which QH. and E295 have pK values close to those in aqueous environments. The fact that the species measured by Cape et al. [30] in wild type at pH 8.0 was the anionic form would favor this interpretation.
The pK ~8.5 might also determine the dissociation constant for QH. in wildtype. If so, the glutamate acceptor group might be expected to have a pK similar to this or higher. In cytochrome aa3, the glutamate at the terminus of the D-channel (water chain on the N-side of the protein) is thought to have a functional pK of ~9.2 [106, 107]. Since E295 in the bc1 complex occupies a similar environment, the scenario in cytochrome aa3 provides an example that might apply to the pK here.
Similar considerations apply to proton exit. Since water chains conduct protons freely, they must approach electrochemical equilibrium with the aqueous phase to which they are connected. At equilibrium, the activity felt within the protein at the P-side terminus must be that of the aqueous P-phase, but in flux, a difference in electrochemical potential along the chain would be present, though likely minimal. In either case, Mitchell’s proton-well effect [108] would be in play, and the distribution of H+ along the path would be determine through fractional contribution of Δψ to the local profile. The proton exit path is better characterized in cyt aa3; both there and in bc1 complex, the proton has eventually to be passed to the P-phase at pH ≤7.0, necessitating delivery from a group with an effective pK lower than this. The pathway in both cases involves a water chain that is also in equilibrium with a heme propionate, which might represent the terminus. It seems possible that coordination of proton and electron transfer, and the redox-linked changes in pK of the heme propionates (cf. [56, 109–112]) would be important in gating both processes. Clearly, mutagenesis of residues along the path from the Qo-site to the aqueous phase, in association with a detailed QM/MD simulation of the changes in electrostatic profile during catalysis, will be instructive, and such studies are in progress (Wilson, C., Harrison, C., Schulten, K. and Crofts, A.R., unpublished).
It is likely that the empirical rate constants would pertain to processes that are gated [44], but details of mechanisms remain to be determined. For the high potential chain, several labs have discussed possible gating mechanisms in terms of conformations of the protein that change with changes in occupancy of the different domains of the Qo-site [104, 113, 114], nicely reviewed by Berry and Huang [115]. In particular, access to the Qo-site occupant from the P-phase (for either O2 or ISP) is likely modulated by closure of the access port through which ISP contacts occupants. Tyr-302 is the putative “trap-door” for closure of the Qo-site access port [104], and is also involved in stabilization of the extrinsic domain of ISP at the Qo-site through H-bonding to the backbone =O of Cys-151 of ISP. Mutation of this residue slows rates for the normal forward reaction [104, 116, 117], and can dramatically increase rates for SO generation [114, 118], so likely this is an important player.
For the low potential chain, the choice of gating mechanism depends on choice of starting point. If a single rate constant for electron transfer pertains, the choices are limited. Osyczka et al. [44], having discounted any role for E295, suggested control by coulombic effects on dipolar orientation of waters to gate H+ pathways. With the necessity for SQ movement, and the essential roles for E295 envisaged here, the scope is greatly enlarged, with the most obvious mechanisms those proposed, - coulombic forces would keep Q.- in the distal domain if heme bL− had a net negative charge, so that only the slow rate constants would be in effect. Similar forces would also modulate displacements of the carboxylate form of E295. In any case, reversal of the reaction would require coordination of electron and proton transfer, with many possibilities for gating, as discussed above.
4.9 A kinetic model for the Qo-site reaction
From the considerations above, a strong case can be made for a mechanism that depends on movement of SQ in the Qo-site to facilitate the second electron transfer. Covian et al. [31] had suggested that such a model was untenable. Their main criticism was that in the previous work [9] we had failed to take account of plausible values for diffusion coefficient of SQ in the Qo-site. We agree that the diffusional rate constant was not properly considered in that study, but we disagree with the more general conclusion they reached, that all models involving the movement of SQ in the site are therefore untenable. In our previous discussions, we had assumed that occupancy of the SQ would be maintained as low as possible so as to minimize the probability of bypass reactions, and used Moser-Dutton values for of 4 × 106 s−1 from the distal domain and 4 × 109s−1 from the proximal domain as primary constraints. If the movement of SQd to SQp was sufficiently rapid, the intrinsic rate would be determined by electron transfer from the proximal domain, allowing a very low occupancy. The implicit ad hoc assumption, that the rate constant for diffusion was faster than from the proximal domain, was not realistic. The original model could be readily extended to demonstrate that reduction of heme b via diffusion of SQ in the site was realistic, but it had omitted a proper representation of other essential features, in particular the competition between the forward and reverse reactions [31]. The model shown here is expanded to include all redox centers of the complex, and established partial processes [8, 9]. The main considerations are SQ occupancy, now determined, and the three pairs of forward and reverse rate constants, - for electron transfer from the distal and proximal domains, and for the diffusional step. For the first electron transfer, partial processes included are formation of ES-complex, the probability for the H+ to be in the configuration needed for electron transfer (the Brønsted barrier for formation of the active state), and the electron transfer step. Values for forward rate constants are taken from the literature or derived from well-established parameters (see Notes to Table 1), and reverse rate constants were adjusted so as to provide values for the overall equilibrium constant consistent with existing data. The value of k2d ~103 s−1 for oxidation of SQ from the distal domain is that derived above from the SQ occupancy and measured rates. We have assumed that this value for rate constant would also be appropriate if Q.- was constrained to the distal domain in wildtype. In this case, the observed rate could be explained only by some additional process increasing the rate constant, simulated here by movement of SQ closer to heme bL. The diffusional step for SQ has been explicitly modeled. The value for diffusional rate constant, 107 <kdiff≤108 s−1, was derived from 〈x2〉 = 2Dt, a 1-D diffusion distance of 5.0 Å, and the diffusion coefficient for ubiquinone-10 [119] or plastoquinone [120] in the membrane. Values measured for D have been in the range 10−7 to 10−9 cm2s−1, depending on protein crowding, and are similar to the value for the diffusion coefficient of phospholipids in membranes (~10−8 cm2s−1). Since diffusion of Q.- would involve an open volume close to heme bL, a value at the high end of the plausible range might be appropriate. In the simulation, SQ occupancy at levels comparable to those observed experimentally requires oxidation of ISPH and ferroheme c1 by an external acceptor to pull the reaction over beyond the first turnover (see discussion of equilibria in section 4.2). The occupancy found depends on the equilibrium constant for this reaction, and hence the Em of the acceptor. The value used in the model, giving K ~ 100, is appropriate for the complex in situ (with RC or cyt oxidase as acceptor). However, as might be expected, this leads to occupancy when the oxidation of SQ is limited (for example by eliminating the diffusional step) that is, at least transiently, higher (>0.2 SQ/site) than that measured using the isolated complex and horse heart cyt c as acceptor. It should also be noted that, because of the competition from back-reaction, the range of values for kdiff is more severely constrained than when the model of [31] was extended (see SI).
Table 1.
Partial processes and kinetic parameters for Qo-site model
| Reaction equation | Rate constants | kforward | kreverse | Keq | ||
|---|---|---|---|---|---|---|
| EFeS.c1.bL.bH + QH2 Ý EFeS.c1.bL.bH .QH2 | konQH | koff | 500×[QH2] | 5×103 | 10 | a |
| EFeS.c1.bL.bH .QH2 Ý EFeS.c1.bL.bH .QH-H | kproton | k−proton | 107 | 1012 | 10−5 | b |
| EFeS.c1.bL.bH .QH-H Ý EFeS−.c1.bL.bH .SQ(d) | k1 | k−1 | 1.65×108 | 3.25×105 | 510 | c |
| EFeS−.c1.bL.bH .SQ(d) Ý EFeS.c1−.bL.bH .SQ(d) | k5 | k−5 | 108 | 3×108 | 0.33 | d |
| EFeS.c1−.bL.bH .SQ(d) + Aox Ý EFeS.c1.bL.bH .SQ(d) + Ared | k3 | k−3 | 107 | 105 | 100 | e |
| EFeS.c1.bL.bH .SQ(d) Ý EFeS.c1.bL−.bH .Q(d) | k2d | k−2d | 1.2×103 | 0.192 | 6250 | f |
| EFeS.c1.bL−.bH .Q(d) Ý EFeS.c1.bL.bH − .Q(d) | k4 | k−4 | 106 | 6250 | 160 | g |
| EFeS.c1.bL.bH .SQ(d) Ý EFeS.c1.bL.bH .SQ(p) | kdiff | k−diff | 107 | 107 | 1 | h |
| EFeS.c1.b− L.b−H .SQ(d) Ý EFeS.c1.b− L.b−H .SQ(p) | kdiff3 | k−diff3 | 106 | 108 | 0.01 | h |
| EFeS.c1.bL.bH .SQ(p) Ý EFeS.c1.bL−.bH .Q(p) | k2p | k−2p | 4.3×109 | 6.9×105 | 6250 | i |
| EFeS.c1.bL−.bH .Q(p) Ý EFeS.c1.bL.bH− .Q(p) | k4 | k−4 | 106 | 6250 | 160 | g |
| EFeS.c1.bL.bH − .Q(p) Ý EFeS.c1.bL.bH− .Q(d) | kdiff | k−diff | 107 | 107 | 1 | h |
| EFeS.c1.bL.bH− .Q(d) Ý EFeS.c1.bL.bH − + Q | koff | konQ | 5×103 | 50×[Q] | 1 | j |
| EFeS.c1.bL.bH − + QH2 Ý EFeS.c1.bL.bH−.QH2 | koff | konQH | 500×[QH2] | 5×103 | 10 | k |
Notes:
The model represents the essential processes of the Qo-site reaction, constrained by antimycin to prevent the Qi-site reaction so that flux out of the b-heme chain is inhibited. The full complement of redox centers is included, and an additional acceptor (by default in 6-fold excess over the complex) of Em,7 420 mV to simulate the driving force available from the rest of the chain. For simplicity, it is assumed that all components retain the Em,7 values determined by redox titration. The reactions shown are appropriate to the first turnover of the Qo-site. The model allows simulation of the second turnover, and uses the same rate constants for similar processes (see Supplementary Information for details).
The value for konQH shown is that suggested in [ 31], with the initial [QH2] given as 200 (no units), to give an apparent first-order rate constant of 5×104 s−1. In chromatophores at saturating QH2, the exchange of Q and QH2 has no appreciable contribution to the kinetics [72], indicating that these reactions are all rapid compared to the rate limiting step. The second-order rate constant determined in situ was ~2 × 105 M−1s−1 [74], estimated from the observed rate at ~10 mM QH2 in the membrane. The diffusion-limited rate constant would be considerably higher. The displacement of the apparent Em due to formation of the ES1-complex shows a tighter binding of QH2 than Q by a factor ~10.
Rates for H+ exchange along H-bonds have been variously estimated on the range 1011 to 1013s−1 [123–125]. We have used a value of 1012 s−1 for k−proton, the more rapid of the two rate constants, but any choice in the range shown experimentally would give the same kinetics overall. The ratio of rate constants gives the probability for finding the H+ in a position favorable for electron transfer (the equilibrium constant for distribution along the H-bond), derived from the difference in pK values between donor (the −OH of QH2, pK >11.5) and acceptor groups (the Nε of His-152 (His-161 in mitochondrial sequences) of the oxidized ISP, with pKox1 ~7.6). At saturating QH2, electron transfer occurs from the ES1-complex, and the value for pKox1 would be less than in the isolated ISP because of the binding free-energy [8]. A value of 6.5 is used in the simulation (a ΔpK of 105).
The forward rate constant is that for electron transfer from the weakly populated configuration in which the H+ is close to the histidine Nε of His-152 (occupancy 10−5 used here, see (b)). The value for rate constant needed is therefore 105-fold greater than the observed value. The value given here is that from a Marcus analysis using the distance dependence of Moser and colleagues [100], and λ = 0.935, distance 7.0 Å, and driving force ΔGo/F = −0.16 V appropriate to the value for Keq shown. The reverse rate constant is chosen to give a SQ occupancy in the range shown by experiment (~0.05 [30] and this work), after taking account of the occupancy of the intermediate state. It is assumed that the reaction is reversible only in the ES1-complex configuration. After electron transfer, the H-bond to SQ would break, releasing the reduced ISP, which would rapidly rotate away from the ES-complex configuration to passes its electron to heme c1.
Removal of ISPH from interaction with Qo-site, and oxidation to ISPox. The forward rate constant given is that for dissociation and movement of ISPH, since this would release SQ for further reaction. The equilibrium constant is that for oxidation of ISPH by cyt c1, which is weakly unfavorable at pH 7.0.
Second-order rate constant to represent oxidation of cyt c1 by a pool of high-potential acceptor.
The forward rate constant for oxidation of SQ in the domain distal from heme bL. The value is that needed to match the observed rate in strains mutated at Glu-295 (v~40–80 QH2/bc1/s at pH 8.5 – 9), with a SQ occupancy ~0.05 [30], using the relation, v = k(occupancy).
The rate constant for electron transfer from heme bL to heme bH. The forward rate constant was calculated using Marcus/Moser-Dutton analysis from the distance given by modeling. The reverse rate constant was chosen so that the equilibrium constant matches that calculated from Em values. The value is similar to that determined from the rate of oxidation of heme bH induced by generation of a membrane potential [126]. The kinetics of heme bH reduction were not significantly changed by varying the value for k4 over the range 104 to 106 s−1, though a transient reduction of heme bL (maximal occupancy 0.08) was seen when the lower value was used.
An approximate value for kdiff is given by k ≈ 1/t = 2D /<x2>. Using x = 5×10−8 cm, D in the range 10−9 to 10−7 cm2 s−1 [119, 120], we find 0.8× 106 <kdiff <0.8 × 108 s−1, assuming a 1- dimensional diffusion path, and diffusion in both directions from a starting position; if movement was constrained to the direction closer to heme bL, the values would scale up. The value used in the simulation could be varied over the range 108 – 107 s−1 without changing the overall rate of reaction. In all the simulations shown in which heme bL was available in the oxidized form, the same value was used in forward and reverse directions. The limited range could be extended by applying a bias to the diffusional process. For example, bias would increase kdiff if there was a coulombic driving force for displacement of Q.- by a positive field around heme bL. Similarly, the occupancy of the distal domain by Q.- could be constrained if k−diff was increased by a negative field around ferroheme bL. In the simulation, we implemented such an effect under conditions in which both b-hemes were reduced, but the only outcome affected was the distribution between SQd and SQp states. In the mechanism outlined in Fig. 8 we speculate that the change in charge on reduction would switch the field from +ve to −ve, but have not otherwise implemented that feature in the present models. For the neutral Q species, the same value for kdiff was used in both reaction directions. The value for kdiff was varied in the plots of Figs. 7–9, as indicated, to simulate impaired oxidation of SQ by mutation of Glu-295.
The forward rate constant for oxidation of SQ in the domain proximal to heme bL. The value was calculated using the Moser-Dutton distance parameter, the distance to heme bL shown by myxothiazol-containing structures, and the Marcus exponential term [8]. Both here, and in estimation of the complementary parameter for the distal domain, the reverse rate constant was calculated so as to be consistent with the equilibrium for the overall reaction and the other values for equilibrium constants used in the simulation.
The values chosen reflect a 10-fold tighter binding for QH2 than Q at the catalytic site. As noted in a), the actual value is conservative.
Because of the 25 Å distance, and the neutral charged state of the binding species, it was assumed that the redox state of heme bH did not affect the binding of Q or QH2 at the Qo-site.
Reactions, rate constants and values for all parameters used are shown in Table 1, and Dynafit scripts are provided in the supplementary materials. Three kinetic models are simulated using the same set of parameters: (i) a single turnover of the Qo-site following addition of QH2 to the oxidized complex inhibited by antimycin, allowing exploration of occupancy of SQ, and the dependence of the rate of heme bH reduction on the diffusional step; (ii) as (i), but with equilibria for additional turnovers to allow the oxidized complex to come to equilibrium with the Q-pool; and (iii) as (ii) but with steps to allow the high potential chain to equilibrate with an acceptor pool. We note that, although all equilibrium constants used are natural, the rate constants for one process, the oxidation of cyt c1 by added acceptor (k3 and k−3) are higher than justified by observed rates, and therefore unnatural. Lowering these values led to slowed kinetics, especially for the second turnover; we tentatively ascribe this discrepancy to gating in the high potential chain that is not included in the current model. The scripts for these models, and an early preliminary model based on [31], are available in the supplementary materials.
4.9.1. Simulation outcome
As shown in Fig. 9A, the kinetics modeled depended strongly on the value chosen for the diffusional rate constant. Rates of heme bH reduction associated with oxidation of the first QH2 reflected the limiting first electron transfer as long as kdiff was in the range >107 s−1, - well within plausible estimates. Kinetics of ISP and cyt c1 reduction summed to follow the kinetics of heme bH reduction in the first turnover, but individual amplitudes (and initial rates) reflected the equilibrium constant between ISP and heme c1. Note that whenever acceptors were available in both chains, the SQ occupancies (for example, with kdiff = 107 s−1, [SQd] was transiently ~1.5·10−4) were below the detection limits currently accessible using EPR (Fig. 9C). The model therefore accounts for the data of Zhu et al. [38], but in the context of our model. The SQp occupancy (~2.5·10−7) was in the range of the minimal value we assumed in previous discussions [9, 10]. When the simulation was allowed to continue into a second turnover, a slow reduction of heme bL occurred, driven by the large excess of QH2, but little additional SQ (~2·10−5) accumulated (Fig. 9B, C). As seen experimentally, reduction of heme bL developed only as reduction of heme bH reached completion, and was slow unless additional acceptor was present. Only when an excess of acceptor for the high potential chain was included did the rates for heme bL reduction approach experimental rates; SQ occupancy lagged heme bL reduction, and approach the experimental range only with the heme substantially reduced (Fig. 9D). If diffusion of SQ was impeded, a high transient level of SQ was detected during the first turnover, but if SQ was able to move rapidly, significant levels developed only during the second turnover.
The changes in kinetics of SQ occupancy on changing the values of kdiff would model those expected between wild-type and the E295W mutant. In the experimental context, the conditions are the same, but the mechanism by which SQ removal is inhibited is different. However, in each case, after inhibition has set in, the SQ has to reach an equilibrium poise consistent with that of other reactants in the reaction. In the case of the antimycin-inhibited wild-type, two quinol equivalents are consumed (in reduction of the two b-hemes) before the inhibition leading to substantial accumulation of SQ sets in; after this delay the SQ would approach equilibrium with other components of the overall reaction. In the mutant, the removal of the SQ is inhibited from the outset, so that the SQ accumulates rapidly on oxidation of the first quinol (on activation via oxidation of the high-potential chain in chromatophores), in transient equilibrium with the reactants of the first electron transfer, but the poise of the second electron transfer reaches equilibrium on a much slower time scale, due to the inhibited delivery of electrons to heme bL. These effects can be seen in the kinetics modeled in Fig. 9F. The consequence of the different time courses is that the occupancy of SQ in the two strains at any particular time will be different, but will approach the same value at longer times. The occupancy in the wild-type approaches equilibrium from below, and that in E295W from above, so that occupancy in the former would be lower than in the latter at any time prior to that at which both had reached equilibrium, consistent with the difference observed experimentally.
When a large pool of acceptor was provided, and an additional reaction step for oxidation of the SQ by O2 was included, the steady-state flux of SO generation could also be modeled, with rate dependent on choice of rate constant (not shown).
The kinetic limitation in the E295 mutants could in principle come from any partial process in the second electron transfer, including the pathway for proton exit, but was modeled here simply by decreasing the rate constant for the diffusional state. Software constraints limit the complexity of the kinetic model, and, apart from provisions for testing coulombic bias, we did not attempt to extend it to include separate partial processes for gating mechanisms, or for the proton exit pathway via E295, which we assume to be rapid in wildtype compared to the rate limiting step.
4.9.2. Inhibition by antimycin
Ransac et al. [102, 121], had pointed out that their stochastic kinetic models allowed the equilibrations that open up the bypass processes discussed by Osyzcka et al. [44], and that as a consequence, inhibition by antimycin could not be modeled without ad hoc additions to the model. Instead, the bifurcated reaction was essentially decoupled, as also emphasized by Moser et al. [122]. In their choice of rate constants, each of these groups had assumed that the second electron transfer involved a single rate constant for SQ oxidation calculated from distance, with SQ constrained to the location of its generation, - the distal domain in [102], some intermediate location in [44]. Kim et al. [101] in an exercise using similar constraints reached a similar conclusion. It might be expected from this that in the more severely crippled E295 mutant strains, a marked decoupling would also be observed. However, the bypass rates in these strains were in the same range as in antimycin inhibited wildtype, which showed almost complete inhibition. As noted above, with a known SQ occupancy under conditions in which bypass reactions occur at a measured rate, empirical values for rate constants of particular bypass processes (reduction of SQ by heme bL −, reduction of O2 or ISPox by SQ) can be calculated, and applied in modeling rates for the states in which SQ accumulates. In the present model, we have not included these processes because we do not know what additional partial processes are involved, but the empirical rate constants discussed would lead to the strongly inhibited rates observed. By introducing the diffusional step in the second electron transfer, we open the possibility of the gating envisaged in [44]. Control of the bifurcated reaction to minimize reduction of SQ by heme bL− might involve a restriction of SQ to the distal domain by the coulombic mechanism previously proposed, and we have included such as a token. However, in the absence of these additional processes, it has no effect. The model simulates the essential features of the forward Qo-site reaction, and provides a starting point for a more complete study in which further complexities associated with control and gating of the reaction in situ can be explored.
5. Conclusions
1) From the work establishing the role of ISPox [27, 62, 81], and from the changes in pH dependence compared to wildtype in ISP and E295 mutants, it is clear that the changed profile in the latter is not a direct consequence of loss of the glutamate sidechain, but reflects a change in the rate limiting process from the first to the second electron transfer. The pK at ~8.5 is due to dissociation of some other group to a form that favors electron transfer. A plausible candidate is the dissociation of the neutral SQ to the anionic form: QH. ⇌ Q.- + H+.
2) Contrary to previous conclusions [55, 56], mutation of E295 severely lowers the rate constant for oxidation of SQ by heme bL. However, the data do not allow an unambiguous distinction between roles in the electron and proton transfer. The pattern of Zn-inhibition is consistent with a role in proton exit [52]. Our original hypothesis [24] was driven by the rotational displacement of the sidechain needed to fulfill this role, clearly demonstrated by the changes in E295 configuration seen in structures with stigmatellin [19, 26] compared to those with myxothiazol (or other MOA-inhibitors) [26, 50]. The structures also showed occupancy of the volume proximal to heme bL by the MOA-class of inhibitors [26, 50]. As noted before [26], the effects seen when residues impinging on this volume are mutated, the differential sensitivity to the two classes of inhibitor, the slowing of electron transfer observed in these mutants, and the lack of effect on binding in the distal domain, strongly hint at a need for occupancy of the proximal domain by an intermediate state. In the absence of inhibitor, rotational displacement of the E295 sidechain would open the volume to SQ if the latter were mobile. The properties revealed by interactions with local nuclear magnets [30] are consistent with such a mobility, and our kinetic modeling demonstrates that, with these factors incorporated, movement of SQ in the site is plausible, and is sufficient to account for the high rates observed in wildtype. In the hypothesis tested, both the direct role in H+ exit, and the indirect role in facilitating electron transfer would be necessary, but choice of which is the step inhibited depends on the dissociation sate of SQ. If Q.- is the form of SQ accumulated in E295 mutants, then dissociation is not rate limiting, and inhibited movement in the site is the best explanation. We believe that in E295W the case is strong, but recognize that the balance could flip in the other direction in other mutants, and our current studies are aimed at resolving this uncertainty.
4) The occupancy measured here in E295W under conditions of favorable driving force, together with the constraint on mobility from the bulk of tryptophan, provides a plausible basis for calculation of a rate constant for oxidation of SQ from the distal domain. The value obtained is >1000-fold lower than that calculated for electron transfer over the distance from the distal domain to heme bL. Starting there, two explanations for the inhibition observed in E295 mutants seem plausible. i) If the SQ species detected in E295W is Q.-, the inhibition must be in some step after dissociation, likely movement of SQ. In that case, the rate constant calculated would be appropriate in the native complex and would exclude explanations for the rapid rates seen in uninhibited systems based on electron transfer from Q.- that remains in the distal domain. We believe the weight of evidence supports this explanation. ii) If the SQ species is QH., then dissociation could well be the inhibited step, the rate constant measured would pertain to that form, and a different value might pertain in the wildtype mechanism. As noted in the previous paragraph, the hypothesis is framed in the context of a straightforward test.
5) With the qualifications above, we can claim that the main features of the hypothesis sketched out a dozen years ago [24] are now supported by experimental data. The results are consistent with a plausible mechanism for this second step of the bifurcated reaction, and suggest a molecular ballet, with E295 as the prima ballerina, whose choreography is directed by coulombic interactions that implement control functions. In the mechanism we propose, and in contrast to previous proposals [56], the participation of E295 lowers energy barriers for the forward reaction as in the conventional view of catalysis, but raises them towards the bypass reactions by exploiting separate pathways for electron and proton, the spatial separation between the distal domain (at which SQ is generated) and the proximal domain (close enough to acceptor heme bL for rapid oxidation), the mobility of the SQ, and the potential for gating in the flexibility of conformation in the Qo-site, and a dance, frozen in the structures, whose step-by-step choreography remains to be explored.
Supplementary Material
Highlights.
In mutants at the -PEWY- glutamate, the second electron transfer becomes limiting.
With kcat~103 s−1, electron transfer from the distal domain is kinetically incompetent.
Bifurcated rate at the Qo-site is pH independent below 8, and increases with pK~8.5.
There is no marked pH-dependence of ES formation over the range <8 in mutants.
E295 catalyzes H+ exit and likely acts as a gate for semiquinone migration in the site.
Acknowledgements
We gratefully acknowledge supported for this work from NIH Grant R01 GM035438. We acknowledge support from NIH Grant R01 GM62954 to Dr. Sergei Dikanov for support of R. Burton, and for purchase of freeze-quench equipment. We thank Colin Wraight for useful discussions, including his suggestion of the mechanistic scenario for coulombic attraction of Q.- by ferriheme bL. We thank Drs. Mark Nilges and Amga Baldansuren for technical help and advice in use of the EPR facilities.
Abbreviations
- SQ
semiquinone (dissociation state unspecified). Occupancy is at the Qo-site unless otherwise specified
- QH.
neutral semiquinone
- Q.-
anionic semiquinone
- QH2
ubihydroquinone-10, ubiquinol, quinol
- Q
ubiquinone-10, quinone
- decyl-QH2
decyl-ubihydroquinone-0
- decyl-Q
decyl-ubiquinone-0
- ISP
Iron-sulfur protein
- SU IV
subunit IV
- ROS
reactive oxygen species
- SO
superoxide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Berry EA, Lee D-W, Huang L-S, Daldal F. Structural and mutational studies of the Cytochrome bc1 Complex. In: Hunter CN, Daldal F, Thurnauer MC, Beatty JT, editors. The Purple Phototrophic Bacteria. Dordrecht, The Netherlands: Springer; 2009. [Google Scholar]
- 2.Crofts AR. The cytochrome bc1 complex – function in the context of structure. Annu. Rev. Physiol. 2004:689–733. doi: 10.1146/annurev.physiol.66.032102.150251. [DOI] [PubMed] [Google Scholar]
- 3.Crofts AR, Meinhardt SW, Jones KR, Snozzi M. The role of the quinone pool in the cyclic electron-transfer chain of Rhodopseudomonas sphaeroides: A modified Q-cycle mechanism. Biochim. Biophys. Acta. 1983;723:202–218. doi: 10.1016/0005-2728(83)90120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell P. Proton motive redox mechanism of the cytochrome b-c1 complex in the respiratory chain: Proton motive ubiquinone cycle. FEBS Lett. 1975;56:1–6. doi: 10.1016/0014-5793(75)80098-6. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell P. Possible molecular mechanisms of the protonmotive function of cytochrome systems. J. Theor. Biol. 1976;62:327–367. doi: 10.1016/0022-5193(76)90124-7. [DOI] [PubMed] [Google Scholar]
- 6.Crofts AR. The Q-cycle, - a personal perspective. Photosynth. Res. 2004;80:223–243. doi: 10.1023/B:PRES.0000030444.52579.10. [DOI] [PubMed] [Google Scholar]
- 7.Hong SJ, Ugulava N, Guergova-Kuras M, Crofts AR. The energy landscape for ubihydroquinone oxidation at the Qo-site of the bc1 complex in Rhodobacter sphaeroides. J. Biol. Chem. 1999;274:33931–33944. doi: 10.1074/jbc.274.48.33931. [DOI] [PubMed] [Google Scholar]
- 8.Crofts AR. Proton-coupled electron transfer at the Qo-site of the bc1 complex controls the rate of ubihydroquinone oxidation. Biochim. Biophys. Acta. 2004;1655:77–92. doi: 10.1016/j.bbabio.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Crofts AR, Lhee S, Crofts SB, Cheng J, Rose S. Proton pumping in the bc1 complex: A new gating mechanism that prevents short circuits. Biochim. Biophys. Acta. 2006;1757:1019–1034. doi: 10.1016/j.bbabio.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Crofts AR, Holland JT, Victoria D, Kolling DR, Dikanov SA, Gilbreth R, Lhee S, Kuras R, Kuras MG. The Q-cycle reviewed: How well does a monomeric mechanism of the bc1 complex account for the function of a dimeric complex? Biochim. Biophys. Acta. 2008;1777:1001–1019. doi: 10.1016/j.bbabio.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denke E, Merbitz-Zahradnik T, Hatzfeld OM, Snyder CH, Link TA, Trumpower BL. Alteration of the midpoint potential and catalytic activity of the Rieske iron-sulfur protein by changes of amino acids forming hydrogen bonds to the iron-sulfur cluster. J. Biol. Chem. 1998;273:9085–9093. doi: 10.1074/jbc.273.15.9085. [DOI] [PubMed] [Google Scholar]
- 12.Schröter T, Hatzfeld OM, Gemeinhardt S, Korn M, Friedrich T, Ludwig B, Link T. Mutational analysis of residues forming hydrogen bonds in the Rieske [2Fe2S]. cluster of the cytochrome bc1 complex of Paracoccus denitrificans. Eur. J. Biochem. 1998;255:100–106. doi: 10.1046/j.1432-1327.1998.2550100.x. [DOI] [PubMed] [Google Scholar]
- 13.Lhee S, Kolling DR, Nair SK, Dikanov SA, Crofts AR. Modifications of protein environment of the [2Fe-2S]. cluster of the bc1 complex: Effects on the biophysical properties of the Rieske iron-sulfur protein and on the kinetics of the complex. J. Biol. Chem. 2009;285:9233–9248. doi: 10.1074/jbc.M109.043505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rottenberg H, Covian R, Trumpower BL. Membrane potential greatly enhances superoxide generation by the cytochrome bc1 complex reconstituted into phospholipid vesicles. J. Biol. Chem. 2009;284:19203–19210. doi: 10.1074/jbc.M109.017376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl. Acad. Sci. (USA) 1993;90:7915–7921. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harman D. Free radical theory of aging. In: Emerit I, Chance B, editors. Free Radicals and Aging. Birkhauser Verlag: Basel; 1992. [Google Scholar]
- 17.Muller FL, Lustgarten MS, Jang Y, Richardson A, Remmen HV. Trends in oxidative aging theories. Free Radical Biology & Medicine. 2007;43:477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 18.Xia D, Yu C-A, Kim H, Xia J-Z, Kachurin AM, Zhang L, Yu L, Deisenhofer J. Crystal structure of the sytochrome bc1 somplex from bovine heart mitochondria. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, Huang L-S, Shulmeister VM, Chi Y-I, Kim K-K, Hung L-W, Crofts AR, Berry EA, Kim S-H. Electron transfer by domain movement in cytochrome bc1. Nature (Lond.) 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 20.Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, Link TA, Ramaswamy S, Jap BK. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 21.Hunte C, Koepke J, Lange C, Roßmanith T, Michel H. Structure at 2.3 Å resolution of the cytochrome bc1 complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 2000;8:669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 22.Berry EA, Huang L-S, Saechao LK, Pon NG, Valkova-Valchanova M, Daldal F. X-ray structure of Rhodobacter capsulatus cytochrome bc1: comparison with its mitochondrial and chloroplast counterparts. Photosynth. Resc. 2004;81:251–275. doi: 10.1023/B:PRES.0000036888.18223.0e. [DOI] [PubMed] [Google Scholar]
- 23.Esser L, Elberry M, Zhou F, Yu C-A, Yu L, Xia D. Inhibitor complexed structures of the cytochrome bc1 complex from the photosynthetic bacterium Rhodobacter sphaeroides. J. Biol. Chem. 2008;283:2846–2857. doi: 10.1074/jbc.M708608200. [DOI] [PubMed] [Google Scholar]
- 24.Crofts AR, Hong SJ, Ugulava N, Barquera B, Gennis R, Guergova-Kuras M, Berry EA. Pathways for proton release during ubihydroquinone oxidation by the bc1 complex. Proc. Natl. Acad. Sci. (U.S.A.) 1999;96:10021–10026. doi: 10.1073/pnas.96.18.10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berry EA, Huang LS. Observations concerning the quinol oxidation site of the cytochrome bc1 complex. FEBS Lett. 2003;555:13–20. doi: 10.1016/s0014-5793(03)01099-8. [DOI] [PubMed] [Google Scholar]
- 26.Crofts AR, Barquera B, Gennis RB, Kuras R, Guergova-Kuras M, Berry EA. Mechanism of ubiquinol oxidation by the bc1 complex: the different domains of the quinol binding pocket, and their role in mechanism, and the binding of inhibitors. Biochemistry. 1999;38:15807–15826. doi: 10.1021/bi990962m. [DOI] [PubMed] [Google Scholar]
- 27.Guergova-Kuras M, Kuras R, Ugulava N, Hadad I, Crofts AR. Specific mutagenesis of the Rieske iron sulfur protein in Rhodobacter sphaeroides shows that both thermodynamic gradient and the pK of the oxidized form determine the rate of quinol oxidation by the bc1 complex. Biochemistry. 2000;39:7436–7444. doi: 10.1021/bi992491+. [DOI] [PubMed] [Google Scholar]
- 28.Hsueh K-L, Westler WM, Markley JL. NMR investigations of the Rieske protein from Thermus thermophilus support a coupled proton and electron transfer mechanism. J. Am. Chem. Soc. 2010;132:7908–7918. doi: 10.1021/ja1026387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zu Y, Couture MM-J, Kolling DRJ, Crofts AR, Eltis LD, Fee JA, Hirst J. The reduction potentials of Rieske clusters: the importance of the coupling between oxidation state and histidine protonation state. Biochemistry. 2003;42:12400–12408. doi: 10.1021/bi0350957. [DOI] [PubMed] [Google Scholar]
- 30.Cape JL, Bowman MK, Kramer DM. A semiquinone intermediate generated at the Qo site of the cytochrome bc1 complex: Importance for the Q-cycle and superoxide production. Proc. Natl. Acad. Sci. (U.S.A.) 2007;104:7887–7892. doi: 10.1073/pnas.0702621104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Covián R, Trumpower BL. The rate-limiting step in the cytochrome bc1 complex is not changed by inhibition of cytochrome b-dependent deprotonation: implications for the mechanism of ubiquinol oxidation at center P of the bc1 complex. J. Biol. Chem. 2009;284:14359–14367. doi: 10.1074/jbc.M109.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Osyczka A, Dutton PL, Moser CC. Exposing the complex III Qo semiquinone radical. Biochim. Biophys. Acta. 2007;1767:883–887. doi: 10.1016/j.bbabio.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crofts AR, Wang Z. How rapid are the internal reactions of the ubiquinol:cytochrome c2 oxidoreductase? Photosynth. Res. 1989;22:69–87. doi: 10.1007/BF00114768. [DOI] [PubMed] [Google Scholar]
- 34.Roberts JA, Kirby JP, Wall ST, Nocera DG. Electron transfer within ruthenium(II) polypyridyl-(salt bridge)-dimethylaniline acceptor-donor complexes. Inorg. Chim. Acta. 1997;263:395–405. [Google Scholar]
- 35.Snozzi M, Crofts AR. Electron transport in chromatophores from Rhodopseudomonas sphaeroides GA fused with liposomes. Biochim. Biophys. Acta. 1984;766:451–463. doi: 10.1016/0005-2728(84)90261-5. [DOI] [PubMed] [Google Scholar]
- 36.Crofts AR, Holland JT, Victoria D, Kolling DR, Dikanov SA, Gilbreth R, Lhee S, Kuras R, Kuras MG. The Q-cycle reviewed: How well does a monomeric mechanism of the bc1 complex account for the function of a dimeric complex? Biochim. Biophys. Acta. 2008;1777:1001–1019. doi: 10.1016/j.bbabio.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Chobot SE, Osyczka A, Wraight CA, Dutton PL, Moser CC. Quinone and non-quinone redox couples in Complex III. J. Bioenerg. Biomembr. 2008;40:493–499. doi: 10.1007/s10863-008-9174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu J, Egawa T, Yeh SR, Yu L, Y CA. Simultaneous reduction of iron-sulfur protein and cytochrome bL during ubiquinol oxidation in cytochrome bc1 complex. Proc Natl Acad Sci USA. 2007;104:4864–4869. doi: 10.1073/pnas.0607812104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim H, Xia D, Yu CA, Xia JZ, Kachurin AM, Zhang L, Yu L, Deisenhofer J. Inhibitor binding changes domain mobility in the iron-sulfur protein of the mitochondrial bc1 complex from bovine heart. Proc. Natl. Acad. Sci. USA. 1998;95:8026–8033. doi: 10.1073/pnas.95.14.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunte C, Palsdottir H, Trumpower BL. Protonmotive pathways and mechanisms in the cytochrome bc1 complex. FEBS Lett. 2003;545:39–46. doi: 10.1016/s0014-5793(03)00391-0. [DOI] [PubMed] [Google Scholar]
- 41.Osyczka A, Moser CC, Daldal F, Dutton PL. Reversible redox energy coupling in electron transfer chains. Nature. 2004;427:607–612. doi: 10.1038/nature02242. [DOI] [PubMed] [Google Scholar]
- 42.Bartoschek S, Johansson M, Geierstanger BH, Okun JG, Lancaster CRD, Humpfer E, Yu L, Yu C-A, Griesinger C, Brandt U. Three molecules of ubiquinone bind specifically to mitochondrial cytochrome bc1 complex. J. Biol. Chem. 2001;276:35231–35234. doi: 10.1074/jbc.C100365200. [DOI] [PubMed] [Google Scholar]
- 43.Ding H, Moser CC, Robertson DE, Tokito MK, Daldal F, Dutton PL. Ubiquinone pair in the Qo site central to the primary energy conversion reactions of cytochrome bc1 complex. Biochemistry. 1995;34:15979–15996. doi: 10.1021/bi00049a012. [DOI] [PubMed] [Google Scholar]
- 44.Osyczka A, Moser CC, Dutton PL. Fixing the Q-cycle. Trends in Biochemical Science. 2005;30:176–182. doi: 10.1016/j.tibs.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Link TA. The role of the ‘Rieske’ iron sulfur protein in the hydroquinone oxidation (Q(P)) site of the cytochrome bc1 complex. The ‘proton-gated affinity change’ mechanism. FEBS Lett. 1997;412:257–264. doi: 10.1016/s0014-5793(97)00772-2. [DOI] [PubMed] [Google Scholar]
- 46.Berry EA, Huang LS. Observations concerning the quinol oxidation site of the cytochrome bc1 complex. FEBS Lett. 2003;555:13–20. doi: 10.1016/s0014-5793(03)01099-8. [DOI] [PubMed] [Google Scholar]
- 47.Crofts AR, Guergova-Kuras M, Huang L-S, Kuras R, Zhang Z, Berry EA. The mechanism of ubiquinol oxidation by the bc1 complex: the role of the iron sulfur protein, and its mobility. Biochemistry. 1999;38:15791–15806. doi: 10.1021/bi990961u. [DOI] [PubMed] [Google Scholar]
- 48.Crofts AR, Guergova-Kuras M, Ugulava N, Kuras R, Hong S. Proton processing at the Qo-site of the bc1 complex of Rhodobacter sphaeroides. Proc. XIIth Congress of Photosynthesis Research; Brisbane, Australia. 2002. p. 6. [Google Scholar]
- 49.Izrailev S, Crofts AR, Berry EA, Schulten K. Steered molecular dynamics simulation of the Rieske subunit motion in the cytochrome bc1 complex. Biophys. J. 1999;77:1753–1768. doi: 10.1016/S0006-3495(99)77022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Esser L, Quinn B, Li Y-F, Zhang M, Elberry M, Yu L, Yu C-A, Xia D. Crystallographic studies of quinol oxidation site inhibitors: A modified classification of inhibitors for the cytochrome bc1 complex. J. Mol. Biol. 2004;341:281–302. doi: 10.1016/j.jmb.2004.05.065. [DOI] [PubMed] [Google Scholar]
- 51.Hunte C, Solmaz S, Palsdóttir H, Wenz T. Bioenergetics. Springer Berlin: Heidelberg; 2008. A structural perspective on mechanism and function of the cytochrome bc1 complex; pp. 253–278. [DOI] [PubMed] [Google Scholar]
- 52.Lee D-W, El Khoury Y, Francia F, Zambelli B, Ciurli S, Venturoli G, Hellwig P, Daldal F. Zinc inhibition of bacterial cytochrome bc1 reveals the role of cytochrome b E295 in proton release at the Qo site. Biochemistry. 2011;50:4263–4272. doi: 10.1021/bi200230e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castellani M, Covian R, Kleinschroth T, Anderka O, Ludwig B, Trumpower BL. Direct demonstration of half-of-the-sites reactivity in the dimeric cytochrome bc1 complex. J. Biol. Chem. 2010;285:502–510. doi: 10.1074/jbc.M109.072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Świerczek M, Cieluch E, Sarewicz M, Borek A, Moser CC, Dutton PL, Osyczka A. An electronic bus bar lies in the core of cytochrome bc1. Science. 2010;329:451–454. doi: 10.1126/science.1190899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wenz T, Hellwig P, MacMillan F, Meunier B, Hunte C. Probing the role of E272 in quinol oxidation of mitochondrial complex III. biochemistry. 2006;45:9042–9052. doi: 10.1021/bi060280g. [DOI] [PubMed] [Google Scholar]
- 56.Osyczka A, Zhang H, Mathé C, Rich PR, Moser CC, Dutton PL. Role of the PEWY glutamate in hydroquinone-quinone oxidation-reduction catalysis in the Qo site of cytochrome bc1. Biochemistry. 2006;45:10492–10503. doi: 10.1021/bi060013a. [DOI] [PubMed] [Google Scholar]
- 57.Guergova-Kuras M, Salcedo-Hernandez R, Bechmann G, Kuras R, Gennis RB, Crofts AR. Expression and one-step purification of a fully active polyhistidine-tagged cytochrome bc1 complex from Rhodobacter sphaeroides. Protein Expression and Purification. 1999;15:370–380. doi: 10.1006/prep.1998.1018. [DOI] [PubMed] [Google Scholar]
- 58.Kuras R, Guergova-Kuras M, Crofts AR. Steps toward constructing a cytochrome b6f complex in the purple bacteria Rhodobacter sphaeroides: An xxample of the structural plasticity of a membrane cytochrome. Biochemistry. 1998;37:16280–16288. doi: 10.1021/bi9813476. [DOI] [PubMed] [Google Scholar]
- 59.Meinhardt SW, Crofts AR. The Role of Cytochrome b566 in the Electron Transfer Chain of Rps. sphaeroides. Biochim. Biophys. Acta. 1983;723:219–230. doi: 10.1016/0005-2728(83)90120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bowyer JR, Meinhardt SW, Tierney GV, Crofts AR. Resolved difference spectra of redox centers involved in photosynthetic electron flow in Rhodopseudomonas capsulata and Rps. sphaeroides. Biochim. Biophys. Acta. 1981;635:167–186. doi: 10.1016/0005-2728(81)90016-5. [DOI] [PubMed] [Google Scholar]
- 61.Meinhardt SW, Crofts AR. Kinetic and thermodynamic resolution of cytochrome c1 and cytochrome c2 from Rps. sphaeroides. FEBS Lett. 1982;149:223–227. [Google Scholar]
- 62.Lhee S, Kolling DR, Nair SK, Dikanov SA, Crofts AR. Modifications of protein environment of the [2Fe-2S]. cluster of the bc1 complex: effects on the biophysical properties of the Rieske iron-sulfur protein and on the kinetics of the complex. J. Biol. Chem. 2010;285:9233–9248. doi: 10.1074/jbc.M109.043505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal. Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 64.Bowyer JR, Tierney GV, Crofts AR. Secondary electron transfer in chromatophores of Rhodopseudomonas capsulata A1a pho− - binary oscillations. FEBS Lett. 1979;101:201–206. doi: 10.1016/0014-5793(79)81326-5. [DOI] [PubMed] [Google Scholar]
- 65.Crofts AR, Wraight CA. The electrochemical domain of photosynthesis. Biochim. Biophys. Acta. 1983;726:149–186. [Google Scholar]
- 66.Holmes NG, Crofts AR. The carotenoid shift in Rhodopseudomonas sphaeroides; the flash-induced change. Biochim. Biophys. Acta. 1977;459:492–505. doi: 10.1016/0005-2728(77)90048-2. [DOI] [PubMed] [Google Scholar]
- 67.Jackson JB, Crofts AR. The high energy state in chromatophores from Rhodopseudomonas spheroides. FEBS Lett. 1969;4:185–189. doi: 10.1016/0014-5793(69)80230-9. [DOI] [PubMed] [Google Scholar]
- 68.Crofts AR. The chromatophore as a functional unit. In: Tager JM, Papa S, Quagliariello E, Slater EC, editors. Electron Transport and Energy Conservation. Bari, Italy: Adriatica Editrice; 1970. pp. 221–228. [Google Scholar]
- 69.Crofts AR, Jackson JB. The high energy state in chromatophores from photosynthetic bacteria. In: Tager JM, Papa S, Quagliariello E, Slater EC, editors. Electron Transport and Energy Conservation. Bari, Italy: Adriatica Editrice; 1970. pp. 383–408. [Google Scholar]
- 70.Crofts AR, Cogdell RJ, Jackson JB. The relation of H+-uptake to electron flow in photosynthetic bacteria. In: Azzone GF, Ernster L, Papa S, Quagliariello E, Siliprandi N, editors. Mechanisms in Bioenergetics. New York and London: Academic Press; 1973. pp. 337–346. [Google Scholar]
- 71.Crofts AR, Jackson JB, Evans EH, Cogdell RJ. The high-energy state in chloroplasts and chromatophores; Proc. IInd. International Cong. on Photosynthesis; Stresa. 1971. 1972. [Google Scholar]
- 72.Crofts AR, Shinkarev VP, Kolling J, DR, Hong S. The modified Q-cycle explains the apparent mismatch between the kinetics of reduction of cytochromes c1 and bH in the bc1 complex. J. Biol. Chem. 2003;278:36191–36201. doi: 10.1074/jbc.M305461200. [DOI] [PubMed] [Google Scholar]
- 73.Jackson JB, Crofts AR. The kinetics of light induced carotenoid changes in Rhodopseudomonas spheroides and their relation to electrical field generation across the chromatophore membrane. Eur. J. Biochem. 1971;18:120–130. doi: 10.1111/j.1432-1033.1971.tb01222.x. [DOI] [PubMed] [Google Scholar]
- 74.Snozzi M, Crofts AR. Electron transport in chromatophores from Rhodopseudomonas sphaeroides Ga fused with liposomes. Biochim Biophys Acta. 1984;766:451–463. doi: 10.1016/0005-2728(84)90261-5. [DOI] [PubMed] [Google Scholar]
- 75.Ugulava NB, Crofts AR. CD-monitored redox titration of the Rieske Fe-S protein of Rhodobacter sphaeroides: pH dependence of the mid-point potential in isolated bc1 complex and in membranes. FEBS Lett. 1998;440:409–413. doi: 10.1016/s0014-5793(98)01493-8. [DOI] [PubMed] [Google Scholar]
- 76.Leggate EJ, Hirst J. Roles of the disulfide bond and adjacent residues in determining the reduction potentials and stabilities of respiratory-type Rieske clusters. Biochemistry. 2005;44:7048–7058. doi: 10.1021/bi050189x. [DOI] [PubMed] [Google Scholar]
- 77.Muller F, Crofts AR, Kramer DM. Multiple Q-cycle bypass reactions at the Qo-site of the cytochrome bc1 complex. Biochemistry. 2002;41:7866–7874. doi: 10.1021/bi025581e. [DOI] [PubMed] [Google Scholar]
- 78.Muller F. The nature and mechanism of superoxide production by the electron transport chain: its relevance to aging. J Am Aging Assoc. 2000;23:227–253. doi: 10.1007/s11357-000-0022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brandt U, Okun JG. Role of deprotonation events in ubihydroquinone: cytochrome c oxidoreductase from bovine heart and yeast mitochondria. Biochemistry. 1997;36:11234–11240. doi: 10.1021/bi970968g. [DOI] [PubMed] [Google Scholar]
- 80.Covián R, Moreno-Sánchez R. Role of protonatable groups of bovine heart bc1 complex in ubiquinol binding and oxidation. Eur. J. Biochem. 2001;268:5783–5790. doi: 10.1046/j.0014-2956.2001.02521.x. [DOI] [PubMed] [Google Scholar]
- 81.Kolling DJ, Brunzelle JS, Lhee S, Crofts AR, Nair SK. Atomic resolution structures of Rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters. Structure. 2007;15:29–38. doi: 10.1016/j.str.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seddiki N, Meunier B, Lemesle-Meunier D, Brasseur G. Is cytochrome b glutamic Acid 272 a quinol binding residue in the bc1 complex of Saccharomyces cerevisiae? Biochemistry. 2008;47:2357–2368. doi: 10.1021/bi701905a. [DOI] [PubMed] [Google Scholar]
- 83.Palsdottir H, Lojero CG, Trumpower BL, Hunte C. Structure of the yeast cytochrome bc1 complex with a hydroxyquinone anion Qo site inhibitor bound. J. Biol. Chem. 2003;278:31303–31311. doi: 10.1074/jbc.M302195200. [DOI] [PubMed] [Google Scholar]
- 84.Berry EA, Huang L-s, Lee D-W, Daldal F, Nagai K, Minagawa N. Ascochlorin is a novel, specific inhibitor of the mitochondrial cytochrome bc1 complex. Biochim. Biophys. Acta. 2010;1797:360–370. doi: 10.1016/j.bbabio.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Crofts AR. The mechanism of ubiquinol:cytochrome c oxidoreductases of mitochondria and of Rhodopseudomonas sphaeroides. In: Martonosi AN, editor. The Enzymes of Biological Membranes. New York: Plenum Publ. Corp.; 1985. pp. 347–382. [Google Scholar]
- 86.Covián R, Gutierrez-Cirlos EB, Trumpower BL. Anti-cooperative oxidation of ubiquinol by the yeast cytochrome bc1 complex. J. Biol. Chem. 2004;279:15040–115049. doi: 10.1074/jbc.M400193200. [DOI] [PubMed] [Google Scholar]
- 87.Covián R, Trumpower BL. Rapid electron transfer between monomers when the cytochrome bc1 complex dimer is reduced through center N. J Biol Chem. 2005;280:22732–22740. doi: 10.1074/jbc.M413592200. [DOI] [PubMed] [Google Scholar]
- 88.Covián R, Trumpower BL. Regulatory interactions in the dimeric cytochrome bc1 complex: The advantages of being a twin. Biochim. Biophys. Acta. 2008;1777:1079–1091. doi: 10.1016/j.bbabio.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lanciano P, Lee D-W, Yang H, Darrouzet E, Daldal F. Intermonomer electron transfer between the low-potential b hemes of cytochrome bc1. Biochemistry. 2011;50:1651–1663. doi: 10.1021/bi101736v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hong S, Victoria D, Crofts AR. Inter-monomer electron transfer is too slow to compete with monomeric turnover in bc1 complex. Biochim. Biophys. Acta. 2012;1817:1053–1062. doi: 10.1016/j.bbabio.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Czapla M, Borek A, Sarewicz M, Osyczka A. Fusing two cytochromes b of Rhodobacter capsulatus cytochrome bc1 using various linkers defines a set of protein templates for asymmetric mutagenesis. Protein Eng. Des. Sel. 2012;25:15–25. doi: 10.1093/protein/gzr055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khalfaoui-Hassani B, Lanciano P, Lee DW, Darrouzet E, Daldal F. Recent advances in cytochrome bc1: Inter monomer electronic communication? FEBS Lett. 2012;586:617–621. doi: 10.1016/j.febslet.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Czapla M, Borek A, Sarewicz M, Osyczka A. Enzymatic activities of isolated cytochrome bc1-like complexes containing fused cytochrome b subunits with asymmetrically inactivated segments of electron transfer chains. Biochemistry. 2012;51:829–835. doi: 10.1021/bi2016316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rajagukguk S, Yang S, Yu CA, Yu L, Durham B, Millett F. Effect of mutations in the cytochrome b ef loop on the electron-transfer reactions of the Rieske iron-sulfur protein in the cytochrome bc1 complex. Biochemistry. 2007;46:1791–1798. doi: 10.1021/bi062094g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Engstrom G, Xiao K, Yu C-A, Yu L, Durham B, Millett F. Photoinduced electron transfer between the Rieske iron-sulfur protein and cytochrome c1 in the Rhodobacter sphaeroides cytochrome bc1 complex: effects of pH, temperature, and driving force. J. Biol.Chem. 2002;277:31072–31078. doi: 10.1074/jbc.M202594200. [DOI] [PubMed] [Google Scholar]
- 96.Wenz T, Covian R, Hellwig P, MacMillan F, Meunier B, Trumpower BL, Hunte C. Mutational analysis of cytochrome b at the ubiquinol oxidation site of yeast complex III. J. Biol. Chem. 2006;282:3977–3988. doi: 10.1074/jbc.M606482200. [DOI] [PubMed] [Google Scholar]
- 97.Zhang L, Li Z, Quinn B, Yu L, Yu C-A. Nonoxidizable ubiquinol derivatives that are suitable for the study of the ubiquinol oxidation site in the cytochrome bc1 complex. Biochim. Biophys. Acta. 2002;1556:226–232. doi: 10.1016/s0005-2728(02)00368-7. [DOI] [PubMed] [Google Scholar]
- 98.Andrews KM, Crofts AR, Gennis RB. Large scale purification and characterization of a highly active four-subunit cytochrome bc1 complex from Rb. sphaeroides. Biochemistry. 1990;29:2645–2651. doi: 10.1021/bi00463a004. [DOI] [PubMed] [Google Scholar]
- 99.Crofts AR, Rose S. Marcus treatment of endergonic reactions: a commentary. Biochim. Biophys. Acta. 2007;1767:1228–1232. doi: 10.1016/j.bbabio.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moser CC, Page CC, Farid R, Dutton PL. Biological electron transfer. J. Bioenerg. Biomembr. 1995;27:263–274. doi: 10.1007/BF02110096. [DOI] [PubMed] [Google Scholar]
- 101.Kim N, Ripple MO, Springett R. Measurement of the mitochondrial membrane potential and pH gradient from the redox poise of the hemes of the bc1 complex. Biophys. J. 2012;102:1194–1203. doi: 10.1016/j.bpj.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ransac S, Parisey N, Mazat J-P. The loneliness of the electrons in the bc1 complex. Biochim. Biophys. Acta. 2008;1777:1053–1059. doi: 10.1016/j.bbabio.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 103.Chobot SE, Zhang H, Moser CC, Dutton PL. Breaking the Q-cycle: finding new ways to study Qo through thermodynamic manipulations. Bioenerg. Biomembr. 2008;40:501–507. doi: 10.1007/s10863-008-9175-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Crofts AR, Guergova-Kuras M, Kuras R, Ugulava N, Li J, Hong S. Proton-coupled electron transfer at the Qo-site: what type of mechanism can account for the high activation barrier? Biochim. Biophys. Acta. 2000;1459:456–466. doi: 10.1016/s0005-2728(00)00184-5. [DOI] [PubMed] [Google Scholar]
- 105.Lin I-J, Chen Y, Fee JA, Song J, Westler WM, Markley JL. Rieske protein from Thermus thermophilus: 15N NMR titration study demonstrates the role of iron-ligated histidines in the pH dependence of the reduction potential. J. Am. Chem. Soc. 2006;128:10672–10673. doi: 10.1021/ja0627388. [DOI] [PubMed] [Google Scholar]
- 106.Namslauer A, Pawate AS, Gennis RB, Brzezinski P. Redox-coupled proton translocation in biological systems: Proton shuttling in cytochrome c oxidase. Proc. Natl. Acad. Sci. USA. 2003;100:15543–15547. doi: 10.1073/pnas.2432106100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wikström M, Verkhovsky MI. Mechanism and energetics of proton translocation by the respiratory heme-copper oxidases. Biochim. Biophys. Acta. 2007;1767:1200–1214. doi: 10.1016/j.bbabio.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 108.Mitchell P. Chemiosmotic Coupling and Energy Transduction. Bodmin, Cornwall: Glynn Research Ltd; 1968. [Google Scholar]
- 109.Rich PR. The quinone chemistry of bc complexes. Biochim. Biophys. Acta. 2004;1658:165–171. doi: 10.1016/j.bbabio.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 110.Belevich I, Bloch DA, Belevich N, Wikström M, Verkhovsky MI. Exploring the proton pump mechanism of cytochrome c oxidase in real time. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2685–2690. doi: 10.1073/pnas.0608794104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Olsson MHM, Siegbahn PEM, Blomberg MRA, Warshel A. Exploring pathways and barriers for coupled ET/PT in cytochrome c oxidase: A general framework for examining energetics and mechanistic alternatives. Biochim. Biophys. Acta. 2007;1767:244–260. doi: 10.1016/j.bbabio.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brändén G, Pawate AS, Gennis RB, Brzezinski P. Controlled uncoupling and recoupling of proton pumping in cytochrome c oxidase. Proc. Natl. Acad. Sci. USA. 2006;103:317–322. doi: 10.1073/pnas.0507734103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Esser L, Gong X, Yang S, Yu L, Yu C-A, Xia D. Surface-modulated motion switch: Capture and release of iron-sulfur protein in the cytochrome bc1 complex. Proc. Natl. Acad. Sci. (U.S.A.) 2006;103:13045–13050. doi: 10.1073/pnas.0601149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee D-W, Selamoglu N, Lanciano P, Cooley JW, Forquer I, Kramer DM, Daldal F. Loss of a conserved tyrosine residue of cytochrome b induces reactive oxygen species production by cytochrome bc1. J. Biol. Chem. 2011;286:18139–18148. doi: 10.1074/jbc.M110.214460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Berry EA, Huang L-S. Conformationally linked interaction in the cytochrome bc1 complex between inhibitors of the Qo site and the Rieske iron–sulfur protein. Biochim. Biophys. Acta. 2011;1807:1349–1363. doi: 10.1016/j.bbabio.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 116.Fisher N, Castleden CK, Bourges I, Brasseur G, Dujardin G, Meunier B. Human disease-related mutations in cytochrome b studied in yeast. J. Biol. Chem. 2004;279:12951–12912. 12958. doi: 10.1074/jbc.M313866200. [DOI] [PubMed] [Google Scholar]
- 117.Fisher N, Meunier B. Re-examination of inhibitor resistance conferred by Qo-site mutations in cytochrome b using yeast as a model system. Pest. Manag. Sci. 2005;61:973–978. doi: 10.1002/ps.1066. [DOI] [PubMed] [Google Scholar]
- 118.Wibrand F, Ravn K, Schwartz M, Rosenberg T, Horn N, Vissing J. Multisystem disorder associated with a missense mutation in the mitochondrial cytochrome b gene. Ann. Neurol. 2001;50:540–543. doi: 10.1002/ana.1224. [DOI] [PubMed] [Google Scholar]
- 119.Rajarathnam K, Hochman J, Schindler M, Ferguson-Miller S. Synthesis, location, and lateral mobility of fluorescently labeled ubiquinone-10 in mitochondrial and artificial membranes. Biochemistry. 1989;28:3168–3176. doi: 10.1021/bi00434a009. [DOI] [PubMed] [Google Scholar]
- 120.Blackwell MF, Whitmarsh J. Effect of integral membrane proteins on the lateral mobility of plastoquinone in phosphatidylcholine proteoliposomes. Biophys. J. 1990;58:1259–1271. doi: 10.1016/S0006-3495(90)82466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ransac S, Mazat J-P. How does antimycin inhibit the bc1 complex? A part-time twin. Biochim. Biophys. Acta. 2010;1797:1849–1857. doi: 10.1016/j.bbabio.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 122.Moser CC, Farid TA, Chobot SE, Dutton PL. Electron tunnelling chains of mitochondria. Biochim. Biophys. Acta. 2006;1757:1096–1109. doi: 10.1016/j.bbabio.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 123.Eigen M. Proton transfer and general acid-base catalysis. In: Claesson S, editor. Fast React. Primary Processes Chem. Kinet., Proc.5th Nobel Symp. Stockholm, Swed.: Almqvist and Wiksell; 1967. pp. 245–252. [Google Scholar]
- 124.Cukier RI. Theory and simulation of proton-coupled electron transfer, hydrogen-atom transfer, and proton translocation in proteins. Biochim. Biophys. Acta. 2004;1655:37–44. doi: 10.1016/j.bbabio.2003.06.011. [DOI] [PubMed] [Google Scholar]
- 125.Rini M, Magnes B-Z, Pines E, Nibbering ETJ. Real-time observation of bimodal proton transfer in acid-base pairs in water. Science. 2003;301:349–352. doi: 10.1126/science.1085762. [DOI] [PubMed] [Google Scholar]
- 126.Shinkarev VP, Crofts AR, Wraight CA. The electric field generated by photosynthetic reaction center induces rapid reversed electron transfer in the bc1 complex. Biochemistry. 2001;40:12584–12590. doi: 10.1021/bi011334j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.