Abstract
Pyroglutamate amyloid-β peptides (pGlu-Aβ) are particularly pernicious forms of amyloid-β peptides (Aβ) present in Alzheimer’s disease (AD) brains. pGlu-Aβ peptides are N-terminally truncated forms of full-length Aβ peptides (flAβ(1-40/42)) in which the N-terminal glutamate is cyclized to pyroglutamate to generate pGlu-Aβ(3-40/42). β-secretase cleavage of amyloid-β precursor protein (AβPP) produces flAβ(1-40/42), but it is not yet known whether the β-secretase BACE1 or the alternative β-secretase cathepsin B (CatB) participate in the production of pGlu-Aβ. Therefore, this study examined the effects of gene knockout of these proteases on brain pGlu-Aβ levels in transgenic AβPPLon mice, which express AβPP isoform 695 and have the wild-type (wt) β-secretase activity found in most AD patients. Knockout or overexpression of the CatB gene reduced or increased, respectively, pGlu-Aβ(3-40/42), flAβ(1-40/42), and pGlu-Aβ plaque load, but knockout of the BACE1 gene had no effect on those parameters in the transgenic mice. Treatment of AβPPLon mice with E64d, a cysteine protease inhibitor of CatB, also reduced brain pGlu-Aβ(3-42), flAβ(1-40/42), and pGlu-Aβ plaque load. Treatment of neuronal-like chromaffin cells with CA074Me, an inhibitor of CatB, resulted in reduced levels of pGlu-Aβ(3-40) released from the activity-dependent, regulated secretory pathway. Moreover, CatB knockout and E64d treatment has been previously shown to improve memory deficits in the AβPPLon mice. These data illustrate the role of CatB in producing pGlu-Aβ and flAβ that participate as key factors in the development of AD. The advantages of CatB inhibitors, especially E64d and its derivatives, as alternatives to BACE1 inhibitors in treating AD patients are discussed.
Keywords: pyroglutamate amyloid-β, cathepsin B, BACE1, AβPP, protease, transgenic AD mice, inhibitor, cysteine protease, secretion
Introduction
The abnormal accumulation of amyloid-β peptides (Aβ) has long been thought to be the likely cause of Alzheimer’s disease (AD) [1-4]. Aβ are heterogeneous peptides differing at the carboxyl (C-terminal) and amino (N-terminal) ends, and in their biophysical and neurotoxic properties [5]. Full-length Aβ (flAβ) have the same N-terminus but different C-termini with the two most prominent forms being flAβ(1-40) and flAβ(1-42), which have 40 and 42 amino acids, respectively, and the longer flAβ(1-42) being more hydrophobic and more neurotoxic than flAβ(1-40) [5]. AD brains also contain N-terminally truncated Aβ species, with notable presence of pyroglutamate amyloid-β peptides (pGlu-Aβ) which lack the first two N-terminal amino acids of flAβ and have a cyclized N-terminal glutamate (pyroglutamate, pGlu) [6-8]. The pGlu-Aβ(3-42) species is a dominant fraction of the Aβ in brains of AD and Down syndrome patients, who develop an age-accelerated AD pathology [9-14]. pGlu-Aβ(3-42) is more stable, neurotoxic, and causes more aggregation of Aβ than does flAβ(1-42) [10, 13, 15-18]. Aβ aggregated into oligomers is thought to be the in vivo neurotoxic form of Aβ and recently pGlu-Aβ(3-42) containing oligomers were found to be more neurotoxic than those lacking pGlu-Aβ(3-42) [19-22]. Structural differences and similarities among these Aβ species are illustrated in Figure 1.
Figure 1. Illustration of flAβ(1-40), flAβ(1-42), N-truncated Aβ(3-40), N-truncated Aβ(3-42), pGlu-Aβ(3-40), and pGlu-Aβ(3-42) indicates the differences and similarities among these Aβ species.
All Aβ species are shown with details of their N- and C-termini. Aβ species having C-terminal residues at position 40 and 42 are colored blue and red, respectively. This study analyzed flAβ(1-40), flAβ(1-42), pGlu-Aβ(3-40), and pGlu-Aβ(3-42) (but not N-truncated Aβ(3-40) and N-truncated Aβ(3-42)).
A. flAβ(1-40). In this Aβ species, aspartic acid (D) is located at the N-terminus, which is known as position 1 of the Aβ, and valine (V) is at the C-terminus located at position 40. The N-terminus of flAβ(1-40) is created by β-secretase cleavage of AβPP.
B. flAβ(1-42). Like flAβ(1-40), this Aβ species begins at the N-terminus position 1 with D but has two additional amino acids (compared to flAβ(1-40)) at the C-terminus, which are isoleucine (I) and alanine (A) with the latter located at position 42. These additional C-terminal residues make flAβ(1-42) more neurotoxic with a greater propensity to aggregate Aβ than flAβ(1-40). The N-terminus of flAβ(1-42) is also created by β-secretase cleavage of AβPP.
C. N-truncated Aβ(3-40). D and A found in flAβ at positions 1 and 2 are not present and the N-terminus begins with glutamate (E) at position 3. This Aβ species has the C-terminal V residue at position 40 as in flAβ(1-40). N-truncated Aβ(3-40) is required for pGlu-Aβ(3-40) formation because E can only be cyclized if it is an N-terminal amino acid. How N-truncated Aβ(3-40) is formed from AβPP is not known.
D. N-truncated Aβ(3-42). This species has features of N-truncated Aβ(3-40) in that the N-terminus is E at position 3 and the C-terminus is residue A at position 42 (like that of flAβ(1-42)). Again, N-truncated Aβ(3-42) is required for pGlu-Aβ(3-42) formation but how that occurs is not known.
E. pGlu-Aβ(3-40). This Aβ species is the same as N-terminal Aβ(3-40) except the N-terminal E residue is cyclized to pyroglutamate (pGlu or pE) at position 3. E is converted to pE by the enzyme glutaminyl cyclase (QC). One aspect of this study was to determine if the established β-secretase, BACE1, or the alternative β-secretase, CatB, affects pGlu-Aβ(3-40) levels.
F. pGlu-Aβ(3-42). This Aβ species is the same as N-terminal Aβ(3-42) except the N-terminal E residue is cyclized to pE at position 3. pGlu-Aβ(3-42) is more neurotoxic, has a greater propensity to aggregate Aβ, and is much more resistant to degradation than flAβ(1-42). pGlu-Aβ(3-42) is thought by some to be the Aβ species which causes AD. Again, a focus of this study was to evaluate the effects of BACE1 and CatB on pGlu-Aβ(3-42) levels.
Significantly, in transgenic AD mice, pGlu-Aβ(3-42) causes age-dependent behavioral deficits and inhibits hippocampal long-term neuronal potentiation, which reflects memory impairment [23-26]. Glutaminyl cyclase (QC) is the enzyme that catalyzes the cyclization of the free N-terminal glutamate on truncated Aβ(3-40/42) to generate pGlu-Aβ(3-40/42) [27, 28]. Importantly, QC inhibitors or passive immunotherapy using pGlu-Aβ antibodies reduce brain pGlu-Aβ, pGlu-Aβ amyloid plaque load, and improve behavioral deficits in transgenic mouse models of AD [29-31]. Thus the pGlu-Aβ may be the “bad actor” among Aβ peptides causing AD and reducing brain pGlu-Aβ, especially pGlu-Aβ(3-42), may be critical to effective treatment of the disease [32].
pGlu-Aβ must be excised from amyloid precursor protein (AβPP) by proteases but the identity of those proteases is not as yet known. It is possible that pGlu-Aβ is formed from flAβ by removal of the N-terminal amino acids followed by QC-mediated glutamate cyclization. Thus, a potential protease candidate is that which cleaves AβPP to generate the N-terminus of flAβ, generically called β-secretase. The aspartyl protease BACE1 is considered by many to be the primary β-secretase [33, 34]. Alternatively, the cysteine protease cathepsin B (CatB) has also been shown to have β-secretase activity [35, 36]. Although BACE1 and CatB inhibitors are being developed as AD therapeutics to reduce flAβ, neither BACE1 nor CatB have been shown to affect pGlu-Aβ in vivo and, thus, if pGlu-Aβ is the true cause of AD, it is not clear if inhibitors of these proteases will affect AD.
This study used a transgenic AD mouse model to evaluate effects of BACE1 or CatB gene knockout on pGlu-Aβ in brain. While many AD mouse models have been developed that overexpress human AβPP transgenes to study Aβ production, most express an AβPP transgene encoding the very rare Swedish (Swe) mutations at the β-secretase cleavage site [37, 38]. However, the vast majority of AD patients have AβPP with the wild-type (wt) β-secretase site, and BACE1 and CatB are known to have very different cleavage efficiencies for these two β-secretase sites [36, 39-43]. The models also express different isoforms of AβPP, but in human brain the AβPP isoform 695 (AβPP-695) is expressed at greater levels than the other isoforms and is the main form expressed in neurons [44-48]. Thus, in order to evaluate the β-secretase production of pGlu-Aβ occurring in the brain neurons of most AD patients, it is critical that the transgenic model express AβPP containing the wt β-secretase site sequence and isoform AβPP-695. Two models having these features are those expressing human wt AβPP (AβPPwt), which is that found in most AD patients [49], and human AβPP-695 containing the London mutation (AβPPLon), which has the wt β-secretase site and a rare familial mutation near the AβPP C-terminal cleavage site of Aβ [50]. AβPPwt and AβPPLon mice overproduce brain flAβ(1-40/42) and develop memory deficits, but AβPPLon mice are also known to produce pGlu-Aβ(3-42) and develop brain Aβ plaque pathology and inflammatory processes, which mimic that seen in AD patients [51, 52]. Thus, the AβPPLon mouse model was used here to evaluate the effects of BACE1 and CatB activity on brain pGlu-Aβ production.
This study also evaluated regulated and constitutive pGlu-Aβ secretion because flAβ and neurotoxic oligomers are formed and secreted by neurons in these pathways [53-56]. Notably, neural activity modulates the formation, secretion, and deposition of Aβ [53-56]. Neuronal-like chromaffin cells (bovine) in primary culture are a well-established model of regulated and constitutive secretion of neurotransmitters by neuronal cells [57, 58]. These cells were used by us to first identify CatB producing flAβ in the regulated secretory pathway [35], which was subsequently validated to also occur in brain neuronal cells in primary culture by others [59]. Importantly, chromaffin cells have recently been found to contain pGlu-Aβ and flAβ in regulated secretory vesicles [60]. Moreover, unlike AβPPLon mice, chromaffin cells are a natural model and, thus, provide a control for potential transgene artifacts. Bovine chromaffin cells express AβPP-695 having the wt β-secretase site and flAβ(1-40/42) sequences found in man [61, 62] and as such model human wt β-secretase activity. Therefore, neuronal-like chromaffin cells were used to study the effects of CatB inhibition on pGlu-Aβ levels released from regulated and constitutive secretory pathways.
Here, the effects of deleting the BACE1 gene and the CatB gene, as well as overexpressing the CatB gene, on brain pGlu-Aβ(3-40/42), flAβ(1-40/42), and pGlu-Aβ amyloid plaque load in AβPPLon mice were studied. Further, the effects of administering the cysteine protease inhibitor E64d, which inhibits CatB but not BACE1, on those parameters in AβPPLon mice having or lacking the CatB gene are presented. In addition, regulated and constitutive secretion of pGlu-Aβ(3-40) was compared in neuronal-like chromaffin cells treated with the CatB inhibitor CA074Me. Results show that CatB, but not BACE1, participates in the production of brain pGlu-Aβ in the AβPPLon mouse model, and that it may do so in the regulated secretory pathway. These data are highly supportive of CatB as a drug target for reducing pGluAβ(3-40/42), flAβ(1-40/42), and pGlu-Aβ plaque load. These findings indicate that compounds, which inhibit CatB activity, have strong potential as AD therapeutics.
Materials and Methods
Transgenic animal methods
Transgenic AβPPLon mice with protease gene knockout or overexpression
The AβPPLon mice were generated by methods previously described [50]. Briefly, the AβPPLon transgene was made by site-directed mutagenesis to insert the V717I London mutation into human AβPP DNA. The transgenic mice were created by transfection with the transgene containing the platelet-derived growth factor beta (PDGFβ) promoter and an SV40 polyadenylation site for expression of AβPP-695. BACE1 deficient knockout (BACE1 KO) mice were obtained from Jackson Laboratory (Bar Harbor, ME) and CatB deficient (CatB KO) mice were obtained from Christoph Peters (Albert Ludwig University, Freiburg, Germany).
Transgenic CatB (CatB TG) mice, which overexpressed CatB, were generated as follows. For generation of the CatB construct, the pEGFP-N1 vector from Clontech was used. The pEGFP-N1 CMV promoter was excised by AseI/BglII cleavage and a 1 kb fragment of the human PDGFβ promoter was cloned into the vector. The PDGFβ promoter fragment was produced by PCR amplification of human genomic DNA using forward and reverse primers consisting of 5′-GGGATTAATGATCCACAGTCTCCTGAGTAGCTG-3′ and 5′-GGGAGATCTGGGAGGCAGGCAGGCCGCTC-3′, respectively. The PDGFβ promoter fragment included a 27 base pair (bp) 5′ untranslated region lacking the ATG codon. Downstream of the PDGFβ promoter fragment, the β-globin intron II with flanking exon sequences (665 bp) was inserted into the XhoI site as a SalI/XhoI fragment. The β-globin sequence was PCR amplified using forward and reverse primers consisting of 5′-GGGGTCGACGATCCTGACAACTTCAGGGTG-3′ and 5′-GGGCTCGAGGCCCTATAGTGAGTCGTATTAC-3′, respectively from the pSG5 vector. For construction of the CatB expression vector, which was a plasmid containing the platelet derived growth factor beta (PDGFβ) and CatB (pPDGFβ/CatB) sequences, the enhanced green fluorescent protein (EGFP) gene was excised from pPDGFβ/EGFP by HindIII/NotI cleavage. The open reading frame of the CatB cDNA was isolated from mouse brain messenger RNA (mRNA) by PCR amplification using forward and reverse primers consisting of 5′-GGGGTCGACAAGCTTGCCACCATGTGGTGGTCCTTGATCC-3′ and 5′-GGGGTCGACGCGGCCGCCTTAGAATCTTCCCCAGTACT-3′, respectively. The PCR primers contained HindIII/NotI sites for cloning into the HindIII/NotI sites. The forward primer sequence also included a consensus Kozak motif for translational initiation in front of the ATG codon.
All mice were maintained on a C57BL/6 background. The transgenic AβPPLon mice were crossed either with the BACE1 KO, CatB KO, or CatB TG mice to generate the following transgenic mice: AβPPLon, AβPPLon x BACE1 KO, AβPPLon x CatB KO, and AβPPLon x CatB TG mice. Polymerase chain reaction (PCR) analysis was utilized to determine the genotype of the animals as previously described [49].
All mice were analyzed at 12 months because at that age AβPPLon mice express pGlu-Aβ(3-42), display pGlu-Aβ plaque load, and develop memory deficits [50, 51]. All experimental mice were male and were given free access to food and water before and during the experiment. The animals were sacrificed by isoflurane overdose and exsanguination.
E64d treatment of transgenic mice
E64d was synthesized and formulated into mouse chow by methods previously described [63]. Briefly, 99% pure E64d was made by American Life Science Pharmaceuticals (San Diego, CA) using methods developed and modified from those described [64]. However, E64d is also commercially available from various suppliers (e.g., Calbiochem, San Diego, CA). E64d-doped mouse chow was made in 2018, Teklad Global 18% Protein Rodent Diet (Harlan Laboratories, Inc., Madison, WI). The chow containing 40 mg/kg of E64d was processed according to the manufacturer’s procedure into standard 12.7 mm pellets and analysis showed that the chow contained that concentration of E64d (data not shown). The mice consumed on average about 5 gm of chow per day and had an average body weight of 20 gm and, thus, the animals received an average calculated daily E64d dose of 10 mg/kg. Animals were fed E64d chow beginning at 9 months of age and continued for 3 months until analysis. All other mice received the same chow but without E64d during that time. No difference was observed in the amount of E64d-doped and normal chow consumed by the different groups (data not shown).
E64d inhibits papain-like cathepsin cysteine proteases and calpain-activated neutral proteases [65] but not BACE1 [36]. To evaluate the relative importance of inhibiting CatB vs. the other proteases, E64d was administered to AβPPLon (E64d) mice and to CatB KO x AβPPLon (E64d CatB KO) mice.
Animal care
The regulations of the National Institutes of Health (NIH) as approved by the Institutional Animal Care and Use Committee (IACUC) at the Medical University of South Carolina and the Ralph H. Johnson Veterans Affairs Medical Center, Charleston, SC were adhered to in conducting the animal experiments.
Brain BACE1 and CatB activities
BACE1 and CatB activities were determined in brain samples prepared and analyzed as previously described using commercial assay kits (ab65357 and ab65300, respectively, Abcam, Cambridge, MA) [50, 63]. Brain tissue was washed twice in ice-cold phosphate buffered saline and homogenized in extraction buffer as per the manufacturer. After 10 minutes incubation on ice, the extract was centrifuged (100,000 g for 5 minutes) and 50 μl of supernatant was mixed with an equal volume of 2X reaction buffer and 2 μl substrate in a 96-well microplate. The plates were kept in the dark at 37°C for 1 hour and the fluorescence was recorded using FLUOstar Optima plate reader (BMG Labtech, Cary, NC). Protein concentration was determined by the bicinchoninic acid (BCA) method (Bio-Rad, Life Science, Hercules, CA). BACE1 and CatB activities were expressed as fluorescent units/mg protein.
Corroborating our previous studies, BACE1 KO and CatB KO mice had no brain BACE1 and CatB activities, respectively, and the same brain CatB and BACE1 activities, respectively, as compared to control mice (data not shown) [50, 63]. In contrast, CatB TG mice had a significant 500% higher brain CatB activity and no change in brain BACE1 activity relative to control mice (data not shown). These data established that the BACE1 KO and CatB KO mice were suitable for evaluating the effects of the absence of these protease activities and that the CatB TG mice were suitable for evaluating the effects of higher CatB activity on the parameters assessed in this study.
E64d-treated mice had a significant 80% lower brain CatB activity and no significant change brain BACE1 activity relative to control untreated mice (data not shown), which reproduced our previous report [63]. As expected, E64d CatB KO mice had no brain CatB activity and no significant change in BACE1 activity relative to control mice (data not shown). These data established that the oral E64d dose regime used was suitable for determining the effect of reducing brain CatB activity on pGlu-Aβ and the other parameters of this study.
Brain pGlu-Aβ and flAβ analysis
Brain Aβ peptides analyses were conducted as previously described [50]. Briefly, animals were sacrificed and brain extracts homogenized (1:3 weight/volume of buffer) in 5 M guanidine HCl in 50 mM Tris-HCl and 150 mM sodium chloride (NaCl) with protease inhibitors (Sigma-Aldrich, St. Louis, MO). Homogenates were diluted to 0.5 M guanidine and centrifuged (200,000g for 20 minutes at 4°C), and supernatant and pellet fractions were collected. The pellet was sonicated in 6 M guanidine and centrifuged (200,000g for 20 minutes at 4°C), and the supernatant was diluted to 0.5 M guanidine. The two supernatants were combined and pGlu-Aβ(3-40), pGlu-Aβ(3-42), Aβ(1-40), and Aβ(1-42) were determined using commercial ELISA kits specific for each peptide (JP27418, JP27716, JP27718, and JP27711, respectively, IBL International Corp, Toronto, Canada). This procedure resulted in analyses of total brain Aβ, of combined insoluble and soluble fractions.
pGlu-Aβ amyloid plaque load analysis
Brain sections were analyzed using a commercial polyclonal antibody (218 003, Synaptic Systems, Goettingen, Germany), which recognizes the truncated N-terminus of pGlu-Aβ and thus detects both pGlu-Aβ(3-40) and pGlu-Aβ(3-42). The tissue preparation and sectioning was conducted as previously described [50]. Briefly, brains were fixed in 4% paraformaldehyde and 30% sucrose for 24 hours at 4°C. Tissues were washed in Tris-buffered saline and transferred to optimum cutting temperature medium; cryosections were then cut and blocked with normal serum, incubated with anti-Aβ, and stained with diaminobenzoic acid (Vector ABC Elite kit, Vector Laboratories, Burlingame, CA). The staining in the sections is referred to here as pGlu-Aβ amyloid plaque load or pGlu-Aβ plaque. Bright field light microscopy images of the sections were taken and pGlu-Aβ plaque load quantitated using image analysis (NIH Image software). Ten sections per mouse were analyzed as previously described [50].
Chromaffin cell methods
Neuronal-like chromaffin cells in primary culture
Neuronal-like chromaffin cells in primary culture were generated from fresh bovine adrenal medulla of the sympathetic nervous system as previously described [35]. Briefly, chromaffin cells were dissected from fresh adrenal glands, dissociated in a collagenase/DNase solution at 37°C, filtered, centrifuged, and cells were plated onto fibronectin-coated dishes (EMD Chemicals, Gibbston, NJ) in media containing Dulbecco’s Modified Eagle Medium (DMEM) (Cellgro, Manassas, VA), 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA), and penicillin/streptomycin. Cells were maintained at 37°C and 6% CO2.
Regulated secretion of pGlu-Aβ from chromaffin cells
After six days in culture, chromaffin cells were subjected to potassium chloride (KCl) depolarization to stimulate regulated, activity-dependent secretion. Cells were incubated in standard release medium (SRM) with 0.25 μg/ml BSA (bovine serum albumin) for 90 minutes at 37° C, followed by removal of media, which represents basal secretion, as previously described [35]. Cells were then incubated with KCl in SRM media (50 mM KCl, 37° C, 90 minutes) to stimulate regulated secretion, followed by collection of media Secretion media was collected with addition of a cocktail of protease inhibitors (1 mM EDTA, 500 μM AEBSF, and 10 μM each of E64c, leupeptin, pepstatin A, and chymostatin). The secretion media was concentrated by ultrafiltration through a 2 kilodalton (kDa) cut-off membrane (Vivaspin 2 2K MWCO Hydrosart, Sartorius, Goettingen, Germany), which retains Aβ. The concentrated sample was subjected to measurements of pGlu-Aβ(3-40) (JP27418, IBL). Assays of pGlu-Aβ(3-42) (JP27716, IBL) were conducted but were too low to be detected, although pGlu-Aβ(3-42) and pGlu-Aβ(3-40) are both present in chromaffin cells [60].
CA074Me treatment of chromaffin cells and levels of secreted pGlu-Aβ(3-40)
Chromaffin cells (1 × 106 cells/10 cm petri dish) were either untreated or incubated with CA074Me, a cell-permeable prodrug that is converted intracellular by esterases to CA074, which inhibits CatB [66, 67]. Cells were treated with 50 μM CA074Me for 24 hours, and were then induced to secrete via regulated secretion, stimulated by adding KCl (50 mM, 90 minutes) to the media. Control incubation without KCl was included. ELISA was used to measure pGlu-Aβ(3-40) in cell culture media (as described above).
Statistics
Animal experiments consisted of 10 mice in each group except the control pGlu-Aβ(3-40), flAβ(3-40) and pGlu-Aβ plaque load groups, which had 20 animals each. Chromaffin cell culture experiments were conducted in quadruplicate wells. Each biochemical analysis consisted of two or three replicates. Statistical analyses and data display were conducted utilizing computer software designed for scientific data analysis (Prism 6, GraphPad Software). Quantitative data are displayed as the mean and standard error of the mean (SEM). Differences between the means were evaluated by ANOVA followed by Dunnett’s multiple comparison test, Bonferroni’s multiple comparison test or by student’s unpaired t-test, p < 0.05) as noted.
Results
Transgenic mouse results
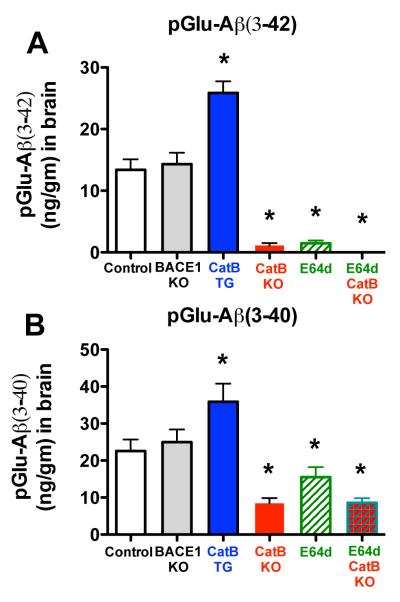
Brain pGlu-Aβ(3-42) levels were reduced by more than 90% by gene knockout (KO) or inhibition of CatB, doubled by over expression of the CatB gene, and not significantly affected by KO of the BACE1 gene
Figure 2A shows that brain pGlu-Aβ(3-42) was about 13 ng/gm in the brains of control mice and was reduced by over 90% to about 1 ng/gm in CatB KO mice and in E64d-treated mice. Furthermore, pGlu-Aβ(3-42) was not detected at all in E64d-treated CatB KO mice relative to control mice. Brain pGlu-Aβ(3-42) levels were statistically equivalent among the CatB KO, E64d, and E64d CatB KO mice (Bonferroni’s multiple comparison test). These data clearly show that endogenous CatB controls almost all of the brain pGlu-Aβ(3-42) and suggests that E64d treatment acted primarily by inhibiting brain CatB activity to reduce brain pGlu-Aβ(3-42) in this model.
Figure 2. pGlu-Aβ(3-40/42) levels were reduced in AβPPLon mice with CatB gene knockout (KO) or with E64d inhibition of CatB activity, increased in animals expressing the CatB gene, and not effected in BACE1 KO animals.
A. pGlu-Aβ(3-42). The mean brain pGlu-Aβ(3-42) levels for AβPPLon mice (control), BACE1 gene knockout AβPPLon mice (BACE1 KO), transgenic CatB AβPPLon mice (CatB TG), CatB knockout AβPPLon mice (CatB KO), AβPPLon mice fed E64d-containing chow (E64d), and CatB knockout AβPPLon mice fed E64d-containing chow (E64d CatB KO) are shown. Brain pGlu-Aβ(3-42) levels were significantly reduced in CatB KO, E64d, and E64d CatB KO mice, significantly increased in CatB TG mice, and the same in BACE1 KO mice as compared to control mice.
B. pGlu-Aβ(3-40). The mean brain pGlu-Aβ(3-40) values for the same animal groups as in Fig. 2A are shown. Brain pGlu-Aβ(3-40) levels were significantly reduced in CatB KO, E64d, and E64d CatB KO mice, significantly increased in CatB TG mice, and the same in BACE1 KO mice as compared to control mice.
(All animal data in this study were from animals that were12 months old at sacrifice. Panels ‘A’ and ‘B,’ ANOVA, one way analysis of variance, p < 0.0001, Dunnett’s multiple comparison with control, *p < 0.05)
The exact opposite occurred in CatB TG mice. Figure 2A shows that CatB TG mice had 97% higher pGlu-Aβ(3-42) levels than control mice. Thus, these data show that exogenous CatB activity clearly drove brain pGlu-Aβ(3-42) production in this model.
In contrast, brain pGlu-Aβ(3-42) levels were not affected in the BACE1 KO mice. Figure 2A shows that BACE1 KO mice had no significant change in levels of brain pGlu-Aβ(3-42) relative to that in control mice. These data demonstrate that BACE1 did not produce pGlu-Aβ(3-42) in this model.
Brain pGlu-Aβ(3-40) levels were reduced by about one-third in E64d-treated mice, reduced by two thirds in CatB KO and E64d-treated CatB KO mice, increased by about 50% in CatB TG mice, and not significantly affected in BACE1 KO mice
As shown in Figure 2B, brain pGlu-Aβ(3-40) was a significant 32% lower in E64d mice, and 65% lower in CatB KO and E64d CatB KO mice, respectively, relative to control mice. There was no significant difference in pGlu-Aβ(3-40) levels among these three experimental groups (Bonferroni’s multiple comparison test). Thus, these data show that endogenous brain CatB activity controlled most of the brain pGlu-Aβ(3-40) in this model and suggests that E64d acted to reduce brain pGlu-Aβ(3-40) by inhibiting CatB activity.
Figure 2B also shows that the opposite effect occurred in CatB TG mice, which had a significant 53% higher brain pGlu-Aβ(3-40) level than control mice. These data show that exogenous CatB activity drove brain pGlu-Aβ(3-40) production in this model.
In contrast, brain pGlu-Aβ(3-40) levels were not affected by deleting the BACE1 gene as shown in Figure 2B. Thus, endogenous BACE1 activity did not produce brain pGlu-Aβ(3-40) in this model.
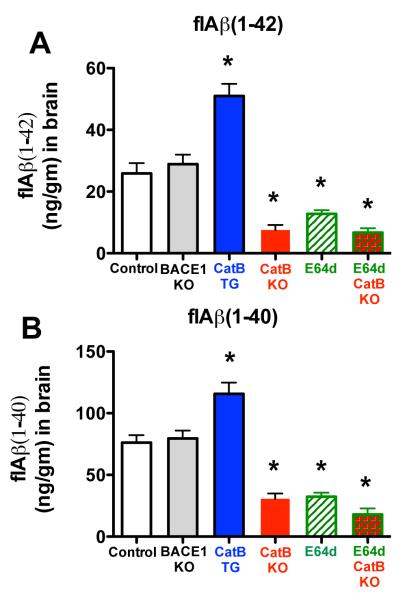
Brain flAβ(1-42) levels were almost doubled in the CatB TG mice, reduced by about half in E64d mice, reduced by over 70% in CatB KO and E64d CatB KO mice, and not significantly affected in BACE1 KO mice
Figure 3A shows that CatB TG mice had a significantly 93% higher flAβ(1-42) level in brain compared to control mice. These new data show that exogenous CatB activity drove brain flAβ(1-42) levels in this transgenic AD model.
Figure 3. flAβ(1-40/42) levels were reduced by CatB gene knockout or inhibition by E64d, increased by CatB gene expression, and not effected by BACE1 knockout in AβPPLon animals.
A. flAβ(1-42). CatB KO, E64d, and E64d CatB KO mice had significantly lower flAβ(1-42), CatB TG mice had significantly higher flAβ(1-42), and BACE1 KO mice had statistically equivalent flAβ(1-42) levels as in control mice.
B. Aβ(1-40). CatB KO, E64d, and E64d CatB KO mice had significantly reduced Aβ(1-40), CatB TG mice had significantly increased Aβ(1-40), and BACE1 KO mice had flAβ(1-40) that were statistically equivalent to that of control mice. (panels ‘A’ and ‘B’, ANOVA, one way analysis of variance, p < 0.0001, Dunnett’s multiple comparison to control,*p < 0.05)
Figure 3A also shows that brain flAβ(1-42) levels were 51% lower in E64d mice and were 71% and 74% lower in CatB KO and E64d CatB KO mice, respectively, vs. control mice. There was no significant difference in the levels among these three experimental groups (Bonferroni’s multiple comparison test). The CatB KO and E64d mouse data reproduce our earlier reports [50, 63], which showed that CatB gene deletion or the same E64d treatment in AβPPLon mice reduced brain flAβ(1-42) by similar amounts. The E64d CatB KO data are new and show that E64d primarily affected brain flAβ(1-42) by inhibiting CatB activity. These data show that endogenous CatB activity produced flAβ(1-42) and that E64d reduced that Aβ species by inhibiting CatB activity in this model.
Figure 3A further shows that BACE1 KO mice had no significant change in brain flAβ(1-42) levels compared to control mice. These data also confirm our previous data showing similar effects due to deleting the BACE1 gene in the AβPPLon model [50]. These data demonstrate that endogenous BACE1 activity did not produce flAβ(1-42) in this model.
Brain flAβ(1-40) levels were 50% higher in CatB TG mice, reduced by over half in CatB KO and E64d mice, reduced by almost three fourths in E64d CatB KO mice, and not significantly affected in BACE1 KO mice
Figure 3B shows that brain flAβ(1-40) levels were 52% higher in CatB TG mice relative to control mice. These new data show that exogenous CatB activity controlled flAβ(1-40) levels in this model.
The opposite occurred with the deletion of the CatB gene or inhibition of CatB activity. Figure 3B shows that brain flAβ(1-40) levels were significantly reduced by 57% in the CatB KO and E64d mice, and by 74% in the E64d CatB KO mice, relative to control mice. There was no significant difference in the levels among these three experimental groups (Bonferroni’s multiple comparison test). The CatB KO and E64d data here confirm our earlier reports [50, 63], which showed that CatB gene deletion or E64d treatment of AβPPLon mice resulted in similar brain flAβ(1-40) reductions. The new E64d CatB KO data suggest that E64d acted primarily by inhibiting CatB to reduce flAβ(1-40) in this model.
Figure 3B also shows that the BACE1 KO mice, on the other hand, had no significant differences in brain flAβ(1-40) levels relative to that of control mice. These data confirmed our previous data [50] showing essentially the same result from deleting the BACE1 gene in this transgenic model. These data show that endogenous BACE1 did not produce flAβ(1-40) in this model.
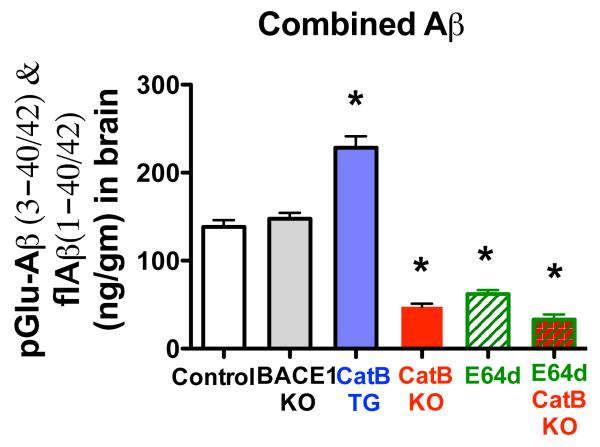
Combined brain Aβ is significantly lower in CatB KO, E64d, and E64d CatB KO mice, higher in CatB TG mice, and not affected in BACE1 KO mice
Figure 4 shows the combined sum of the pGlu-Aβ(3-42), pGlu-Aβ(3-40), flAβ(1-42), and flAβ(1-40) levels for the animal groups. The combined Aβ was significantly decreased by 55%, 65%, and 76% in E64d, CatB KO, and E64d CatB KO mice, respectively. Furthermore, the combined Aβ was significantly increased by 65% for CatB TG mice, and not statistically different for BACE1 KO mice relative to control mice. There was no significant difference in the combined Aβ levels for the E64d, CatB KO, and E64d CatB KO mice (Bonferroni’s multiple comparison test). These data show that endogenous and exogenous CatB activities had major effects on the total measured Aβ whereas endogenous BACE1 activity had no significant effect in this model.
Figure 4. Total measured Aβ was lower in AβPPLon mice with CatB knockout or inhibition, higher in animals having expressing the CatB gene, and not effected in animals with BACE1 gene knockout.
The combined sum of the pGlu-Aβ(3-42), pGlu-Aβ(3-40), flAβ(1-42), and flAβ(1-40) is displayed for each animal group. The total measured Aβ was significantly reduced in CatB KO, E64d, and E64d CatB KO mice, significantly increased in CatB TG mice, and unaffected in BACE1 KO mice compared to control mice. (ANOVA, one way analysis of variance, p < 0.0001, Dunnett’s multiple comparison to control, *p < 0.05)
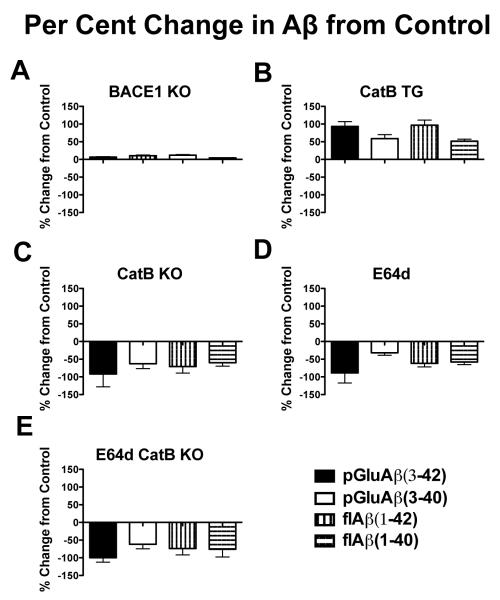
All Aβ species were consistently lower in animals in which CatB activity was inhibited or eliminated, consistently increased for those in which CatB activity was increased, and not significantly affected in animals lacking BACE1 activity
Figures 5A, B, C, D, and E are graphs of the percent change for each measured Aβ species in the BACE1 KO, CatB TG, CatB KO, E64d, and E64d CatB KO mice, respectively, relative to the corresponding Aβ levels in control mice. The figure shows that all the Aβ species in the BACE1 KO mice had little or no percent change, whereas all the Aβ species in the CatB TG, CatB KO, E64d, and E64d CatB KO mice had major percent changes relative to control mice. In the CatB TG mice all the Aβ species had positive percent changes, and all the Aβ species in the CatB KO, E64d, and E64d CatB KO mice had negative percent changes.
Figure 5. The percent changes in pGlu-Aβ(3-42), pGlu-Aβ(3-40), flAβ(1-42), and flAβ(1-40) were all greatly reduced in AβPPLon mice CatB knockout or inhibition, all greatly increased in animals expressing the CatB gene, and not significantly changed in animals with BACE1 knockout relative to the levels in control mice.
The panels show for each experimental animal group the percent changes in pGlu-Aβ(3-42), pGlu-Aβ(3-40), flAβ(1-42), and flAβ(1-40) relative to the corresponding species in control mice. The ordinate axis is the same scale in all panels so that direct comparisons among the panels can be made.
A. BACE1 KO mice. The Aβ species had very small positive and negative percent changes.
B. CatB TG mice. All Aβ species had large positive percent changes with pGlu-Aβ(3-40) and flAβ(1-40) being about 50% higher, and pGlu-Aβ(3-42) and flAβ(1-42) being about 100% higher than controls.
C. CatB KO mice. All Aβ species had large negative percent changes with pGlu-Aβ(3-40) and flAβ(1-40) being more than 50% and 60% lower respectively, and pGlu-Aβ(3-42) and flAβ(1-42) being more than 90% and 70% lower, respectively.
D. E64d mice. All Aβ species had large negative percent changes with pGlu-Aβ(3-40) and flAβ(1-40) being about 35% and 55% lower, respectively, and pGlu-Aβ(3-42) and flAβ(1-42) being more than 90% and 50% lower, respectively.
E. E64d CatB KO mice. All Aβ species had large negative percent changes with pGlu-Aβ(3-40) and flAβ(1-40) being 66% and 75% lower, respectively, and pGlu-Aβ(3-42) and flAβ(1-42) being 100% and 80% lower, respectively.
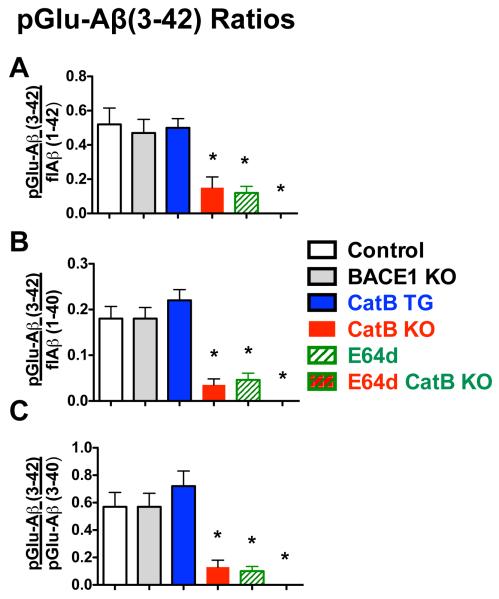
pGlu-Aβ(3-42) to flAβ and pGlu-Aβ(3-40) ratios were significantly different in animals in which CatB activity was inhibited or eliminated, but were not changed in the CatB TG or BACE1 KO mice from that of control animals
The ratios of the Aβ species were calculated and analyzed among the animal groups. The flAβ(1-42) to flAβ(1-40) ratio for control mice was 0.36, which was not significantly different from any of the experimental animal groups (data not shown). Similarly, the pGlu-Aβ(3-40) to flAβ(1-40) and flAβ(1-42) for control mice were 0.32 and 0.90, respectively, and neither differed significantly among all experimental groups (data not shown).
In contrast, as shown in Figures 6A, B, and C, all pGlu-Aβ(3-42) ratios in CatB KO, E64d, and E64d CatB KO mice were significantly lower than the corresponding ratios in control mice. E64d CatB KO mice had zero pGlu-Aβ(3-42) measured, which was not significantly different from that of E6d and CatB KO mice (Bonferroni’s multiple comparison test). The pGlu-Aβ(3-42) ratios in the CatB TG and BACE1 KO mice did not significantly differ from the respective ratios in control mice.
Figure 6. Ratios of pGlu-Aβ(3-42) to flAβ(1-40/42) and pGlu-Aβ(3-40) were significantly lower in AβPPLon mice with CatB gene knockout or with E64d treatment relative to those in control mice.
Of all the ratios of Aβ species, only the pGlu-Aβ(3-42) ratios differed among the animal groups and those ratios are shown here. The E64d CatB KO mouse data is in the far right column position, but because the pGlu-Aβ(3-42) level was zero, all the pGlu-Aβ(3-42) ratios were zero for those animals and thus no column appears at that position.
A. pGlu-Aβ(3-42)/flAβ(1-42). This ratio was about 3.7 – 4.2 fold lower in CatB KO, E64d mice, and E64d CatB KO mice as compared to that ratio in control mice. The ratio did was not significantly different for the BACE1 KO and CatB TG mice relative to control mice.
B. pGlu-Aβ(3-42)/flAβ(1-40). This ratio was about 4.5 - 5 fold lower in the CatB KO, E64d, and E64d CatB KO mice than control mice. There were no significant difference in the ratio of the BACE1 KO and CatB TG relative to control mice but the CatB TG mice tended to have a higher ratio.
C. pGlu-Aβ(3-42)/pGlu-Aβ(3-40). This ratio was a significant 4.8 -5 times lower in the CatB KO, E64d and E64d CatB KO mice relative to control mice. There was no significant difference in this ratio in the BACE1 KO and CatB TG relative to that of the control mice but the CatB mice tended to have a higher ratio.
(ANOVA, one way analysis of variance, p < 0.0001, Dunnett’s multiple comparison to control using standard deviations of the ratios, *p < 0.05)
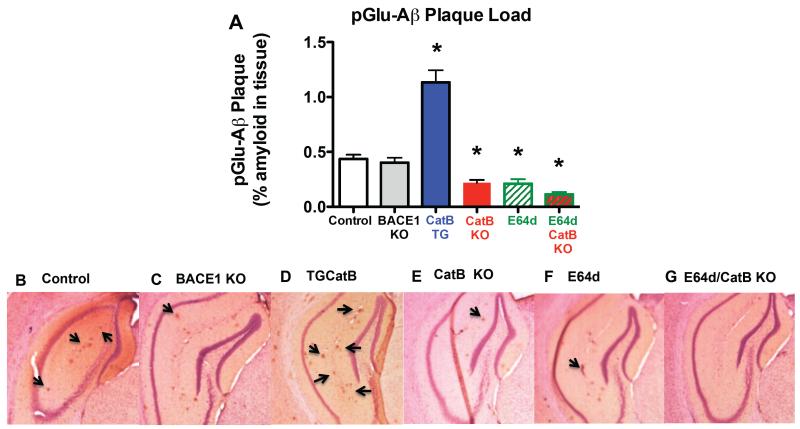
Brain pGlu-Aβ amyloid plaque load was about 50% lower due to eliminating or inhibiting CatB activity, about 175% higher due to increased CatB activity, and not significantly affected by eliminating BACE1 activity
Figure 7A shows the quantitative immunohistological analyses of pGlu-Aβ plaque load in brain sections from the animal groups studied. The antibody used to identify brain pGlu-Aβ plaque load recognized the N-terminus of pGlu-Aβ and, thus, detected both pGlu-Aβ(3-40) and pGlu-Aβ(3-42) but not flAβ.
Figure 7. pGlu-Aβ amyloid plaque load was about 50% lower in CatB KO, E64d, and E64d CatB KO mice, much higher in CatB TG mice, and the same in BACE1 KO mice.
A. Quantitation of pGlu-Aβ amyloid plaque load. Quantitative image analysis data are shown for immunohistological brain sections analyzed for plaques detected with anti-pGlu-Aβ antibody that recognizes both pGlu-Aβ(3-40) and pGlu-Aβ(3-42) in all animal groups. The pGlu-Aβ plaque load was significantly lower in CatB KO, E64d, and E64d CatB KO mice and significantly higher in CatB TG animals, but the same in BACE1 KO mice relative to that in control mice. (ANOVA, one way analysis of variance, p < 0.0001, Dunnett’s multiple comparison to control, *p < 0.05)
B-G. Exemplary micrographs of brain sections. Representative micrographs of the hippocampus from control (B), BACE1 KO (C), CatB TG (D), CatB KO (E), E64d (F), and E64d CatB KO mice (G) are shown. Arrows point to examples of individual pGlu-Aβ plaque deposits within the section.
The data show that the brain pGlu-Aβ plaque load was 46% lower in the E64d and CatB KO mice, and 61% lower in the E64d CatB KO mice relative to control. There was no significant difference in the levels among the three experimental groups (Bonferroni’s multiple comparison test). Thus, endogenous CatB activity controls brain pGlu-Aβ plaque load levels and E64d reduced those levels by inhibiting CatB activity in this model.
Figure 7A also shows that the brain pGlu-Aβ plaque load was 178% higher in the CatB TG mice relative to controls. Thus exogenous CatB activity controlled brain pGlu-Aβ plaque load in this model.
However, Figure 7A also shows that BACE1 KO mice had statistically the same brain pGlu-Aβ plaque load as control. Thus endogenous BACE1 activity did not affect brain pGlu-Aβ plaque load.
Figures 7B-G show exemplary brain micrographs from control, BACE1 KO, CatB TG, CatB KO, E64d, and E64d/CatB KO mice, respectively. The quantitative data was obtained over entire brain sections, whereas the exemplary micrographs display just the hippocampus region of the brain to clearly display pGlu-Aβ plaques.
Summary of the AβPPLon mice data
Table 1 summarizes the mean and SEM values for the animal groups and parameters analyzed.
Table 1.
Quantitative Summary of the AβPPLon Mouse Data
| Mouse model |
Chow | Mean (SEM) | ||||
|---|---|---|---|---|---|---|
| pGlu- Aβ(3-40) (ng/g) |
flAβ(1-40) (ng/g) |
pGlu- Aβ(3-42) (ng/g) |
flAβ(1-42) (ng/g) |
pGlu-Aβ plaque load (% area) |
||
| AβPPLon (control) |
Normal | 23.4 (2.4) | 72.9 (4.4) | 13.4 (1.7) | 25.9 (3.4) | 0.41 (0.05) |
| BACE1 KO AβPPLon |
Normal | 25.0 (3.4) | 79.5 (6.4) | 14.3 (1.8) | 28.9 (3.1) | 0.40 (0.05) |
| CatB TG AβPPLon |
Normal | 35.9 (4.9) | 115.6 (9.2) | 25.9 (1.9) | 51.0 (4.0) | 1.14 (0.11) |
| CatB KO AβPPLon |
Normal | 8.4 (1.4) | 31.0 (4.3) | 1.1 (0.4) | 7.5 (1.7) | 0.22 (0.02) |
| AβPPLon | E64d | 15.4 (2.7) | 32.3 (3.3) | 1.5 (0.5) | 12.8 (1.2) | 0.21 (0.04) |
| CatB KO AβPPLon |
E64d | 8.6 (1.3) | 17.9 (15.6) | 0.0 (0.0) | 6.7 (1.4) | 0.11 (0.02) |
This table shows the mean values, and SEM, for the various species of pGlu-Aβ and flAβ peptides measured in the different transgenic AβPPLon mouse models studied here (Control, BACE1 KO, CatB TG, CatB KO, E64d, and E64d CatB KO mice).
Chromaffin cell results
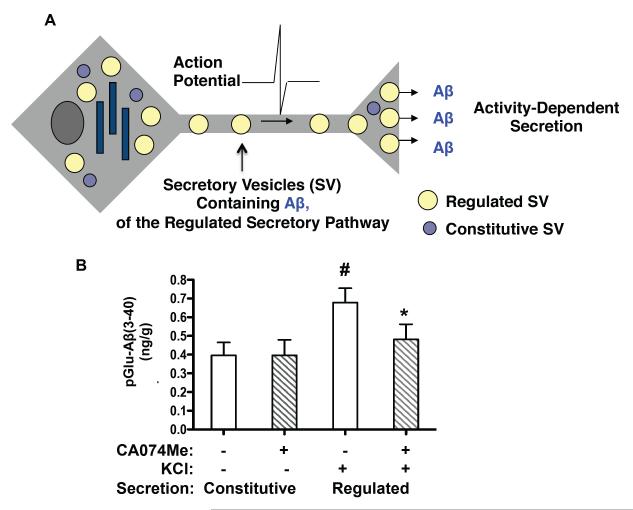
The majority of pGlu-Aβ(3-40) is released via the activity-dependent regulated secretory pathway, and CatB controls pGlu-Aβ(3-40) in that pathway in neuronal-like chromaffin cells
As illustrated in Figure 8A, Aβ may be secreted by neurons via constitutive or activity-dependent regulated secretion. The regulated secretory pathway of neurons provides activity-dependent release of neurotransmitters [68-73]. Notably, neural activity modulates the formation, secretion, and deposition of Aβ [55, 56]. Thus, pGlu-Aβ secretion by these two pathways was investigated in neuronal-like chromaffin cells (bovine) in primary cultures. As shown in Figure 8B, the chromaffin cells produce and release pGlu-Aβ(3-40) by constitutive and regulated secretory pathways. Constitutive secretion is represented by basal secretion, and activity-dependent regulated secretion is modeled by high KCl that depolarizes the cell and stimulates regulated secretion. While both pathways secrete pGlu-Aβ(3-40), significantly more is released by KCl stimulation of the regulated secretory pathway. Thus, the regulated secretory pathway produces most of the released pGlu-Aβ(3-40) in this model.
Figure 8. The CatB inhibitor CA-074Me reduces pGlu-Aβ(3-40) generated in the regulated secretory pathway of neuronal-like chromaffin cells.
A. Illustration of neuronal activity-dependent regulated secretion and constitutive (basal) secretion. Regulated and constitutive secretory pathways represent two distinct secretory vesicle systems in which neurons produce bioactive molecules [68-73]. Notably activity-dependent secretion of neurotransmitters and bioactive molecules utilize the regulated secretory pathway, composed of regulated secretory vesicles that release molecules in an activity-dependent manner [68-73]. Both pathways are illustrated.
B. pGlu-Aβ(3-40) is produced and secreted from the regulated secretory pathway, and is reduced by CA074Me, an inhibitor of CatB. In cultured neurons, activity-dependent secretion is modeled by KCl depolarization (high KCl in the medium). Constitutive, basal secretion was conducted without KCl. Neuronal-like chromaffin cells (in primary culture, prepared from the sympathetic nervous system) were utilized to assess pGlu-Aβ released from the regulated and constitutive secretory pathways, and to assess the effects of the CatB inhibitor CA074Me.
KCl significantly stimulated the regulated secretion of pGlu-Aβ(3-40) by ~2-fold above basal constitutive secretion (open bars). Importantly, treatment of cells with CA074Me (striped bars) reduced the amount of pGluAβ(3-40) in the regulated compared to the constitutive secretory pathway. (Statistically significant for KCl compared to no KCl, student’s t-test, #p < 0.05; statistically significant for CA074Me and KCl condition, compared to KCl condition, student’s t-test, * p < 0.05)
The role of CatB in pGlu-Aβ(3-40) production via the constitutive and regulated secretory pathways was investigated in the chromaffin cell model using the CatB inhibitor CA074Me. Figure 8B shows that CA074Me treatment of chromaffin cells reduced the amount of pGlu-Aβ(3-40) released from the regulated secretory pathway. CA074Me treatment had no effect on levels of constitutively secreted pGlu-Aβ(3-40) (released in the absence of KCl). It is noted that levels of pGlu-Aβ(3-42) were too low to detect in the secretion media, although it can be measured in purified secretory vesicles of chromaffin cells [60]. The data show that the constitutive and regulated secretory pathways produce pGlu-Aβ(3-40) but that the regulated pathway produced substantially more. Further, CatB inhibition essentially eliminated pGlu-Aβ(3-40) released from the regulated secretory pathway, but has no effect on constitutively secreted pGlu-Aβ(3-40) levels in this model. Thus, these data argue that CatB only produces pGlu-Aβ(3-40) in the regulated secretory pathway of neuronal cells.
Discussion
Important findings of this study are that the CatB gene deletion in AβPPLon mice eliminated over 90% of the brain pGlu-Aβ(3-42) and two thirds of the brain pGlu-Aβ(3-40) relative to sufficient AβPPLon mice. Thus, these data firmly establish that endogenous CatB can control pGlu-Aβ(3-40/42) levels in vivo. Complementing those results are the CatB TG mouse data, which show that overexpressing CatB doubled brain pGlu-Aβ(3-42) and increased pGlu-Aβ(3-40) by 50% relative to controls, demonstrating that exogenous CatB activity can also drive pGlu-Aβ(3-40/42) levels in this model. N-truncated and modified pGlu-Aβ peptides, especially pGlu-Aβ(3-42), are key for the initiation of Aβ oligomerization leading to neurodegeneration and memory deficits in AD [17-19]. This hypothesis is reinforced by our earlier results showing that CatB knockout in these same AβPPLon mice resulted in substantial improvement in memory deficits [50]. These data support CatB as a target for reducing pGlu-Aβ neurotoxic peptides, especially pGlu-Aβ(3-42), and brings to the field a new therapeutic target to effectively treat AD. To the best of our knowledge, CatB is the only protease target shown to affect pGlu-Aβ.
The E64d mouse data demonstrated the value of the CatB target for reducing brain pGlu-Aβ because those mice had 80% lower brain CatB activity, 90% less brain pGlu-Aβ(3-42), and one third less brain pGlu-Aβ(3-40) relative to that in the same transgenic mice fed normal chow. E64d treatment of CatB KO mice completely eliminated pGlu-Aβ(3-42) and further reduced pGlu-Aβ(3-40) to the level of CatB KO mice, which was two thirds lower than control mice. While E64d inhibits all papain-like cysteine proteases and calcium-activated neutral proteases [65], these data show that E64d acts primarily through inhibiting CatB because E64d CatB KO mice had equivalent levels of pGlu-Aβ(3-42) and pGlu-Aβ(3-40) as that found in CatB KO mice.
While deleting the CatB gene eliminated most of the brain pGlu-Aβ, CatB KO mice nonetheless had low residual brain pGlu-Aβ levels. Thus, other proteases may also produce pGlu-Aβ. As discussed below, BACE1 was not that other protease because the BACE1 KO mice had the same pGlu-Aβ(3-40/42) levels as control mice in the AβPPLon mouse model. E64d treatment of the CatB KO mice eliminated pGlu-Aβ(3-42) but caused no further reduction in pGlu-Aβ(3-40) beyond that achieved in the CatB KO mice, which suggests that the other proteases may be different for pGlu-Aβ(3-42) and pGlu-Aβ(3-40). For pGlu-Aβ(3-42), cathepsin L and cathepsin S could be candidates because E64d inhibits these papain-like cysteine proteases and they have very efficient wt β-secretase activity [40]. A candidate for pGlu-Aβ(3-40), on the other hand, may be meprin β because it is a metalloprotease not likely inhibited by E64d, has wt β-secretase activity, and produces N-truncated Aβ(2-40), which could be an intermediate in pGlu-Aβ(3-40) production [74].
Brain Aβ amyloid plaque is a hallmark of AD. CatB KO and CatB TG mice had 46% less and 178% more brain pGlu-Aβ plaque load, respectively, than control mice. Our previous study showed that deleting the CatB gene in AβPPLon mice reduced flAβ plaque load by 78% relative to controls [50]. CatB is the only protease drug target we know of that has been shown to affect both brain pGlu-Aβ and flAβ plaque load and, thus, is a drug target with new AD therapeutic potential.
Since both E64d and E64d CatB KO mice had about half the pGlu-Aβ plaque load of control mice, these data suggest that E64d acts primarily by inhibiting CatB activity to reduce pGlu-Aβ plaque load. We previously showed that E64d treatment of AβPPLon mice also reduced flAβ plaque load by about half relative to control mice [63]. Since E64d has been shown to reduce both pGlu-Aβ and flAβ plaque loads, it is a potential AD therapeutic having new capabilities for reducing this hallmark of AD pathology.
Overexpression of the CatB gene in AβPPLon mice doubled brain flAβ(1-42) levels and increased by more than 50% flAβ(1-40) levels relative to controls. That shows exogenous CatB activity can drive flAβ(1-40/42) production in this model. These data and the data showing that deleting the CatB KO gene reduces flAβ(1-40/42) levels are consistent with the hypothesis that CatB produces flAβ. These data showing that CatB overexpression increased flAβ further establishes that CatB can produce flAβ.
The E64d mice had less than half the brain flAβ(1-40/42) levels of control mice, which reproduced our earlier work showing that the same treatment of AβPPLon mice produced similar reductions in flAβ(1-40/42) [63] and that E64d treatment of guinea pigs, which are a natural model of human AβPPwt processing [75], also had similar reductions in brain flAβ(1-40/42) levels as compared to untreated animals [63, 76]. The new data here is that E64d CatB KO mice had the same low levels of flAβ(1-40/42) as in E64d or CatB KO mice, showing that E64d likely acts by inhibiting CatB to reduce flAβ(1-40/42). Recently, others have shown that treating nontransgenic senescence-accelerated mice, which have AβPP containing the wt β-secretase site, with an extract of Ginkgo biloba leaves, bilobalide, also reduced brain flAβ through CatB β-secretase inhibition [77].
The flAβ(1-42) to flAβ(1-40) ratio did not significantly change as a result of eliminating, inhibiting, or overexpressing CatB activity, which suggests that CatB activity may affect both flAβ species equally. The ratio of flAβ(1-42) to flAβ(1-40) is known to increase with age in AβPPLon mice [78] and, although different extraction methods were used, a similar ratio as that found here was previously reported for brain flAβ from AβPPLon mice based interpolation of the data [78].
On the other hand, CatB elimination or inhibition significantly lowered the ratios of pGlu-Aβ(3-42) to flAβ(1-42), flAβ(1-40), and pGlu-Aβ(3-40) relative to the corresponding ratios in the control mice, suggesting that endogenous CatB activity may differentially affect pGlu-Aβ(3-42) to a larger extent than the other species. Overexpressing CatB did not result in a significant difference in those ratios relative to that of control mice although the pGlu-Aβ(3-42) to flAβ(1-40) and pGlu-Aβ(3-40) ratios tended to be higher than that of controls. Further research will be required to resolve if there is a differential effect on pGlu-Aβ(3-42) due to exogenous CatB activity. The pGlu-Aβ(3-40) to flAβ(1-40/42) ratios, in contrast, were equivalent among all animal groups, suggesting the differential effect of CatB elimination or inhibition on pGlu-Aβ(3-42) may be specific to this pGlu-Aβ species.
The ratio of pGlu-Aβ(3-42) to flAβ(1-42) found in control mice here is much greater than previously reported (by about 15- to 60-fold depending on extraction conditions) for AβPPLon mice of approximately the same 12 month age [51]. These differences may be due to the different Aβ extraction methods and analytical antibodies used in the two studies. For example, guanidine Aβ extraction was used here whereas SDS or formic acid extraction was used in the prior study [51]. Also, the previous study used a pan-antibody detecting all Aβ(x-42) species, whereas this study used a specific antibody which detected only flAβ(1-42). The pan-antibody of the previous study detected much more flAβ than we did here and thus the resulting ratio of pGlu-Aβ(3-42) to flAβ was much lower in the previous study [51] than that found here.
Turning to the chromaffin cell studies, the data showed that pGlu-Aβ(3-40) undergoes constitutive and activity-dependent regulated secretion, modeled by KCl depolarization (high KCl in the media) to stimulate regulated secretion. Constitutive secretion (no KCl) provided a low amount of pGlu-Aβ(3-40) released to the media, whereas activity-dependent secretion caused significantly higher pGlu-Aβ(3-40) media levels. CA074Me treatment abolished levels of activity-dependent pGlu-Aβ(3-40) released, but had no effect on constitutive levels secreted; these data suggest that CatB may produce pGlu-Aβ(3-40) in the regulated but not constitutive secretory pathway. Neuronal-like chromaffin cells (bovine) are an excellent model of human AβPPwt processing because they express AβPP-695 having the wt β-secretase and wt Aβ sequences identical to that in man [61, 62]. These chromaffin cell data, which are from an entirely natural model of human AβPPwt processing, are consistent with that we obtained here in the unnatural transgenic AβPPLon mouse model. Thus, evidence from independent natural and artificial transgenic models support the hypothesis that CatB controls pGlu-Aβ and flAβ production.
Chromaffin cells are a predictive model of brain neuronal flAβ secretion because the CatB control of flAβ we showed to occur in the regulated secretory pathway of chromaffin cells [35] was subsequently found by others to also occur in brain neurons [59]. Inhibition of CatB reduced flAβ secretion via the regulated secretory pathway in chromaffin cells [35], as did CatB inhibition or Cat B siRNA silencing in rodent brain neurons [59]. Thus, these data suggest that CatB may control pGlu-Aβ and flAβ production in the regulated secretory pathway of brain neurons, which is consistent with brain studies showing that neural activity modulates Aβ secretion and deposition [55, 56].
The BACE1 gene knockout, on the other hand, had no significant effect on pGlu-Aβ(3-40/42) in AβPPLon mice, which argues that BACE1 may not produce pGlu-Aβ. That is consistent with reports showing that BACE1 inhibitor treatment of SH-SY5Y cells expressing human AβPPwt had no effect on N-terminal truncated Aβ(2-40) or Aβ(3-40) [79] and that BACE1 did not produce pGlu-Aβ(3-x) in kidney cells expressing human AβPPwt [80]. These data suggest that BACE1 inhibitors may not reduce pGlu-Aβ in most AD patients, who have wt β-secretase AβPP processing. That may be a significant limitation to BACE1 inhibitor efficacy given the pathological potential of pGlu-Aβ.
In that regard, an important question is whether pGlu-Aβ is actively produced or a byproduct of extracellular Aβ plaque aging because, if pGlu-Aβ is actively produced, it is more likely to be causing AD than if it is merely a result of plaque aging. Strong support for active pGlu-Aβ production came from the report showing that passive immunization with an anti-pGlu-Aβ antibody during development prevented pGlu-Aβ plaque deposition in young transgenic AD mice [30]. However, another study using different transgenic AD mice found that such treatment did not prevent pGlu-Aβ plaque deposition in young transgenic AD mice (although it did remove it from old mice) from which the by-product theory was hypothesized [31]. The data here clearly show that pGlu-Aβ levels can be regulated in CatB in transgenic mice and cell cultures and, thus, are direct evidence that pGlu-Aβ is actively produced and not merely a by-product of extracellular Aβ aging.
Our hypothesis is that CatB may control pGlu-Aβ by cleaving the wt β-secretase site in AβPP to produce flAβ, which is subsequently processed to pGlu-Aβ. Data has shown that CatB has wt β-secretase activity and readily cleaves the AβPP sequence, SEVK↓M↓D↓AE, at the positions marked by the arrows and produces flAβ [35, 40, 81]. The Aβ species resulting from those cleavages may serve as intermediates in the biosynthesis of pGlu-Aβ(3-40/42) as they could subsequently undergo N-terminal truncation by one or more proteases yet to be identified to form Aβ(3-40/42) species followed by QC conversion to pGlu-Aβ(3-40/42).
Differences in the β-secretase site sequence in AβPP transgenes used in different models may explain some apparently contradictory results in the literature regarding CatB AβPP processing. For example, others found slightly increased flAβ(1-42) resulted from deleting the CatB gene in mice expressing the AβPPSwe transgene [82] whereas we showed here and previously that the CatB deletion caused large reductions in flAβ(1-40/42) in transgenic AβPPLon and AβPPwt mice [49, 50]. That discrepancy may be resolved by the fact that CatB does not cleave the Swe β-secretase site sequence [36] and, thus, deleting the CatB gene in transgenic mouse models expressing AβPPSwe does not reduce flAβ(1-40/42) as has been shown by us and others [49, 50, 82]. But CatB readily cleaves the wt β-secretase site sequence [36] and, thus, its deletion in transgenic models expressing AβPPLon or AβPPwt, which have the wt β-secretase site, caused large reductions in flAβ [49, 50]. Since most AD patients have the wt β-secretase activity, CatB is an important therapeutic target for lowering flAβ.
Others have also shown that activation of CatB with Z-Phe-Ala-diazomethylketone (PADK), which is a non-specific lysosomal protease activator, in transgenic AβPPSwe/Ind and AβPPSwe/PS1ΔE9 mice reduces flAβ [83], whereas we show that specific overexpression of CatB increases flAβ in AβPPLon mice. That difference can also be explained by the differences in the ability of CatB to cleave Swe and wt β-secretase sites, as discussed above. Thus, the PADK activation of CatB did not produce more flAβ because the transgenic animals expressed AβPP containing the Swe mutation, which CatB does not cleave. The reduction in flAβ caused by PADK in those animals is likely not due to CatB because CatB knockout in the AβPPSwe/Ind mouse model (used in the PADK study) had no effect on flAβ levels or β-secretase biomarkers relative to controls [49]. Thus, the reduction in flAβ caused by PADK is likely due to other targets than CatB, which PADK is known to have [84].
As discussed above, AβPP-695 is the major AβPP isoform in human brain and is primarily expressed in neurons [44-48]. However, AβPP-751 and AβPP-770 isoforms are also present in human brain but in lower and trace amounts, respectively, and are primarily expressed in glia cells and not in neurons [44]. AβPP-751 and AβPP-770 structurally differ from AβPP-695 by containing the kunitz protease inhibitor (KPI) domain and with AβPP-770 also containing the MRC and OX-2 domains, all of which are functional domains that are not present in AβPP-695 [44-48]. The AβPPLon mouse model used here expressed AβPP-695 and not AβPP-751 or AβPP-770. Table 2 summarizes these aspects of the three AβPP isoforms.
Table 2.
Comparison of AβPP Isoforms 695, 751, and 770
| AβPP-695 | AβPP -751 | AβPP-770 | |
|---|---|---|---|
| Relative Brain Levels | Major | Minor | Trace |
| Neuronal Expression | Major | Minor | Minor |
| Glial Expression | Minor | Major | Major |
| KPI Domain | No | Yes | Yes |
| MRC & OX-2 Domains | No | No | Yes |
The differences in AβPP isoforms used in different models may resolve apparent conflicts in data obtained from overexpression of CatB activity in different transgenic mice. Others have shown that transgenic mice expressing PDGF-AβPP, which contains the wt β-secretase site sequence (I63 mice), had slightly reduced flAβ(1-42) as a result of CatB overexpression [85]; but we show here that CatB overexpression in AβPPLon mice dramatically increased flAβ(1-40/42). The difference in that prior study [85] is that the PDGF-AβPP mice expressed the AβPP-751 and AβPP-770 isoforms (AβPP-751/770) in brain [47, 86], but in this study the AβPPLon mice expressed AβPP-695, the major AβPP isoform present in human brain neurons [44-48]. In the prior study [85], the altered APP gene structure of the PDGF-AβPP construct had deletions in introns 6 and 8 as well as insertion of four nucleotides in intron 7, which resulted in unusual mRNA splicing to result in AβPP-751/770 containing the KPI domain (exon 7) [47], which contrasts with normal neurons that produce AβPP-695 (lacking the KPI domain) by microRNA mechanisms that allow RNA splicing to produce AβPP-695 [47, 87]. These studies represent neuronal expression since the PDGF promoter was utilized in our study and the other study [85]. This study shows that CatB expression in mice expressing AβPP-695, the main AβPP in human brain neurons [44-48], resulted in increased flAβ(1-40/42).
A similar analysis could also explain apparent conflicting BACE1 knockout data. Others have reported that deleting the BACE1 gene caused a decrease in flAβ in PDGF-AβPP [88] or AβPP51/16 mice [89], which express AβPP-751/770 and AβPP-751, respectively, containing the wt β-secretase site sequence, whereas we show here and previously that the BACE1 gene deletion in AβPPLon mice expressing AβPP-695 had no effect on flAβ [49, 50]. Clearly, there are differences in PDGF-AβPP and AβPP51/16 mouse models expressing AβPP-751/770 [88, 89] compared to our study here of AβPPLon mice expressing AβPP-695, the major isoform of AβPP expressed in human brain [44-48].
In wt mice (not expressing a transgenic AβPP transgene), a group reported that BACE1 KO significantly reduced flAβ(1-40) in mouse brain [90], but that same group also reported that BACE1 overexpression had no effect on mouse flAβ(1-40) from which they concluded that BACE1 has a minimal effect on flAβ levels and that other factors modulate flAβ levels [91]. However, BACE1 gene expression in transgenic mice expressing human APP[V717I] resulted in elevation of Aβ species [92]. Studies of BACE1 KO in wt mice (no AβPP transgene) have reported the absence of wt β-secretase activity [93]; however, the β-secretase assay used in that study contained E64, which inhibits CatB activity, and thus those data do not exclude CatB as also having wt β-secretase activity. Brain neurons from BACE1 KO wt mice have been shown to produce less flAβ(1-40/42) in “conditioned media” relative to neurons of sufficient animals [93, 94]. However, “conditioned media” reflects constitutive and not regulated flAβ secretion and, therefore, these data also do not exclude CatB production of flAβ via regulated neuronal cell secretion. Thus, it may be that BACE1 and CatB both produce flAβ, BACE1 possibly doing so via the constitutive secretory pathway and CatB could do so via the regulated secretory pathway.
Much more effort has been given to developing BACE1 inhibitors than CatB inhibitors to reduce flAβ. As a result, BACE1, but not CatB, inhibitors are now in the clinic where they have been shown to reduce cerebrospinal fluid flAβ(1-40/42) in volunteers. However, there is a real risk that BACE1 inhibitors may have insufficient efficacy or excessive toxicity. As shown here, BACE1 does not seem to control pGlu-Aβ, which may be the Aβ species causing the disease. Thus, BACE1 inhibitors may not succeed because they may fail to lower pGlu-Aβ. And toxicity issues may limit clinical use of BACE1 inhibitors. Two major BACE1 inhibitor clinical trials have been stopped due to toxicity problems [95, 96]. And while it is not clear whether the toxicity is due to on or off BACE1 target effects, BACE1 is a problematic target because animals lacking BACE1 develop neuropathology and muscle pathology [97-100].
CatB inhibitors on the other hand, are potentially more effective and a safer alternative to BACE1 inhibitors. As shown here, CatB inhibitors reduced both flAβ and pGlu-Aβ and, thus, will likely be effective if either of these Aβ species cause the disease. Moreover, CatB inhibitors are potent neuroprotectants in models of traumatic brain injury and ischemia, which are risk factors for AD [101, 102]. And CatB knockout studies show that the absence of CatB prevents tumor necrosis factor (TNF) induced cell death and interleukin-1β (IL1β) inflammation, both of which occur in AD [103-107]. Thus, CatB inhibitor therapeutics would reduce flAβ and pGlu-Aβ, as well as provide neuroprotection and reduce apoptosis and inflammation.
CatB knockout mice are normal and indistinguishable from sufficient littermates [108] and thus the CatB as a drug target does not seem to pose the toxicity target risks of BACE1. Importantly, E64d has demonstrated to be safe to use in man. Originally developed in the 1980’s for treatment of muscular dystrophy, E64d (also known as EST and Loxistatin) completed chronic Phase III trials but did not advance as a drug because of insufficient efficacy for treating muscular dystrophy [109]. Nonetheless, published clinical and toxicology data show that E64d was safe during administration to adult volunteers and pediatric patients on a chronic basis without adverse events and has a wide therapeutic window [110-124]. Thus, CatB inhibitors, generally, and E64d in particular have theoretical and demonstrated safety advantages over BACE1 inhibitors.
In summary, this study supports CatB as a protease target, which produces brain pGlu-Aβ(3-40/42), illustrated by CatB gene knockout and expression. Importantly, pharmacological inhibition of CatB by E64d reduces pGlu-Aβ(3-42) as well as flAβ(1-40/42). Inhibitors of CatB, proposed as an alternative β-secretase, have potential as new AD therapeutics based on their ability to reduce both the pGlu-Aβ and flAβ produced from AβPP containing the wt β-secretase site expressed in the majority of AD patients.
Acknowledgements
The authors wish to thank Christoph Peters, MD, of the Albert Ludwig University, Freiburg, Germany for providing the cathepsin B deficient mice and to Steven Jacobsen at AstraZeneca Neuroscience for his critical thinking and insightful comments on the work. This work was supported in part by grants from the NIH, composed of R44AG032784 to American Life Science Pharmaceuticals (ALSP) (GH), R01ES016774-02 (MSK), and R21AG0428 (VH), as well as a VA Merit Review 1I01RX000331-01 (MSK) and an Alzheimer’s Association award (VH). G. Hook has equity interest in ALSP. V. Hook is the Chair of ALSP’s Scientific Advisory Board and holds equity in ALSP, and the relationship has been disclosed to the Univ. of Calif., San Diego.
References
- [1].Selkoe DJ. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- [2].Gandy S, Martins RN, Buxbaum J. Molecular and cellular basis for anti-amyloid therapy in Alzheimer disease. Alzheimer Dis Assoc Disord. 2003;17:259–266. doi: 10.1097/00002093-200310000-00011. [DOI] [PubMed] [Google Scholar]
- [3].Crews L, Masliah E. Molecular mechanisms of neurodegeneration in Alzheimer’s disease. Hum Mol Genet. 2010;19:R12–20. doi: 10.1093/hmg/ddq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].De Kimpe L, Scheper W. From alpha to omega with Abeta: targeting the multiple molecular appearances of the pathogenic peptide in Alzheimer’s disease. Curr Med Chem. 2010;17:198–212. doi: 10.2174/092986710790149765. [DOI] [PubMed] [Google Scholar]
- [6].Mori H, Takio K, Ogawara M, Selkoe DJ. Mass spectrometry of purified amyloid beta protein in Alzheimer’s disease. J Biol Chem. 1992;267:17082–17086. [PubMed] [Google Scholar]
- [7].Saido TC, Iwatsubo T, Mann DM, Shimada H, Ihara Y, Kawashima S. Dominant and differential deposition of distinct beta-amyloid peptide species, A beta N3(pE), in senile plaques. Neuron. 1995;14:457–466. doi: 10.1016/0896-6273(95)90301-1. [DOI] [PubMed] [Google Scholar]
- [8].Iwatsubo T, Saido TC, Mann DM, Lee VM, Trojanowski JQ. Full-length amyloid-beta (1-42(43)) and amino-terminally modified and truncated amyloid-beta 42(43) deposit in diffuse plaques. Am J Pathol. 1996;149:1823–1830. [PMC free article] [PubMed] [Google Scholar]
- [9].Lemere CA, Blusztajn JK, Yamaguchi H, Wisniewski T, Saido TC, Selkoe DJ. Sequence of deposition of heterogeneous amyloid beta-peptides and APO E in Down syndrome: implications for initial events in amyloid plaque formation. Neurobiol Dis. 1996;3:16–32. doi: 10.1006/nbdi.1996.0003. [DOI] [PubMed] [Google Scholar]
- [10].Russo C, Violani E, Salis S, Venezia V, Dolcini V, Damonte G, Benatti U, D’Arrigo C, Patrone E, Carlo P, Schettini G. Pyroglutamate-modified amyloid beta-peptides--AbetaN3(pE)--strongly affect cultured neuron and astrocyte survival. J Neurochem. 2002;82:1480–1489. doi: 10.1046/j.1471-4159.2002.01107.x. [DOI] [PubMed] [Google Scholar]
- [11].Harigaya Y, Saido TC, Eckman CB, Prada CM, Shoji M, Younkin SG. Amyloid beta protein starting pyroglutamate at position 3 is a major component of the amyloid deposits in the Alzheimer’s disease brain. Biochem Biophys Res Commun. 2000;276:422–427. doi: 10.1006/bbrc.2000.3490. [DOI] [PubMed] [Google Scholar]
- [12].Hosoda R, Saido TC, Otvos L, Jr., Arai T, Mann DM, Lee VM, Trojanowski JQ, Iwatsubo T. Quantification of modified amyloid beta peptides in Alzheimer disease and Down syndrome brains. J Neuropathol Exp Neurol. 1998;57:1089–1095. doi: 10.1097/00005072-199811000-00012. [DOI] [PubMed] [Google Scholar]
- [13].Piccini A, Russo C, Gliozzi A, Relini A, Vitali A, Borghi R, Giliberto L, Armirotti A, D’Arrigo C, Bachi A, Cattaneo A, Canale C, Torrassa S, Saido TC, Markesbery W, Gambetti P, Tabaton M. beta-amyloid is different in normal aging and in Alzheimer disease. J Biol Chem. 2005;280:34186–34192. doi: 10.1074/jbc.M501694200. [DOI] [PubMed] [Google Scholar]
- [14].Portelius E, Bogdanovic N, Gustavsson MK, Volkmann I, Brinkmalm G, Zetterberg H, Winblad B, Blennow K. Mass spectrometric characterization of brain amyloid beta isoform signatures in familial and sporadic Alzheimer’s disease. Acta Neuropathol. 2010;120:185–193. doi: 10.1007/s00401-010-0690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Saido TC, Yamao-Harigaya W, Iwatsubo T, Kawashima S. Amino- and carboxyl-terminal heterogeneity of beta-amyloid peptides deposited in human brain. Neurosci Lett. 1996;215:173–176. doi: 10.1016/0304-3940(96)12970-0. [DOI] [PubMed] [Google Scholar]
- [16].He W, Barrow CJ. The A beta 3-pyroglutamyl and 11-pyroglutamyl peptides found in senile plaque have greater beta-sheet forming and aggregation propensities in vitro than full-length A beta. Biochemistry. 1999;38:10871–10877. doi: 10.1021/bi990563r. [DOI] [PubMed] [Google Scholar]
- [17].Schilling S, Lauber T, Schaupp M, Manhart S, Scheel E, Bohm G, Demuth HU. On the seeding and oligomerization of pGlu-amyloid peptides (in vitro) Biochemistry. 2006;45:12393–12399. doi: 10.1021/bi0612667. [DOI] [PubMed] [Google Scholar]
- [18].Schlenzig D, Manhart S, Cinar Y, Kleinschmidt M, Hause G, Willbold D, Funke SA, Schilling S, Demuth HU. Pyroglutamate formation influences solubility and amyloidogenicity of amyloid peptides. Biochemistry. 2009;48:7072–7078. doi: 10.1021/bi900818a. [DOI] [PubMed] [Google Scholar]
- [19].Nussbaum JM, Schilling S, Cynis H, Silva A, Swanson E, Wangsanut T, Tayler K, Wiltgen B, Hatami A, Ronicke R, Reymann K, Hutter-Paier B, Alexandru A, Jagla W, Graubner S, Glabe CG, Demuth HU, Bloom GS. Prion-like behaviour and tau-dependent cytotoxicity of pyroglutamylated amyloid-beta. Nature. 2012;485:651–655. doi: 10.1038/nature11060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, Selkoe DJ, Ashe KH. Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- [21].Klein WL. Synaptotoxic amyloid-beta oligomers: a molecular basis for the cause, diagnosis, and treatment of Alzheimer’s disease? J Alzheimers Dis. 2013;33(Suppl 1):S49–65. doi: 10.3233/JAD-2012-129039. [DOI] [PubMed] [Google Scholar]
- [22].Larson ME, Lesne SE. Soluble Abeta oligomer production and toxicity. J Neurochem. 2012;120(Suppl 1):125–139. doi: 10.1111/j.1471-4159.2011.07478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wittnam JL, Portelius E, Zetterberg H, Gustavsson MK, Schilling S, Koch B, Demuth HU, Blennow K, Wirths O, Bayer TA. Pyroglutamate amyloid beta (Abeta) aggravates behavioral deficits in transgenic amyloid mouse model for Alzheimer disease. J Biol Chem. 2012;287:8154–8162. doi: 10.1074/jbc.M111.308601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schlenzig D, Ronicke R, Cynis H, Ludwig HH, Scheel E, Reymann K, Saido T, Hause G, Schilling S, Demuth HU. N-Terminal pyroglutamate formation of Abeta38 and Abeta40 enforces oligomer formation and potency to disrupt hippocampal long-term potentiation. J Neurochem. 2012;121:774–784. doi: 10.1111/j.1471-4159.2012.07707.x. [DOI] [PubMed] [Google Scholar]
- [25].Wirths O, Breyhan H, Cynis H, Schilling S, Demuth HU, Bayer TA. Intraneuronal pyroglutamate-Abeta 3-42 triggers neurodegeneration and lethal neurological deficits in a transgenic mouse model. Acta Neuropathol. 2009;118:487–496. doi: 10.1007/s00401-009-0557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wirths O, Breyhan H, Schafer S, Roth C, Bayer TA. Deficits in working memory and motor performance in the APP/PS1ki mouse model for Alzheimer’s disease. Neurobiol Aging. 2008;29:891–901. doi: 10.1016/j.neurobiolaging.2006.12.004. [DOI] [PubMed] [Google Scholar]
- [27].Schilling S, Hoffmann T, Manhart S, Hoffmann M, Demuth HU. Glutaminyl cyclases unfold glutamyl cyclase activity under mild acid conditions. FEBS Lett. 2004;563:191–196. doi: 10.1016/S0014-5793(04)00300-X. [DOI] [PubMed] [Google Scholar]
- [28].Cynis H, Scheel E, Saido TC, Schilling S, Demuth HU. Amyloidogenic processing of amyloid precursor protein: evidence of a pivotal role of glutaminyl cyclase in generation of pyroglutamate-modified amyloid-beta. Biochemistry. 2008;47:7405–7413. doi: 10.1021/bi800250p. [DOI] [PubMed] [Google Scholar]
- [29].Schilling S, Zeitschel U, Hoffmann T, Heiser U, Francke M, Kehlen A, Holzer M, Hutter-Paier B, Prokesch M, Windisch M, Jagla W, Schlenzig D, Lindner C, Rudolph T, Reuter G, Cynis H, Montag D, Demuth HU, Rossner S. Glutaminyl cyclase inhibition attenuates pyroglutamate Abeta and Alzheimer’s disease-like pathology. Nat Med. 2008;14:1106–1111. doi: 10.1038/nm.1872. [DOI] [PubMed] [Google Scholar]
- [30].Frost JL, Liu B, Kleinschmidt M, Schilling S, Demuth HU, Lemere CA. Passive immunization against pyroglutamate-3 amyloid-beta reduces plaque burden in Alzheimer-like transgenic mice: a pilot study. Neurodegener Dis. 2012;10:265–270. doi: 10.1159/000335913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Demattos RB, Lu J, Tang Y, Racke MM, Delong CA, Tzaferis JA, Hole JT, Forster BM, McDonnell PC, Liu F, Kinley RD, Jordan WH, Hutton ML. A Plaque-Specific Antibody Clears Existing beta-amyloid Plaques in Alzheimer’s Disease Mice. Neuron. 2012;76:908–920. doi: 10.1016/j.neuron.2012.10.029. [DOI] [PubMed] [Google Scholar]
- [32].Jawhar S, Wirths O, Bayer TA. Pyroglutamate amyloid-beta (Abeta): a hatchet man in Alzheimer disease. J Biol Chem. 2011;286:38825–38832. doi: 10.1074/jbc.R111.288308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Vassar R. BACE1: the beta-secretase enzyme in Alzheimer’s disease. J Mol Neurosci. 2004;23:105–114. doi: 10.1385/JMN:23:1-2:105. [DOI] [PubMed] [Google Scholar]
- [34].Evin G, Barakat A, Masters CL. BACE: Therapeutic target and potential biomarker for Alzheimer’s disease. Int J Biochem Cell Biol. 2010;42:1923–1926. doi: 10.1016/j.biocel.2010.08.017. [DOI] [PubMed] [Google Scholar]
- [35].Hook V, Toneff T, Bogyo M, Medzihradszky K, F., Nevenu J, Lane W, Hook G, Reisine T. Inhibition of cathepsin B reduces ®-amyloid production in regulated secretory vesicles of neuronal chromaffin cells: evidence for cathepsin B as a candidate ®-secretase of Alzheimer’s disease. Biological Chemistry. 2005;386:931–940. doi: 10.1515/BC.2005.108. [DOI] [PubMed] [Google Scholar]
- [36].Hook VY, Kindy M, Hook G. Inhibitors of cathepsin B improve memory and reduce Abeta in transgenic Alzheimer’s Disease mice expressing the wild-type, but not the Swedish mutant, beta -secretase APP site. J Biol Chem. 2008;283:7745–7753. doi: 10.1074/jbc.M708362200. [DOI] [PubMed] [Google Scholar]
- [37].Ashe KH, Zahs KR. Probing the biology of Alzheimer’s disease in mice. Neuron. 2010;66:631–645. doi: 10.1016/j.neuron.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Masliah E, Rockenstein E. Genetically altered transgenic models of Alzheimer’s disease. J Neural Transm Suppl. 2000;59:175–183. doi: 10.1007/978-3-7091-6781-6_20. [DOI] [PubMed] [Google Scholar]
- [39].Schechter I, Ziv E. Kinetic properties of cathepsin D and BACE 1 indicate the need to search for additional beta-secretase candidate(s) Biol Chem. 2008;389:313–320. doi: 10.1515/BC.2008.025. [DOI] [PubMed] [Google Scholar]
- [40].Schechter I, Ziv E. Cathepsins S, B and L with aminopeptidases display beta-secretase activity associated with the pathogenesis of Alzheimer’s disease. Biological Chemistry. 2011;392:555–569. doi: 10.1515/BC.2011.054. [DOI] [PubMed] [Google Scholar]
- [41].Lin X, Koelsch G, Wu S, Downs D, Dashti A, Tang J. Human aspartic protease memapsin 2 cleaves the beta-secretase site of beta-amyloid precursor protein. Proc Natl Acad Sci U S A. 2000;97:1456–1460. doi: 10.1073/pnas.97.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gruninger-Leitch F, Schlatter D, Kung E, Nelbock P, Dobeli H. Substrate and inhibitor profile of BACE (beta-secretase) and comparison with other mammalian aspartic proteases. J Biol Chem. 2002;277:4687–4693. doi: 10.1074/jbc.M109266200. [DOI] [PubMed] [Google Scholar]
- [43].Bohme L, Hoffmann T, Manhart S, Wolf R, Demuth HU. Isoaspartate-containing amyloid precursor protein-derived peptides alter efficacy and specificity of potential beta-secretases. Biol Chem. 2008;389:1055–1066. doi: 10.1515/BC.2008.125. [DOI] [PubMed] [Google Scholar]
- [44].Rohan de Silva HA, Jen A, Wickenden C, Jen LS, Wilkinson SL, Patel AJ. Cell-specific expression of beta-amyloid precursor protein isoform mRNAs and proteins in neurons and astrocytes. Brain Res Mol Brain Res. 1997;47:147–156. doi: 10.1016/s0169-328x(97)00045-4. [DOI] [PubMed] [Google Scholar]
- [45].Kang J, Muller-Hill B. Differential splicing of Alzheimer’s disease amyloid A4 precursor RNA in rat tissues: PreA4(695) mRNA is predominantly produced in rat and human brain. Biochem Biophys Res Commun. 1990;166:1192–1200. doi: 10.1016/0006-291x(90)90992-v. [DOI] [PubMed] [Google Scholar]
- [46].Jacobsen JS, Blume AJ, Vitek MP. Quantitative measurement of alternatively spliced amyloid precursor protein mRNA expression in Alzheimer’s disease and normal brain by S1 nuclease protection analysis. Neurobiol Aging. 1991;12:585–592. doi: 10.1016/0197-4580(91)90090-7. [DOI] [PubMed] [Google Scholar]
- [47].Rockenstein EM, McConlogue L, Tan H, Power M, Masliah E, Mucke L. Levels and alternative splicing of amyloid beta protein precursor (APP) transcripts in brains of APP transgenic mice and humans with Alzheimer’s disease. J Biol Chem. 1995;270:28257–28267. doi: 10.1074/jbc.270.47.28257. [DOI] [PubMed] [Google Scholar]
- [48].Golde TE, Estus S, Usiak M, Younkin LH, Younkin SG. Expression of beta amyloid protein precursor mRNAs: recognition of a novel alternatively spliced form and quantitation in Alzheimer’s disease using PCR. Neuron. 1990;4:253–267. doi: 10.1016/0896-6273(90)90100-t. [DOI] [PubMed] [Google Scholar]
- [49].Hook VY, Kindy M, Reinheckel T, Peters C, Hook G. Genetic cathepsin B deficiency reduces beta-amyloid in transgenic mice expressing human wild-type amyloid precursor protein. Biochem Biophys Res Commun. 2009;386:284–288. doi: 10.1016/j.bbrc.2009.05.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kindy MS, Yu J, Zhu H, El-Amouri SS, Hook V, Hook GR. Deletion of the Cathepsin B Gene Improves Memory Deficits in a Transgenic Alzheimer’s Disease Mouse Model Expressing AbetaPP Containing the Wild-Type beta-Secretase Site Sequence. J Alzheimers Dis. 2012;29:827–840. doi: 10.3233/JAD-2012-111604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Tanghe A, Termont A, Merchiers P, Schilling S, Demuth HU, Scrocchi L, Van Leuven F, Griffioen G, Van Dooren T. Pathological Hallmarks, Clinical Parallels, and Value for Drug Testing in Alzheimer’s Disease of the APP[V717I] London Transgenic Mouse Model. Int J Alzheimers Dis. 2010;2010 doi: 10.4061/2010/417314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Moechars D, Dewachter I, Lorent K, Reverse D, Baekelandt V, Naidu A, Tesseur I, Spittaels K, Haute CV, Checler F, Godaux E, Cordell B, Van Leuven F. Early phenotypic changes in transgenic mice that overexpress different mutants of amyloid precursor protein in brain. J Biol Chem. 1999;274:6483–6492. doi: 10.1074/jbc.274.10.6483. [DOI] [PubMed] [Google Scholar]
- [53].Nitsch RM, Farber SA, Growdon JH, Wurtman RJ. Release of amyloid beta-protein precursor derivatives by electrical depolarization of rat hippocampal slices. Proc Natl Acad Sci U S A. 1993;90:5191–5193. doi: 10.1073/pnas.90.11.5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Jolly-Tornetta C, Gao ZY, Lee VM, Wolf BA. Regulation of amyloid precursor protein secretion by glutamate receptors in human Ntera 2 neurons. J Biol Chem. 1998;273:14015–14021. doi: 10.1074/jbc.273.22.14015. [DOI] [PubMed] [Google Scholar]
- [55].Bero AW, Yan P, Roh JH, Cirrito JR, Stewart FR, Raichle ME, Lee JM, Holtzman DM. Neuronal activity regulates the regional vulnerability to amyloid-beta deposition. Nat Neurosci. 2011;14:750–756. doi: 10.1038/nn.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kamenetz F, Tomita T, Hsieh H, Seabrook G, Borchelt D, Iwatsubo T, Sisodia S, Malinow R. APP processing and synaptic function. Neuron. 2003;37:925–937. doi: 10.1016/s0896-6273(03)00124-7. [DOI] [PubMed] [Google Scholar]
- [57].Toneff T, Funkelstein L, Mosier C, Abagyan A, Ziegler M, Hook V. Beta-amyloid peptides undergo regulated co-secretion with neuropeptide and catecholamine neurotransmitters. Peptides. 2013;46:126–135. doi: 10.1016/j.peptides.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Carmichael SW, Winkler H. The adrenal chromaffin cell. Sci Am. 1985;253:40–49. doi: 10.1038/scientificamerican0885-40. [DOI] [PubMed] [Google Scholar]
- [59].Klein DM, Felsenstein KM, Brenneman DE. Cathepsins B and L differentially regulate amyloid precursor protein processing. J Pharmacol Exp Ther. 2009;328:813–821. doi: 10.1124/jpet.108.147082. [DOI] [PubMed] [Google Scholar]
- [60].Cynis H, Funkelstein L, Toneff T, Mosier C, Ziegler M, Kotch B, Demuth HU, Hook V. PyroGlu-Abeta and glutaminy cyclase are co-localized with Abeta in secretory vesicles and undergo activity-dependent secretion. Neurodegenerative Diseases. 2014 doi: 10.1159/000358430. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Johnstone EM, Chaney MO, Norris FH, Pascual R, Little SP. Conservation of the sequence of the Alzheimer’s disease amyloid peptide in dog, polar bear and five other mammals by cross-species polymerase chain reaction analysis. Brain Res Mol Brain Res. 1991;10:299–305. doi: 10.1016/0169-328x(91)90088-f. [DOI] [PubMed] [Google Scholar]
- [62].Takeda M, Tanaka S, Kido H, Daikoku S, Oka M, Sakai K, Katunuma N. Chromaffin cells express Alzheimer amyloid precursor protein in the same manner as brain cells. Neurosci Lett. 1994;168:57–60. doi: 10.1016/0304-3940(94)90415-4. [DOI] [PubMed] [Google Scholar]
- [63].Hook G, Hook V, Kindy M. The Cysteine Protease Inhibitor, E64d, Reduces Brain Amyloid-beta and Improves Memory Deficits in Alzheimer’s Disease Animal Models by Inhibiting Cathepsin B, but not BACE1, beta-Secretase Activity. J Alzheimers Dis. 2011;26:387–408. doi: 10.3233/JAD-2011-110101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Tamai M, Yokoo C, Murata M, Oguma K, Sota K, Sato E, Kanaoka Y. Efficient synthetic method for ethyl (+)-(2S,3S)-3-[(S)-3-methyl- 1-(3-methylbutylcarbamoyl)butylcarbamoyl]-2-oxiranecarb oxylate (EST), a new inhibitor of cysteine proteinases. Chem Pharm Bull (Tokyo) 1987;35:1098–1104. [PubMed] [Google Scholar]
- [65].Tamai M, Matsumoto K, Omura S, Koyama I, Ozawa Y, Hanada K. In vitro and in vivo inhibition of cysteine proteinases by EST, a new analog of E-64. J Pharmacobiodyn. 1986;9:672–677. doi: 10.1248/bpb1978.9.672. [DOI] [PubMed] [Google Scholar]
- [66].Yamamoto A, Kaji T, Tomoo K, Ishida T, Inoue M, Murata M, Kitamura K. Crystallization and preliminary X-ray study of the cathepsin B complexed with CA074, a selective inhibitor. J Mol Biol. 1992;227:942–944. doi: 10.1016/0022-2836(92)90234-b. [DOI] [PubMed] [Google Scholar]
- [67].Towatari T, Nikawa T, Murata M, Yokoo C, Tamai M, Hanada K, Katunuma N. Novel epoxysuccinyl peptides. A selective inhibitor of cathepsin B, in vivo. FEBS Lett. 1991;280:311–315. doi: 10.1016/0014-5793(91)80319-x. [DOI] [PubMed] [Google Scholar]
- [68].Lodish H, Berk A, Matsudaira P, Kaiser C, Krieger M, Scott M. Molecular Cell Biology. W.H. Freeman and Company; New York: 2004. [Google Scholar]
- [69].Holz R, Fisher S. In: Synaptic transmission and cellular signaling: an overview In Basic neurochemistry, principles of molecular, cellular and medical neurobiology. Brady S, Siegel G, Albers RW, Price DL, editors. Academic Press; New York: 2012. pp. 235–258. [Google Scholar]
- [70].Gondre-Lewis MC, Park JJ, Loh YP. Cellular mechanisms for the biogenesis and transport of synaptic and dense-core vesicles. Int Rev Cell Mol Biol. 2012;299:27–115. doi: 10.1016/B978-0-12-394310-1.00002-3. [DOI] [PubMed] [Google Scholar]
- [71].Martin TF. Stages of regulated exocytosis. Trends Cell Biol. 1997;7:271–276. doi: 10.1016/S0962-8924(97)01060-X. [DOI] [PubMed] [Google Scholar]
- [72].Sollner T, Rothman JE. Neurotransmission: harnessing fusion machinery at the synapse. Trends Neurosci. 1994;17:344–348. doi: 10.1016/0166-2236(94)90178-3. [DOI] [PubMed] [Google Scholar]
- [73].Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- [74].Bien J, Jefferson T, Causevic M, Jumpertz T, Munter L, Multhaup G, Weggen S, Becker-Pauly C, Pietrzik CU. The metalloprotease meprin beta generates amino terminal-truncated amyloid beta peptide species. J Biol Chem. 2012;287:33304–33313. doi: 10.1074/jbc.M112.395608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Beck M, Bigl V, Rossner S. Guinea pigs as a nontransgenic model for APP processing in vitro and in vivo. Neurochem Res. 2003;28:637–644. doi: 10.1023/a:1022850113083. [DOI] [PubMed] [Google Scholar]
- [76].Hook V, Kindy M, Hook G. Cysteine protease inhibitors effectively reduce in vivo levels of brain beta-amyloid related to Alzheimer’s disease. Biol Chem. 2007;388:247–252. doi: 10.1515/BC.2007.027. [DOI] [PubMed] [Google Scholar]
- [77].Shi C, Zheng DD, Wu FM, Liu J, Xu J. The phosphatidyl inositol 3 kinaseglycogen synthase kinase 3beta pathway mediates bilobalide-induced reduction in amyloid beta-peptide. Neurochem Res. 2012;37:298–306. doi: 10.1007/s11064-011-0612-1. [DOI] [PubMed] [Google Scholar]
- [78].Dewachter I, Van Dorpe J, Smeijers L, Gilis M, Kuiperi C, Laenen I, Caluwaerts N, Moechars D, Checler F, Vanderstichele H, Van Leuven F. Aging increased amyloid peptide and caused amyloid plaques in brain of old APP/V717I transgenic mice by a different mechanism than mutant presenilin1. J Neurosci. 2000;20:6452–6458. doi: 10.1523/JNEUROSCI.20-17-06452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Schieb H, Weidlich S, Schlechtingen G, Linning P, Jennings G, Gruner M, Wiltfang J, Klafki HW, Knolker HJ. Structural design, solid-phase synthesis and activity of membrane-anchored beta-secretase inhibitors on Abeta generation from wild-type and Swedish-mutant APP. Chemistry. 2010;16:14412–14423. doi: 10.1002/chem.201002878. [DOI] [PubMed] [Google Scholar]
- [80].Huse JT, Liu K, Pijak DS, Carlin D, Lee VM, Doms RW. Beta-secretase processing in the trans-Golgi network preferentially generates truncated amyloid species that accumulate in Alzheimer’s disease brain. J Biol Chem. 2002;277:16278–16284. doi: 10.1074/jbc.M111141200. [DOI] [PubMed] [Google Scholar]
- [81].Hook VYH, Toneff T, Aaron W, Yasothornsrikul S, Bundey R, Reisine T. ®-Amyloid peptide in regulated secretory vesicles of chromaffin cells: evidence for multiple cysteine proteolytic activities in distinct pathways for ®-secretase activity in chromaffin vesicles. J Neurochem. 2002;81:237–256. doi: 10.1046/j.1471-4159.2002.00794.x. [DOI] [PubMed] [Google Scholar]
- [82].Mueller-Steiner S, Zhou Y, Arai H, Roberson ED, Sun B, Chen J, Wang X, Yu G, Esposito L, Mucke L, Gan L. Antiamyloidogenic and neuroprotective functions of cathepsin B: implications for Alzheimer’s disease. Neuron. 2006;51:703–714. doi: 10.1016/j.neuron.2006.07.027. [DOI] [PubMed] [Google Scholar]
- [83].Butler D, Hwang J, Estick C, Nishiyama A, Kumar SS, Baveghems C, Young-Oxendine HB, Wisniewski ML, Charalambides A, Bahr BA. Protective effects of positive lysosomal modulation in Alzheimer’s disease transgenic mouse models. PLoS One. 2011;6:e20501. doi: 10.1371/journal.pone.0020501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Zheng X, Gessel MM, Wisniewski ML, Viswanathan K, Wright DL, Bahr BA, Bowers MT. Z-Phe-Ala-diazomethylketone (PADK) disrupts and remodels early oligomer states of the Alzheimer disease Abeta42 protein. J Biol Chem. 2012;287:6084–6088. doi: 10.1074/jbc.C111.328575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Wang C, Sun B, Zhou Y, Grubb A, Gan L. Cathepsin B degrades amyloid-beta in mice expressing wild-type human amyloid precursor protein. J Biol Chem. 2012;287:39834–39841. doi: 10.1074/jbc.M112.371641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Mucke L, Masliah E, Yu GQ, Mallory M, Rockenstein EM, Tatsuno G, Hu K, Kholodenko D, Johnson-Wood K, McConlogue L. High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci. 2000;20:4050–4058. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Smith S, Hashimi AA, Girard J, Delay C, Hebert SS. In vivo regulation of amyloid precursor protein neuronal splicing by microRNAs. J Neurochem. 2011;116:240–247. doi: 10.1111/j.1471-4159.2010.07097.x. [DOI] [PubMed] [Google Scholar]
- [88].McConlogue L, Buttini M, Anderson JP, Brigham EF, Chen KS, Freedman SB, Games D, Johnson-Wood K, Lee M, Zeller M, Liu W, Motter R, Sinha S. Partial reduction of BACE1 has dramatic effects on Alzheimer plaque and synaptic pathology in APP Transgenic Mice. J Biol Chem. 2007;282:26326–26334. doi: 10.1074/jbc.M611687200. [DOI] [PubMed] [Google Scholar]
- [89].Rabe S, Reichwald J, Ammaturo D, de Strooper B, Saftig P, Neumann U, Staufenbiel M. The Swedish APP mutation alters the effect of genetically reduced BACE1 expression on the APP processing. J Neurochem. 2011;119:231–239. doi: 10.1111/j.1471-4159.2011.07412.x. [DOI] [PubMed] [Google Scholar]
- [90].Nishitomi K, Sakaguchi G, Horikoshi Y, Gray AJ, Maeda M, Hirata-Fukae C, Becker AG, Hosono M, Sakaguchi I, Minami SS, Nakajima Y, Li HF, Takeyama C, Kihara T, Ota A, Wong PC, Aisen PS, Kato A, Kinoshita N, Matsuoka Y. BACE1 inhibition reduces endogenous Abeta and alters APP processing in wild-type mice. J Neurochem. 2006;99:1555–1563. doi: 10.1111/j.1471-4159.2006.04178.x. [DOI] [PubMed] [Google Scholar]
- [91].Hirata-Fukae C, Sidahmed EH, Gooskens TP, Aisen PS, Dewachter I, Devijver H, Van Leuven F, Matsuoka Y. Beta-site amyloid precursor protein-cleaving enzyme-1 (BACE1)-mediated changes of endogenous amyloid beta in wild-type and transgenic mice in vivo. Neurosci Lett. 2008;435:186–189. doi: 10.1016/j.neulet.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Willem M, Dewachter I, Smyth N, an Dorren T, Borghgraef P, Haass C, Van Leuven F. Beta-site precursor protein cleaving enzyme increases amyloid deposition in brain parenchyma but reduces cerebrovascular amyloid angiopathy in aging BACE x APP[V717] double-transgenic mice. Amer J Path. 2004;165:1621–1631. doi: 10.1016/s0002-9440(10)63419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Roberds SL, Anderson J, Basi G, Bienkowski MJ, Branstetter DG, Chen KS, Freedman SB, Frigon NL, Games D, Hu K, Johnson-Wood K, Kappenman KE, Kawabe TT, Kola I, Kuehn R, Lee M, Liu W, Motter R, Nichols NF, Power M, Robertson DW, Schenk D, Schoor M, Shopp GM, Shuck ME, Sinha S, Svensson KA, Tatsuno G, Tintrup H, Wijsman J, Wright S, McConlogue L. BACE knockout mice are healthy despite lacking the primary beta-secretase activity in brain: implications for Alzheimer’s disease therapeutics. Hum Mol Genet. 2001;10:1317–1324. doi: 10.1093/hmg/10.12.1317. [DOI] [PubMed] [Google Scholar]
- [94].Cai H, Wang Y, McCarthy D, Wen H, Borchelt DR, Price DL, Wong PC. BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat Neurosci. 2001;4:233–234. doi: 10.1038/85064. [DOI] [PubMed] [Google Scholar]
- [95].May PC, Dean RA, Lowe SL, Martenyi F, Sheehan SM, Boggs LN, Monk SA, Mathes BM, Mergott DJ, Watson BM, Stout SL, Timm DE, Smith Labell E, Gonzales CR, Nakano M, Jhee SS, Yen M, Ereshefsky L, Lindstrom TD, Calligaro DO, Cocke PJ, Greg Hall D, Friedrich S, Citron M, Audia JE. Robust central reduction of amyloid-beta in humans with an orally available, non-peptidic beta-secretase inhibitor. J Neurosci. 2011;31:16507–16516. doi: 10.1523/JNEUROSCI.3647-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Lillly voluntarily terminates Phase II study of LY2886721, a beta secretase inhibitor, being investigated as a treatment for Alzheimer’s disease. Press Release, PRNewswire. Jun 13, 2013. release ELp.
- [97].Rajapaksha TW, Eimer WA, Bozza TC, Vassar R. The Alzheimer’s beta-secretase enzyme BACE1 is required for accurate axon guidance of olfactory sensory neurons and normal glomerulus formation in the olfactory bulb. Mol Neurodegener. 2011;6:88. doi: 10.1186/1750-1326-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Willem M, Garratt AN, Novak B, Citron M, Kaufmann S, Rittger A, DeStrooper B, Saftig P, Birchmeier C, Haass C. Control of peripheral nerve myelination by the beta-secretase BACE1. Science. 2006;314:664–666. doi: 10.1126/science.1132341. [DOI] [PubMed] [Google Scholar]
- [99].Hu X, Hicks CW, He W, Wong P, Macklin WB, Trapp BD, Yan R. Bace1 modulates myelination in the central and peripheral nervous system. Nat Neurosci. 2006;9:1520–1525. doi: 10.1038/nn1797. [DOI] [PubMed] [Google Scholar]
- [100].Cheret C, Willem M, Fricker FR, Wende H, Wulf-Goldenberg A, Tahirovic S, Nave KA, Saftig P, Haass C, Garratt AN, Bennett DL, Birchmeier C. Bace1 and Neuregulin-1 cooperate to control formation and maintenance of muscle spindles. EMBO J. 2013;32:2015–2028. doi: 10.1038/emboj.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Hook G, Yu J, Sipes N, Pierschbacher M, Hook V, Kindy MS. The Cysteine Protease Cathepsin B is a Key Drug Target and Cysteine Protease Inhibitors are Potential Therapeutics for Traumatic Brain Injury. J Neurotrauma. 2013 doi: 10.1089/neu.2013.2944. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Yamashima T, Kohda Y, Tsuchiya K, Ueno T, Yamashita J, Yoshioka T, Kominami E. Inhibition of ischaemic hippocampal neuronal death in primates with cathepsin B inhibitor CA-074: a novel strategy for neuroprotection based on ’calpain-cathepsin hypothesis’. Eur J Neurosci. 1998;10:1723–1733. doi: 10.1046/j.1460-9568.1998.00184.x. [DOI] [PubMed] [Google Scholar]
- [103].Guicciardi ME, Deussing J, Miyoshi H, Bronk SF, Svingen PA, Peters C, Kaufmann SH, Gores GJ. Cathepsin B contributes to TNF-alpha-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J Clin Invest. 2000;106:1127–1137. doi: 10.1172/JCI9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Foghsgaard L, Wissing D, Mauch D, Lademann U, Bastholm L, Boes M, Elling F, Leist M, Jaattela M. Cathepsin B acts as a dominant execution protease in tumor cell apoptosis induced by tumor necrosis factor. J Cell Biol. 2001;153:999–1010. doi: 10.1083/jcb.153.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Terada K, Yamada J, Hayashi Y, Wu Z, Uchiyama Y, Peters C, Nakanishi H. Involvement of cathepsin B in the processing and secretion of interleukin-1beta in chromogranin A-stimulated microglia. Glia. 2010;58:114–124. doi: 10.1002/glia.20906. [DOI] [PubMed] [Google Scholar]
- [106].Sun L, Wu Z, Hayashi Y, Peters C, Tsuda M, Inoue K, Nakanishi H. Microglial cathepsin B contributes to the initiation of peripheral inflammation-induced chronic pain. J Neurosci. 2012;32:11330–11342. doi: 10.1523/JNEUROSCI.0677-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, Fitzgerald KA, Latz E, Moore KJ, Golenbock DT. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Reinheckel T, Deussing J, Roth W, Peters C. Towards specific functions of lysosomal cysteine peptidases: phenotypes of mice deficient for cathepsin B or cathepsin L. Biol Chem. 2001;382:735–741. doi: 10.1515/BC.2001.089. [DOI] [PubMed] [Google Scholar]
- [109].Satoyoshi E. Therapeutic trials on progressive muscular dystrophy. Intern Med. 1992;31:841–846. doi: 10.2169/internalmedicine.31.841. [DOI] [PubMed] [Google Scholar]
- [110].Miyahara T, Shimojo S, Toyohara K, Imai T, Miyajima M, Honda H, Kamegai M, Ohzeki M, Kokatsu J. Clinical Phase I trial of thiol protease inhibitor (Report 2): safety and pharmacokinetics in continuous administration. Rinsho Yakuri. 1985;16:537–546. [Google Scholar]
- [111].Miyahara T, Shimojo S, Toyohara K, Imai T, Miyajima M, Honda H, Kamegai M, Ohzeki M, Kokatsu J. Phase I clinical trial of thiol protease inhibitor EST (Report 1): Safety and pharmacokinetics with single administration. Rinsho Yakuri. 1985;16:357–365. [Google Scholar]
- [112].Ohshima T, Watanabe T, Nagato C, Kimura M, Tsuchida T, Nakane S. Toxicological studies on ethyl(+)-(2S,2S)-3[(S)-3-methyl-1-(3-methylbutylcarbamoyl)butylcarbamoyl]-2-oxiranecarboxylate (EST)(Report VI) Chronic toxicity study in rats. Iyakuhin Kenkyu. 1986;17:781–801. [Google Scholar]
- [113].Abe S, Aida S, Nakane S. Toxicological studies on ethyl(+)-(2S,2S)-3[(S)-3-methyl-1-(3-methylbutylcarbamoyl)butylcarbamoyl]-2-oxiranecarboxylate (EST)(Report X) Effect on Vision and hearing in rats. Iyakuhin Kenkyu. 1986;17:826–834. [Google Scholar]
- [114].Kimura M, Yagi K, Fujinuma S, Tsuchida T, Tarumoto Y, Noda K, Nakane S. Toxicological studies on ethyl(+)-(2S,2S)-3[(S)-3-methyl-1-(3-methylbutylcarbamoyl)butylcarbamoyl]-2-oxiranecarboxylate (EST) (Report III) Subacute toxicity in rats. Iyakuhin Kenkyu. 1986;17:744–767. [Google Scholar]
- [115].Setoyama K, Koike M, Abe S, Tsutsui Y, Tarumoto Y, Nakane S. Toxicological Studies of Ethyl (+)-(2S,3S)-3-[(S)-3-methyl-1-(3-methylbutylcarbamoyl)butylcarbamoyl]-2-oxiranecarboxylate (EST) (Report 1): Acute Toxicity Studies of EST and Metabolite and By-Product of EST. Iyakuhin Kenkyu. 1986;17:736–743. [Google Scholar]
- [116].Ueki S, Watanabe S, Fujiwara M, Shibata S, Shi-yu L, Iwazaki K, Ohta H, Shimazoe T. Effect of Cysteine Proteinase Inhibitor EST on Central Nervous System. Yakuri to Chiryo. 1986;14:725–735. [Google Scholar]
- [117].Tarumoto Y, Sakagawa T, Tsutsui Y, Kawanishi M, Kimura M, Nakane S. Toxicological studies on ethyl(+)-(2S,2S)-3[(S)-3-methyl-1-(3-methylbutylcarbamoyl)butylcarbamoyl]-2-oxiranecarboxylate (EST)(Report V) Subacute toxicity in dogs. Iyakuhin Kenkyu. 1986;17:768–780. [Google Scholar]
- [118].Yasui H, Goto H, Suzuki H, Sakai S, Takamura T, Nakane S. Toxicological studies on ethyl(+)-(2S,2S)-3[(S)-3-methyl-1-(3-methylbutylcarbamoyl)butylcarbamoyl]-2-oxiranecarboxylate (EST)(Report IX) Mutagenicity study. Iyakuhin Kenkyu. 1986;17:815–825. [Google Scholar]
- [119].Yamada T, Nishiyama T, Nakane S. Reproduction studies of ethyl(+)-(2S,2S)- 3[(S)-3-methyl-1-(3-methylbutylcarbamoyl)butylcarbamoyl]-2-oxiranecarboxylate (EST) (Report I): Study of administration to rats prior to and in early stage of gestation. Iyakuhin Kenkyu. 1986;17:609–616. [Google Scholar]
- [120].Yamada T, Nishiyama T, Ohno H, Nakane S. Reproduction studies of ethyl(+)-(2S,2S)-3[(S)-3-methyl-1-(3-methylbutylcarbamoyl)butylcarbamoyl]-2-oxiranecarboxylate (EST) (Report III): Study of administration to rabbits during organogenesis. Iyakuhin Kenkyu. 1986;17:632–638. [Google Scholar]
- [121].Yamada T, Uchida H, Ohno H, Matsuzawa N, Nakane S. Reproduction studies of ethyl(+)-(2S,2S)-3[(S)-3-methyl-1-(3-methylbutylcarbamoyl)butylcarbamoyl]-2-oxiranecarboxylate (EST) (Report IV): Study of administration to rats during perinatal and postnatal periods. Iyakuhin Kenkyu. 1986;17:639–651. [Google Scholar]
- [122].Fukushima K, Kono Y, Osabe W, Shinozaki F, Kudo K, Arai M, Taigawa K, Suwa T. Pharmacokinetics of EST (Report 2): Tissue Distribution of 14C-EST. Kiso to Rinsho. 1986;20:328–341. [Google Scholar]
- [123].Fukushima K, Yoshida H, Osabe W, Shinozaki F, Kudo K, Arai M, Suwa T. Pharmacokinetics of EST (Report 1): Absorption and Excretion of 14C-EST. Kiso to Rinsho. 1986;20:319–327. [Google Scholar]
- [124].Watanabe T, Fukushima K, Ushiyama Y, Noda K, Suwa T. Pharmacokinetics of EST (Report 5): Pharmacokinetics of EST ih Humans. Kiso to Rinsho. 1986;20:362–366. [Google Scholar]