Summary
Circulating tumor cells (CTCs) originate from the primary tumor mass and enter into the peripheral bloodstream. CTCs hold the key to understanding the biology of metastasis and also play a vital role in cancer diagnosis, prognosis, disease monitoring, and personalized therapy. However, CTCs are rare in blood and hard to isolate. Additionally, the viability of CTCs can easily be compromised under high shear stress while releasing them from a surface. The heterogeneity of CTCs in biomarker expression makes their isolation quite challenging; the isolation efficiency and specificity of current approaches need to be improved. Nanostructured substrates have emerged as a promising biosensing platform since they provide better isolation sensitivity at the cost of specificity for CTC isolation. This review discusses major challenges faced by CTC isolation techniques and focuses on nanostructured substrates as a platform for CTC isolation.
Keywords: Circulating tumor cells, Nano surface, Cancer, Diagnosis, Biomarkers, Heterogeneity
Introduction
Tumor cells shed from the primary tumor mass, circulate through peripheral blood in the early stage, and establish distant metastatic lesions in other organs [1]. Therefore, circulating tumor cell (CTC) analysis as a “liquid biopsy” can provide valuable information on prognosis, monitor systemic anticancer therapy, and helps identify the proper therapeutic targets [2]. For example, patients with metastatic breast and prostate cancer have a lower survival rate (10.1 months vs. >18 months, P < 0.001), if their CTC count is more than 5 CTCs per 7.5 mL of whole blood [1,3]. Analysis of molecular characteristics might be valuable for noninvasive serial monitoring of tumor genotypes during treatment [4]. Moreover, for routine clinical analysis, instead of collecting disseminated tumor cells from bone marrow samples, enriching CTCs from peripheral blood is more acceptable to breast cancer patients because of compliance issues associated with repeated bone marrow collection [5]. Thus, platforms that can detect and isolate CTCs are highly desirable for routine tumor analysis.
The detection and isolation of CTCs hold great promise but remain technically challenging [3]. First, CTCs are rare and patients with early stage cancer can have as few as 1 CTC per 1 mL peripheral blood [1]. Second, isolated CTCs should be kept viable for subsequent cell culture and molecular analysis [6]. Tumor cells are vulnerable and the extremely high detachment forces applied when detaching captured cells may harm the intactness of cell structure and disturb the cell microenvironment [7,8,158]. Insufficient biomarkers and heterogeneity of CTCs pose additional challenges. Currently molecular affinity based approaches that rely on anti-EpCAM antibody are widely used for CTC isolation. However, CTCs may have little or no expression of detection biomarkers on the cell membrane [9,10]. Finally, the isolation efficiency and specificity of current techniques must be improved as the mean capture yield using anti-EpCAM antibodies in CTC-microchip has been reported to be around 60%, and purity is about 50% [11].
To efficiently isolate rare CTCs in a viable state, a number of approaches have been reported, including devices that rely on mechanical forces [12], dielectrophoresis [13,14], microscale optical interactions [9,15], magnetic cell sorting [16], flow cytometry [17,18], and microfluidic devices [11]. The advantages and disadvantages of these CTC isolation and detection technologies are summarized in Table 1. Previous reviews have discussed these CTC isolation approaches in detail [3,19]. Although these isolation approaches have various advantages, there is a need to improve CTC isolation techniques for routine clinical analysis.
Table 1.
Advantages and disadvantages of various CTC isolation/detection technologies.
| Device | Method | Sensitivity | Purity | Cell viability | Recovery | Throughput | Sample volume |
Advantages | Limitations |
|---|---|---|---|---|---|---|---|---|---|
| CellSearch [138–144] | Immunomagnetic and fluorescence imaging | High | 0.1–1.4% | Fixed for counting and molecular profiling | 42–85% | ~90 min per detection round | 7.5 mL | High intra- and inter-assay precision; standardized and semi- automated | Unable to detect antigen (−) cancer cells; detection only; expensive facility |
| MagSweeper [145] | Immunomagnetic based | 100% | 51–100% | 94% | ~50% | 120 min per detection round | ~9 mL | Simple; easy to handle; fast | Require additional antibody functionalized magnetic microbeads |
| ISET [146] | Size based | High | N/A | 85–90%, or fixed for counting and molecular profiling; high retrieval efficiency | 80–90% | Filtration for 3 min | 10 mL | Label free; not relying on capture antibodies; simple; fast; satisfies cell lysis, cell | Unable to capture small size cancer cells; poor specificity |
| Cellsievo [147] | ~80% (cancer cell lines) | N/A | 1–2 mL blood per hour | 1 mL | collection, and molecular profiling | ||||
| CTChip [148] | ~80% (cancer cell lines) | N/A | 1 mL blood per 25 min | 2 mL | |||||
| Micropore Chip [18] | Size, elasticity | High | N/A | N/A | ~40–70% | ~0.1 mL per hour | ~0.1–0.2 mL | Not requiring tumor-specific target molecules; label-free | Low throughput. Low specificity |
| Inertial force [149] | Size, weight and shape based | 68–90% (cancer cell lines) [150] | N/A | ~95% [151] | N/A | ~1 million cells per min | N/A | Label free; not relying on capture antibody; simple; fast; inexpensive | Low specificity |
| Dielectrophoresis [152] | Dielectric property based | 1 cancer cell (cancer cell lines) in 1000 mononuclear cells | N/A | 70–90% | 10–92% | Handle 30 million cells within 30 min | 0.25–4.5 mL | Not requiring tumor-specific target molecules; label-free | Strict requirements on cell number, ratio, frequency of electric field, and cell type |
| CTC-Chip [11] | Bioaffinity based | ~65% | ~50% | ~98%, allows for reliable molecular analysis | ~60% | 1–2 mL blood per hour | 0.9–5.1 mL | Satisfies cell lysis, cell collection, molecular profiling; rapid | High cost; unable to capture antigen (−)cancer cells |
| CTC-iChip [153] | N/A | >0.1% for posCTC- iChip | N/A | 77–98% | 10 million cells per sec | 8 mL | May or may not require tumor-specific target molecules | Low purity in the case of negCTC-iChip | |
| Microvortex Chip [154] | 93% | 14% | 95% | 92% | 1–2 mL blood per hour | 4 mL | Satisfies cell lysis, cell collection, molecular profiling; rapid | Unable to detect antigen (−) cancer cells | |
| MicroGEDI Chip [155] | 94% | 62–74% | N/A | 80–100% | 1 mL blood per hour | 1 mL | Satisfy cell lysis, cell collection, molecular profiling; rapid | Unable to detect antigen (−) cancer cells | |
| Micropillar [156] | N/A | N/A | N/A | 71% | 1 mL blood per hour | 1 mL | Satisfies cell lysis, cell collection, molecular profiling; rapid | Unable to detect antigen (−) cancer cells | |
| MicroeDAR cytometer [157] | 100% | 10–50% | N/A | 93% | 3 mL blood per hour | 2 mL | Satisfies cell lysis, cell collection, molecular profiling; rapid | Requires fluorescence labeling; Unable to detect antigen (−) cancer cells | |
| Nanostructured substrates [25,87,92,100] | Bioaffinity based or surface structure based | 80–95%; normally ~90% | N/A | 91–95% | 94–99% | Capture cells within 30 min | 0.1–0.5 mL | With or without antibody for cell isolation; satisfies cell lysis, cell collection, molecular profiling; rapid | The increased sensitivity decreases specificity to a certain degree |
Nanostructured scaffolds and substrates with nanoscale topography that mimic the natural extracellular matrix (ECM) or basement membrane have been extensively employed in tissue engineering and regenerative medicine [20]. Natural ECM proteins are structurally bundled together to form nanostructures with diameters ranging from 260 to 410 nm [21]. The basement membrane also shows a complex network of pores and fibers with dimensions in the range of 30–400 nm [22]. Nanostructures can promote cellular organization, cell matrix and cell–cell interactions, cellular proliferation, and ECM synthesis [23]. The major advantage of nanostructured scaffolds is that a larger biomimetic surface area facilitates firm cellular attachment and this feature is also helpful for isolation and detection of rare cells [24]. Although CTC detection and sorting based on affinity interactions yield higher efficiency and greater specificity in contrast to mechanical and electrical sorting techniques, harvesting pure and viable CTCs still remains difficult [19]. The combination of affinity interactions and biomimetic nanostructured surfaces further improve CTC isolation and detection efficiency. The nanotextured substrates provide more surface area for antibody immobilization and lower the rolling velocity of cells in microfluidic channels. It has been recently reported that the cancer cells preferred the nanotextured surface even without surface functionalization because of dynamic arrangement of integrin-mediated focal adhesions (FA) [25]. Nanostructured substrates emerge as a promising bio-platform for CTC and/or rare cell isolation.
In this review, we first briefly introduce different type of nanostructured substrates available and their preparation methods followed by their applications in CTC and rare cell isolation. The potential cell capture mechanisms of nanostructured substrates, advantages and disadvantages of various substrates, grand challenges for CTC isolation, and future directions are also discussed.
Nanostructure preparation techniques
A brief background on various tools and processes available for fabricating nanostructured surfaces with varying pore sizes, dimensions and lengths is necessary. Modified or improved protocols can also be used for achieving customized characteristics. In general, top-down and bottom-up are the two approaches that can be used for fabricating nanostructured substrates.
In the top-down fabrication methods, a bulk material is laterally patterned by a series of subtractive steps. (I) Lithography is the process of transferring a pattern from a mask onto a substrate such as in optical lithography, X-ray lithography and E-beam lithography. Controllable geometries and patterns can be created using lithography except in colloidal lithography in which geometries depend on a random dispersal of coated nanoparticles [26–32]. The process flow to fabricate quartz nanowire arrays using colloidal lithography and chemical etching is shown in Fig. 1. Lithography and mask fabrication methods have also been adapted to tissue engineering and biological systems [33]. (II) Nano-embossing is a promising method of nanostructure preparation, enabling inexpensive and high throughput nanofabrication. It uses a nanostructured stamp to transfer the pattern to the substrate surface via an embossing process [34]. (III) Etching is a subtractive method that selectively removes materials from the substrate. Nano-structured surfaces can be obtained by etching a number of polymers, such as PDMS, PLGA, poly-L-lactic acid (PLLA), polyethylene terephthalate (PET), and polyether urethane (PU) [35]. Hydrofluoric acid (HF) based electrochemical etching of silicon wafers can also produce random nanostructures. HF concentration, current density, etching time, and wafer type can affect the size of silicon crystallites [36,37]. If an aluminum template is applied in the etching process, controllable geometries and precise pore sizes can be obtained [38]. With deep reactive ion etching, 20–100 nm high-aspect-ratio nanostructures can be produced on the wafer surface [39].
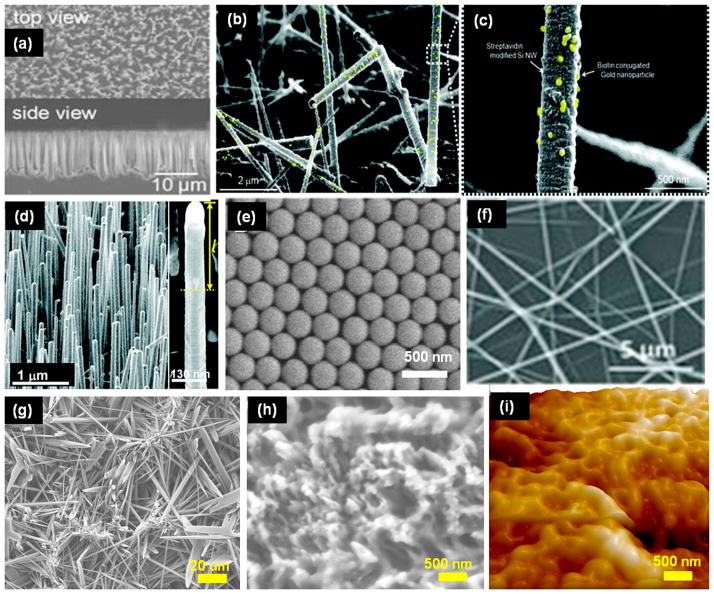
Figure 1.
Fabrication of quartz nanowire (QNW) arrays. (a) Polystyrene nanoparticles (PS NPs) of ~100 nm diameter were spin coated onto quartz surface. (b) Oxygen plasma was used to reduce the size of PS NPs. (c) A thin layer of Cr metal (25 nm) was deposited followed by lift-off of PS NPs with N-methy1-2-pyrrolidone. (d) Ni metal was deposited and used as an etch mask. (e, f) Top- and tilted-view after second plasma etching. Wet chemical etchant was used to completely remove Ni metal. Scale bar is 200 nm in all images.
Reprinted (adapted) with permission from (Lee et al.; Nano Lett. 12 (2012) 2697–2704). Copyright (2012) American Chemical Society.
In the bottom-up fabrication methods, building blocks are brought in close proximity to form the desired structures. The assembly of the building blocks is usually manipulated by physical aggregation and chemical reaction. Adaptation of bottom-up approaches to biological systems has already resulted into various biotechnology applications, for instance in tissue engineering using self assembly [40], magnetic [41] and acoustic assembly methods [42], and bio-printing [43]. (I) Electrospinning utilizes electrical forces to synthesize nanometer scale polymer fibers. More than 20 polymers, including polyethylene oxide, nylon, polyimide, polyaramid, and polyaniline have been used for nanofiber fabrication. This technique can produce long polymer fibers with diameters ranging from 40 to 2000 nm. Many parameters including polymer type, solvent, applied voltage, and the distance between the nozzle and the collector determine both the width and the length of the nanofibers. These nanofibers can be regularly or irregularly arranged on a substrate to form a nanostructure surface [44]. (II) Chemical vapor deposition (CVD) is a process in which materials react and/or decompose on the substrate surface to produce the desired deposition. Silicon nanowires (SiNWs) can be fabricated by employing the CVD process [45–48]. (III) Organic film coating with vapor-phase utilizes the volatility of hydrophilic and hydrophobic monomers at lower vapor tension and results in deposition of biocompatible organic nanofilms on substrate [49]. Using this technique, polymer layers of 300 nm thickness can easily be fabricated. The deposited polymeric film is nanoporous with pore diameters ranging from 100 to 500 nm. (IV) Gas foaming is a technique that uses gas as a porogen. High pressure CO2 is used for generating a 0.1–100 nm sponge like structure [50]. (V) Thermally induced phase separation is a technique similar to gas foaming. Phase separation utilizes a solvent with a low melting point that can easily sublime to remove a certain chemical and leave nanostructured caves in the substrates. This process produces an uncontrolled random pattern with dimensions of few nanometers [51]. The advantages and disadvantages of these techniques are summarized in Table 2.
Table 2.
Nanostructure preparation techniques and their minimum feature size.
| Fabrication method | Min. feature size | Advantages/limitations |
|---|---|---|
| Optical lithography [29,31,32,52] | ~50 nm | Precise patterns can be created. |
| X-ray lithography [30,52] | ~20 nm | Requires expensive optical lenses. |
| E-beam lithography [27,52–55] | 5–10 nm | Controlled geometries can be achieved. Serial, time consuming process. |
| Colloidal lithography [52,56] | ~300 nm | Random geometries. |
| Nano-embossing [34] | ~100 nm | Controllable geometries and patterns. |
| Etching polymer [35] | >1 nm | Inexpensive method but produces random nanostructures. |
| Etching silicon wafer [36–38] | 2–3 nm | No control on dimensions and pore sizes unless using aluminum template. |
| Reactive ion etching [39] | 20–100 nm | High aspect ratio patterns. |
| Electrospinning [44] | 40–2000 nm | Over 20 polymers can be used; limited to only fiber formation. |
| Chemical vapor deposition [45–48] | ~2 nm | Require high vacuum furnaces, slow process especially in the case of epitaxy but layer thickness can be well controlled. |
| Vapor-phase coating [49] | 100–500 nm | Easy fabrication method but the coatings are not very stable. |
| Gas foaming [50] | 0.1–100 nm | Low-cost method but not widely applicable. |
| Phase separation [51] | >1 nm | Nanofeatures and pore sizes cannot be controlled precisely. |
Nanostructured substrates for CTC isolation
Recently, researchers have extensively explored cell proliferation, migration, differentiation and alignment on nanostructured surfaces [57–61]. Examples of such surfaces include random nanofibers, nanopits and nanogrooves. Nanostructured substrates are also employed in tissue engineering for better understanding of mammalian cell adhesion mechanisms and for the restoration of function to damaged tissues [33,43,62,65–70]. Nanostructures facilitate protein (such as fibronectin and vitronectin) adherence to surfaces due to a larger surface area [71]. Protein epitopes are recognized by integrin heterodimers on the cell membrane; integrins assemble in focal adhesions which initiate cell adhesion [72–77]. Therefore, the nanostructure itself facilitates cell attachment. The adhesive forces between cells and substrate are significantly improved by antigen–ligand binding. Various nanostructure substrates are employed for rare cell isolation including nanopillars, nanowires, and nanotextured polydimethylsiloxane (PDMS). These substrates are discussed thoroughly in the following sections.
Nanopillars and nanowires
Micron/nano sized silicon pillars in an array were used for studying cell adhesion, migration, and morphology [78]. On nanostructured substrates, as the number of nanopillars or nanowires per unit surface area increases, the surface area to volume ratio increases resulting in an increase of van der Waals adhesion forces [79–81]. It was reported that biomimetic nanowires adhere to silicon probe with forces 0.04–200 times higher than the adhesive strength of geckos foot-hairs [78,79,81–85]. Thus, by decreasing the diameter of nanopillars or nanowires, adhesive forces might increase due to geometric features alone. On the other hand, under higher surface area to volume ratio, bioaffinity interactions between capturing reagents and target molecules also increase by grafting more capturing reagents on the surface of the nanopillars or nanowires.
Silicon nanopillars (SiNP) with diameters in the range of 100–200 nm on a silicon wafer have been used for cancer cell isolation (Fig. 2(a)). Capture yield of MCF7 cells in culture medium was 45–65% on SiNP; 10 times more than what was achieved on flat silicon. About 84–91% of the captured cells were viable [87,88]. In another report, SiNPs were integrated into a microfluidic device with chaotic micromixers to increase cell-SiNP contact frequency by making a vortex with a serpentine chaotic mixing channel [89]. Three different cancer cell lines, MCF7, PC3, and T24, were spiked in concentrations ranging from 50 to 1000 cells per mL into culture medium, whole blood, lysed blood and PBS. The isolation efficiency in all cases was more than 95% when flow rate did not exceed 2 mL/h. In another report, silicon nanowires (SiNWs) of 100–200 nm in size were used for T lymphocyte separation, and 88% capture efficiency was reported (Fig. 2(b) and (c)) [90]. SiNWs covered with uniform gold nanoclusters (Au NCs) were also synthesized (diameter: 50–160 nm; length: 5–10 μm, Fig. 2(d)) [91]. Antibody attachment and cell capture efficiency can be controlled by adjusting the size and dispersal of Au NCs. A 25 μL MCF7 cell suspension (105 cells per mL of PBS) was introduced onto a SiNW sample and allowed to incubate for 5, 20, and 40 min, respectively. It was reported that MCF7 cancer cells were captured with up to 40% efficiency in 5 min, and 95% of captured cells were viable. Extending incubation time to 40 min for target cells to have sufficient time to contact with nanostructured substrate surfaces, further improved the capture efficiency to 88%. In another report, quartz nanowires (QNWs) were fabricated with sizes in the range of 80–100 nm [92]. Average capture efficiency for A549 cells was found to be 89% on QNWs when cancer cells are suspended in PBS, compared to 23% achieved on flat glass surfaces. Mean purity for captured A549 cells from A549/U937 cell mixture was 83.4% when cells were mixed in 1:1 ratio in PBS. The purity reduced significantly once the nonspecific cells (PBMCs) are present at densities comparable to clinical samples, i.e., ~1 million PBMCs/mL. In order to demonstrate the applicability of QNW platform to capture cancer cells from clinical samples, 15 A549 cancer cells were spiked into peripheral blood mononuclear cells (PBMC) in culture medium and QNWs successfully captured 9 cells. Further, ~10 A549 cells were spiked into 1 mL peripheral blood followed by red blood cell lysis and the remaining cells were resuspended into 1 mL buffer. Using QNWs, ~6 out of ~10 A549 cells were captured from the lysed blood samples. Recently, bare nanorough glass surfaces with root-mean-square (Rq) value of 150 nm were synthesized [25]. These nanorough glass surfaces (without antibody functionalization) were utilized to capture 93.3% MCF7 and 95.4% MDA-MB231 cells spiked into 500 μL lysed blood, regardless of their EpCAM expression [25]. For comparison with nanorough glass surfaces (Rq = 150 nm), only 22% MCF7 and 13.9% MDA-MB231 cells were captured on smooth glass surfaces (Rq = 1 nm). By observing the fluidic shear stress applied to captured cells, it was demonstrated that non-functionalized nanorough glass surfaces could significantly enhance adhesion of cancer cells to the surfaces (i.e., MCF7, MDA-MB231, and PC3). Moreover, cancer cells showed distinct FA formation on nanorough surfaces. They also compared non-functionalized and anti-EpCAM coated nanorough glass surfaces and found no significant difference in cancer cell capture efficiency when Rq value was greater than 50 nm [25]. Although it is reported that cancer cells can be captured selectively on nanorough surfaces without antibody functionalization, high nonspecific binding due to PBMCs was observed. When mixing ratio of MDA-MB231 and PBMCs was changed from 1:1 to 1:200, capture purity of MDA-MB231 cells was significantly reduced from 84% to 14% [25].
Figure 2.
Nanostructured surfaces fabricated by various processes. (a) SEM images of densely packed silicon nanopillars with diameters of 100–200 nm. These were produced using wet chemical etching by Ag+ and hydrofluoric acid. Reprinted with permission from John Wiley and Sons (Wang et al. Angew. Chem. Int. Ed. Engl. 2009; 48(47): 8970–8973). (b) SEM image of silicon nanowires (SiNWs) produced by vapor–liquid–solid method (SiNW diameter ~ 200 nm). (c) Magnified image of SiNWs bound to biotin-gold nanoparticles. Gold nanoparticles are highlighted as yellow for better presentation. Reprinted (adapted) with permission from (Kim et al. Nano Lett., 10 (8) (2010) 2877–2883). Copyright (2010) American Chemical Society. (d) SEM image of Au nanoclusters coated SiNWs produced by rapid thermal chemical vapor deposition (diameter ~100 nm). Inset shows the magnified images of Au coated SiNWs. Reprinted (adapted) with permission from (Park et al. Nano Lett. 12 (2012) 1638–1642). Copyright (2012) American Chemical Society. (e) SEM image of close-packed arrays of silica nanobeads with uniform diameter of 319 nm. The nanobeads were deposited on glass to control the substrate topography. Reprinted (adapted) with permission from (Wang et al. Langmuir 27 (2011) 11229–11237). Copyright (2011) American Chemical Society. (f) SEM image of TiO2 nanofibers with diameter ranging from 100 to 300 nm. These nanofibers were fabricated and coated on silicon wafer using electrospinning and calcination. Reprinted with permission from John Wiley and Sons (Zhang et al. Adv. Mater. 24 (20) (2012) 2756–2760). (g, h) Micro needles and nanotextured surfaces were synthesized by chemical etching of chicken eggshell with sulfuric and hydrochloric acid. Micro needles and nanotextured surfaces were made of calcium sulphate crystals and calcium carbonate, respectively. (i) AFM image of PDMS surface that was cured on nanotextured chicken eggshell. Reprinted with permission from Institute of Physics (Asghar et al. Nanotechnology 23 (2012) 475601).
Nanotextured PDMS
Nanotextured substrates have also received much attention lately for cancer cell enrichment [8]. The basement membrane can anchor cancer cells through cell adhesion molecules. Therefore, it can improve cell adhesion and growth [93]. In metastasis, before cancer cells enter into peripheral blood, they first attach to the basement membrane, proliferate, and then break through the barrier [94]. In one report, aptamer functionalized nanotextured PDMS substrates mimicking nanostructure of basement membrane were used for cancer cell isolation [95]. NaOH etched poly(lactic-co-glycolic acid) (PLGA) polymer with nanostructured surface (310 nm roughness) was used as a master for preparing a nanotextured PDMS substrate. Availability of larger surface area allows abundant aptamer immobilization, which favors tumor cell isolation. The total number of captured human glioblastoma cells on nanotextured PDMS was twice that of on a glass surface.
Surface coating with nanomaterials
Nanostructured surfaces can be obtained by coating nanomaterials on flat surfaces [159,160]. Capture efficiency of KG1a cells was improved using the nanoscale surface coating [96,97]. Capillary channels were coated with poly-L-lysine (PLL) which carried positively charged ammonium groups. Negatively charged P-selectin grafted silica was then deposited on PLL surface via electrostatic interactions. The total amount of P-selectin on silica, measured by immunofluorescence quantification, was increased up to 35% on nanoscale surface compared to planar surface, which enhanced cell binding. Therefore, rolling velocity of cells on a silica coated microtube surface decreased by around 50% [97]. The total number of cells captured on the nanoscale surface was double that of the planar control surface. Titanium (IV) Butoxide and halloysite nanotubes (smaller than 450 nm) were also investigated, and similar results were reported [96,97]. In another report, glass surface with a layer of indium tin oxide was homogeneously coated with anti-EpCAM grafted poly(3,4-ethylenedioxy)thiophenes polymer (PEDOT) with various dot diameter (feature size) ranging from 98 nm to 333 nm for cancer cell isolation [98]. Capture efficiency of MCF7 (EpCAM+) cells were strongly related to polymer dot size. In particular, MCF7 cells were efficiently captured on 232 nm nanostructured surface, reaching 4–5 times higher than on a flat surface. It was further reported that capture efficiency of EpCAM+ cells (MCF7 and T24) was 10 times more than capture efficiency of EpCAM− (negative) cells (HeLa). The finding indicates that anti-EpCAM coated surfaces cannot be used for capturing EpCAM− (negative) cancer cells. In another report, surface nanotopography of glass slides was carefully controlled over a large range by coating with silica beads and its influence on Jurkat cell capture was also studied [99]. Antibody modified silica beads ranging from 100 to 1150 nm in diameter were deposited on hydrophobically treated glass slides (Fig. 2(e)). At a lower flow rate (2 μL/min), cell capture efficiency on nanostructured surface was 1.2–1.6 times higher than that on a smooth surface. It was also found that cell capture efficiency did not scale linearly with increased nanostructured surface area. It could have been due to mechanical deformation of cells at the cellular and subcellular levels. Electrospinning was employed to coat silicon wafer with TiO2 nanofibers (100–300 nm in size) followed by antibody immobilization (Fig. 2(f)) [100]. Over 80% of HCT116 cells were captured from spiked culture medium (20–250 cells per mL). It was reported that cell capture efficiency decreased to ~45% when cells were spiked into human blood samples as the antigen binding area of capture antibody might be partially covered. They further performed CTC isolation studies on 0.5 mL peripheral blood samples from colorectal and gastric cancer patients. In colorectal cancer patients’ samples, 0–2 CTCs were captured from 2 out of 3 samples; in gastric cancer patients’ samples, 3–19 CTCs were captured from all 7 samples. However, the exact number of CTCs in each clinical blood samples was not identified; the respective isolation efficiency was not reported either.
Underlying isolation mechanisms on nanostructured substrates
The nanostructured substrates discussed above significantly increase cell isolation efficiency compared to flat surfaces. Four important features of nanostructured substrates contributing higher capture efficiency are discussed here. First, one common feature of nanostructured surfaces is to provide larger surface area for biomolecule immobilization. For example, aptamer and laminin immobilization on nanotextured PDMS was putatively 20 and 2 times more than that on a planar surface, respectively [95,101]. It is well established that the number of immobilized capturing reagents can affect the cell isolation efficiency [102]. The larger the area for immobilization, the higher the number of capturing reagents that can be immobilized. Although the receptor and capturing reagents have a one-to-one instantaneous binding mechanism, more capturing reagents undoubtedly can increase the odds of binding. Second, nanostructured substrates provide lower rolling velocity that facilitates cell attachment. In microfluidic devices, cells experience rolling adhesive dynamics [96,97,103,104]. During rolling, cells move forward by forming new adhesive bonds at the leading edge and decelerate as force develops at the trailing edge. An overall tensile force due to adhesive bonds and detachment of bonds at the trailing edge primarily determines the velocity of a rolling cell [105]. On nanostructured substrates abundant capturing reagents can increase total binding forces that result in a decrease of cell rolling velocity. The cells are finally stopped by higher adhesive forces that can withstand higher shear stress. It is important to highlight here that cell rolling velocity also depends on the cell shape and elastic properties [105–107]. Third, pseudopodia formation might also be related to cell capture yield (Fig. 3) [108]. The morphology of captured cells was found to be flatter on nanostructured substrates than on smooth substrates; cells changed from a globular shape to a semielliptical one [109]. The flatter shape of captured cell and pseudopodia formation indicated that more receptors on the cell membrane may have contacted with immobilized capturing reagents. Therefore, higher binding forces assist cell attachment on the surface. Moreover, decreased cell height further helps them from being eluted under high shear stress [110]. Fourth, the nanostructure itself might assist in cell attachment and improve cell-substrate adhesion force [111]. For example, five kinds of cancer cells, i.e., HeLa, PC3, SUM149, MCF7 and MDA-MB231 cells were captured on non-functionalized nanostructured glass surfaces, indicating nanostructured surfaces were preferable to these cancer cells as compared to flat substrates [25,112]. The molecular arrangement and dynamic organization of integrin-mediated focal adhesions were reported to be sensitive and responsive to nanostructured substrates [25,112].
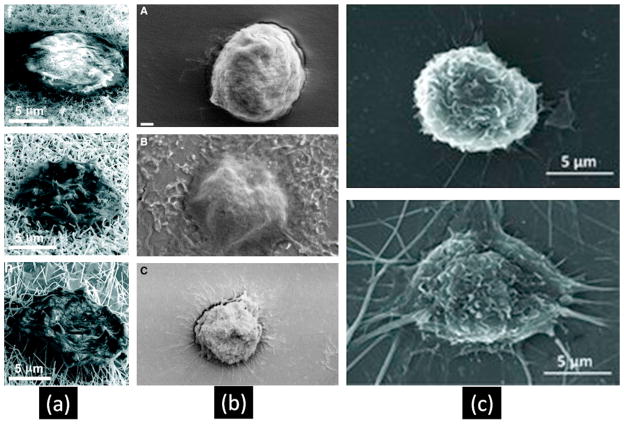
Figure 3.
Pseudopodia formation on nanostructured substrates and cells show flatter shapes. (a) SEM images of captured cells on different size of SiNWs (Park et al., Nano Lett. 12 (3) (2012) 1638–1642). Copyright (2010) American Chemical Society. (b) SEM images of captured cells on plane glass, nanotextured PDMS and plane PDMS surface (top to down) (Wan et al., Cancer 18 (4) (2012) 1145–1154). Copyright (2012) Wiley. (c) SEM images of captured cells on plane surface and nanofibers coated surface (Zhang et al., Adv Mater. 24 (20) (2012) 2756–2760). Copyright (2012) Wiley.
Advantages and current limitations of nanostructured substrates
CellSearch™, the only FDA approved CTC diagnostics tool, immunomagnetically isolates tumor cells from whole blood [113]. Magnetic nanoparticles are functionalized with monoclonal antibody against EpCAM and mixed with blood samples. The functionalized magnetic nanoparticles specifically bind with CTCs and can be isolated using an external magnet. On average 7.5 mL of blood sample is required for analysis in CellSearch™ system; additionally, its multiple washing and processing steps might result in loss of CTCs for detection. In order to detect a few CTCs in billions of other cells in vivo diagnostic tools or ultrasensitive techniques are needed. For an in vivo diagnostic tool, a sensor must be implanted across blood vessels to analyze flowing cells. Nanostructured substrates can be used as an ultrasensitive tool for enriching rare CTCs from blood samples. Moreover, nanostructured substrates can keep most captured cells viable for subsequent molecular analysis and/or cell culture. Rather than just CTC enumeration (standard CellSearch™), further molecular analysis and characterization of captured cells might guide personal therapy and track tumor cell genotype during treatment. It is important to highlight here that the CellSearch™ platform can also be potentially updated for molecular and genetic analysis. Thus, viability of captured cells would benefit various diagnostic modalities, such as fluorescent in situ hybridization, phosphoproteomics, DNA mutation, and mRNA profiling. In addition, nanostructured substrates can easily be integrated into microfluidic devices for CTC recognition, isolation, transport and diagnostic modalities. Currently the nanostructure based CTC isolations techniques lack the large scale clinical testing. On the other hand, general trends of strengthened cell adhesion onto nanostructured surfaces were reported. Therefore, nanostructured substrates are potentially promising tools for cell isolation. A new class of highly-functional, highly-integrated microfluidic devices are needed that require lesser volume of analyte for multiple analysis, while reducing physical contamination and improving reaction speed and sensitivity [114]. In brief, the combination of nanostructured substrates and microfluidic devices might facilitate cell isolation.
On the other hand, nanostructured substrates still need to overcome the following limitations before becoming a reliable rare cell isolation tool. In the case of nanopillars and nanowires, the fabrication processes normally involve multiple photolithography, lift-off, wet/dry etching, chemical vapor deposition, and sputtering steps. Although a non-functionalized nanorough glass substrate could enrich cancer cells from lysed whole blood with capture efficiency of more that 90%, further testing on clinical samples is necessary before reaching conclusions [25]. Higher nonspecific binding was also observed on non-functionalized nanorough glass substrates [25]. In nanotextured PDMS, it is a challenge to maintain a stable, cell-adhesive layer because the generated hydroxyl groups undergo a dehydration reaction, and high chain mobility pulls the hydrophobic methyl groups to the surface [115]. This process can significantly inhibit cell attachment and therefore decrease cell isolation efficiency. Nanomaterial coated surfaces can be easily synthesized, however, the higher aspect ratio which might affect cell isolation efficiency cannot be obtained [74]. It was reported that on 3D nanostructured silicon material, the aspect ratio of the nanostructure was critical and efficient cell capture of MCF7 cells only occurred when the aspect ratio is greater than 30 [89]. In some cases non-covalent bonds make the surface coating layer unstable [73]. Furthermore, the most important limitation of nanotexture based cell isolation techniques is that they can only process limited amount of blood at a time. The technique should be capable for processing about ~5–10 mL of blood especially when there are only very few CTCs present in blood.
Major challenges and future directions
Despite the promising results obtained from nanostructured substrates in terms of capture efficiency and cell viability of cancer cell lines, none of these nanostructured substrates have unequivocally shown clinical validity or utility and most of these methods remain in the laboratory settings. The critical distinction between cancer cell lines and true CTCs from tumor patients’ made the true CTC isolation more challenging as true CTCs have heterogeneity in biomarker expression. In general, there are two major challenges faced by CTC isolation technologies. (I) CTC heterogeneity is the primary issue. CTCs can differ between different cancers, even within patients. In brief, CTCs have variable expression of one or more biomarkers on the cell membrane, which further affects morphology and characteristics of CTCs [116]. Due to heterogeneity, not a single cell surface biomarker can confidently be used for cancer cell isolation. Currently, EpCAM is a widely targeted cell membrane receptor for cancer cell isolation. However, some malignant cells do not express EpCAM, resulting in false negatives or positives [117]. Although CD146, CD24/44, ALD-1 and other biomolecules as alternative biomarkers have been used to detect these cancer cells, new markers are needed [118]. (II) The other pressing issue is releasing captured CTCs from the substrate, especially releasing them from a nanostructured substrate. Although the above mentioned techniques can keep the captured CTCs viable on device, cell viability might be reduced after release. It was reported that enzymatic and mechanical lift-off methods can significantly damage ECM proteins [7]. The strong adhesion forces between the captured cells and functionalized surface need to be overcome to detach cells. Methods like physical elution with fluidic flow, thermodynamic release, electrochemical desorption, proteolytic enzyme degradation, endonuclease clearance, oligonucleotide hybridization, removable glass beads array, and negative selection strategy have been used for cell detachment [8,87,119–126]. However, these methods have certain limitations requiring elaborate device design; extremely high shear stress can physically rupture cell membrane; or enzymes can easily lose activity. Therefore, it poses another challenge in detaching captured cells from a microfluidic device, while keeping cells viable for following molecular examination. Recently stimuli-responsive polymer (poly(N-isopropylacrylamide), PNIPAAm) was used to release captured cells without using high shear stress or enzymes [127,128]. When the temperature of the polymer is reduced below lower critical solution temperature (LCST), the polymer swells and helps in efficient release of captured cells. The captured CD4+ and CD34+ cells were released with 59% efficiency and 94% of the cells were viable after release [127]. In the future, PNIPAAm polymer coated surfaces could also be employed in CTC isolation and release studies. The integration of thermally responsive polymer with a nanostructured substrate inside a microfluidic channel would further improve CTC capture and release efficiencies.
Although numerous studies have shown that many cell types react strongly to nanostructured substrates, nanostructured substrates might not be suitable for all cell types; a decrease in cell attachment, proliferation and spreading on nanostructured substrates has also been reported [129–133]. Therefore, in the future, optimization of topographical design and selection of target cells should be studied in detail. Second, preferred nanostructure roughness value and topography is still not clear for cancer cell isolation. In recent studies, nanostructures ranging from 30 to 1150 nm size were employed and higher sensitivity was reported in all cases. It is noted that different cancer cells may show dissimilar response on the same nanostructured substrates. For example, PC3 cells showed stronger focal adhesive forces on a nanostructured surface (Rq = 100 nm) than that of MCF7 and MDA-MB231 cells [112]. Third, mechanical properties of employed materials might also affect cell isolation; however, the exact relationship between mechanical properties of the material and cell capturing efficiency needs to be investigated [98]. Soft biomaterials, such as biocompatible polymers, which mimic native tissues, might facilitate cell isolation in vitro. Finally, it is possible to consider integrating nanostructured substrates with other isolation techniques, such as dielectrophoresis, cell sieve, and negative enrichment and embody them into a device [14,134,135]. A combination of different isolation techniques might help in achieving reliable CTCs isolation results in the future.
Conclusion
CTC detection potentially represents a reliable modality to predict the outcomes of cancer patients and cancer therapy [3,117,136,137]. Hence, it has become increasingly important to develop efficient and reliable systems for CTC isolation. The efficient isolation and accurate identification of CTCs pose some major challenges. Multifaceted expression of cancer renders it even more difficult to use one method for all types of cancer. Finally, CTC isolation involves many factors including cell elasticity, cell type, density and quantity of adhesive elements, immunoaffinity interactions, substrate material, surface topography, flow rate, channel dimensions, and various flow conditions. Nanostructured substrates have emerged as an ultrasensitive platform in CTC isolation, and various fabrication techniques are now available for preparing different types of nanostructured substrates. The utilization of cell release techniques along with nanostructured substrates would help to enhance cell capture and release technologies.
Acknowledgments
We would like to acknowledge partial support from U54 EB015408, R21 HL112114, R01 AI093282, R01 AI081534, R21 AI087107, R21 HL095960, R01 EB015776 and NSF CAREER Award Number 1150733.
Biographies
 Lixue Wang, MD, graduated from Department of Clinical Medicine, Southeast University Medical School, China in 2004, and took a comprehensive study and training of tumor chemotherapy in Xuzhou Medical College, China from 2005 to 2008. In the same year, he joined the Department of Oncology in the Affiliated Hospital of Southeast University. His interests include circulating tumor cell detection and isolation, and their prognostic value in cancer.
Lixue Wang, MD, graduated from Department of Clinical Medicine, Southeast University Medical School, China in 2004, and took a comprehensive study and training of tumor chemotherapy in Xuzhou Medical College, China from 2005 to 2008. In the same year, he joined the Department of Oncology in the Affiliated Hospital of Southeast University. His interests include circulating tumor cell detection and isolation, and their prognostic value in cancer.
 Waseem Asghar, PhD, is a Postdoctoral Research Fellow in Medicine at Harvard Medical School and Division of Biomedical Engineering and Division of Infectious Diseases at Brigham and Women’s Hospital. He earned his Ph.D. from University of Texas at Arlington, Texas, USA in April 2012. His current research interests are in the field of disease diagnosis and prognosis, cancer detection, biosensors, point of care devices and biomaterials. He has published more than 20 peer reviewed journal and full length conference papers. He has also contributed two book chapters published by Springer. Dr. Asghar’s research achievements have been recognized by numerous awards and fellowships: Nanofab Best Graduate Student Award, IEn-gage Mentoring Fellowship, and Science, Technology, Engineering or Mathematics (STEM) Fellowship.
Waseem Asghar, PhD, is a Postdoctoral Research Fellow in Medicine at Harvard Medical School and Division of Biomedical Engineering and Division of Infectious Diseases at Brigham and Women’s Hospital. He earned his Ph.D. from University of Texas at Arlington, Texas, USA in April 2012. His current research interests are in the field of disease diagnosis and prognosis, cancer detection, biosensors, point of care devices and biomaterials. He has published more than 20 peer reviewed journal and full length conference papers. He has also contributed two book chapters published by Springer. Dr. Asghar’s research achievements have been recognized by numerous awards and fellowships: Nanofab Best Graduate Student Award, IEn-gage Mentoring Fellowship, and Science, Technology, Engineering or Mathematics (STEM) Fellowship.
 Utkan Demirci is an Assistant Professor of Medicine and Health Sciences and Technology at the Harvard Medical School (HMS), Brigham and Women’s Hospital (BWH). He received his bachelor’s degree in Electrical Engineering (Summa Cum Laude) in 1999 from the University of Michigan, Ann Arbor, his master’s degrees in Electrical Engineering in 2001 and in Management Science and Engineering in 2005 from Stanford University, and his doctorate in Electrical Engineering in 2005 also from Stanford University. His current work involves applying nano- and micro-scale technologies to manipulate cells in nanoliter volumes, with applications in infectious disease diagnostics and monitoring, cell encapsulation and assembly for cryobiology, tissue engineering, and regenerative medicine. His research interests include the applications of microelectromechanical systems (MEMS) and acoustics in medicine. Dr. Demirci has authored over 75 peer-reviewed journal publications in journals including Nature Materials, PNAS, Advanced Materials, Biomaterials, and Lab on a Chip, more than 100 conference abstracts and proceedings, 12 book chapters and edited a book on point of care diagnostics. His work has been highlighted in Wired Magazine, Nature Photonics, Nature Medicine, MIT Technology Review Magazine, AIP News, BioTechniques, and Biophotonics. He has given over 100 national and international presentations including invited keynotes at various academic, governmental, and industrial institutions.
Utkan Demirci is an Assistant Professor of Medicine and Health Sciences and Technology at the Harvard Medical School (HMS), Brigham and Women’s Hospital (BWH). He received his bachelor’s degree in Electrical Engineering (Summa Cum Laude) in 1999 from the University of Michigan, Ann Arbor, his master’s degrees in Electrical Engineering in 2001 and in Management Science and Engineering in 2005 from Stanford University, and his doctorate in Electrical Engineering in 2005 also from Stanford University. His current work involves applying nano- and micro-scale technologies to manipulate cells in nanoliter volumes, with applications in infectious disease diagnostics and monitoring, cell encapsulation and assembly for cryobiology, tissue engineering, and regenerative medicine. His research interests include the applications of microelectromechanical systems (MEMS) and acoustics in medicine. Dr. Demirci has authored over 75 peer-reviewed journal publications in journals including Nature Materials, PNAS, Advanced Materials, Biomaterials, and Lab on a Chip, more than 100 conference abstracts and proceedings, 12 book chapters and edited a book on point of care diagnostics. His work has been highlighted in Wired Magazine, Nature Photonics, Nature Medicine, MIT Technology Review Magazine, AIP News, BioTechniques, and Biophotonics. He has given over 100 national and international presentations including invited keynotes at various academic, governmental, and industrial institutions.
 Yuan Wan, MD, PhD, majored in Clinical Medicine and Genetics in Southeast University Medical School, China from 1999 to 2007. In 2012, after he graduated from Department of Bioengineering, University of Texas at Arlington, USA, he moved to Adelaide, South Australia. Currently, he is working as a Research Associate in Mawson Institute and Ian Wark Research Institute. His research interests include micro-/nano-devices fabrication, detection and enrichment of biomolecules, and nanomaterials for drug delivery.
Yuan Wan, MD, PhD, majored in Clinical Medicine and Genetics in Southeast University Medical School, China from 1999 to 2007. In 2012, after he graduated from Department of Bioengineering, University of Texas at Arlington, USA, he moved to Adelaide, South Australia. Currently, he is working as a Research Associate in Mawson Institute and Ian Wark Research Institute. His research interests include micro-/nano-devices fabrication, detection and enrichment of biomolecules, and nanomaterials for drug delivery.
Footnotes
Conflict of interest
The authors declare no competing financial interest.
References
- 1.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LWMM, Hayes DF. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 2.Pantel K, Alix-Panabieres C. Trends Mol Med. 2010;16:398–406. doi: 10.1016/j.molmed.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Dotan E, Cohen SJ, Alpaugh KR, Meropol NJ. Oncologist. 2009;14:1070–1082. doi: 10.1634/theoncologist.2009-0094. [DOI] [PubMed] [Google Scholar]
- 4.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slade MJ, Payne R, Riethdorf S, Ward B, Zaidi SAA, Stebbing J, Palmieri C, Sinnett HD, Kulinskaya E, Pitfield T, McCormack RT, Pantel K, Coombes RC. Br J Cancer. 2008;100:160–166. doi: 10.1038/sj.bjc.6604773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.den Toonder J. Lab Chip. 2011;11:375–377. doi: 10.1039/c0lc90100h. [DOI] [PubMed] [Google Scholar]
- 7.Canavan HE, Cheng X, Graham DJ, Ratner BD, Castner DG. J Biomed Mater Res Pt A. 2005;75A:1–13. doi: 10.1002/jbm.a.30297. [DOI] [PubMed] [Google Scholar]
- 8.Wan Y, Liu Y, Allen PB, Asghar W, Mahmood MAI, Tan J, Duhon H, Kim Y-t, Ellington AD, Iqbal SM. Lab Chip. 2012;12:4693–4701. doi: 10.1039/c2lc21251j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, Doyle GV, Matera J, Allard WJ, Miller MC. J Clin Oncol. 2005;23:1420–1430. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 10.Went PTH, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, Dirnhofer S. Hum Pathol. 2004;35:122–128. doi: 10.1016/j.humpath.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 11.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang LR, Cox EC, Austin RH, Sturm JC. Science. 2004;304:987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 13.Shafiee H, Caldwell JL, Sano MB, Davalos RV. Biomed Microdev. 2009;11:997–1006. doi: 10.1007/s10544-009-9317-5. [DOI] [PubMed] [Google Scholar]
- 14.Shafiee H, Sano MB, Henslee EA, Caldwell JL, Davalos RV. Lab Chip. 2010;10:438–445. doi: 10.1039/b920590j. [DOI] [PubMed] [Google Scholar]
- 15.Dochow S, Krafft C, Neugebauer U, Bocklitz T, Henkel T, Mayer G, Albert J, Popp J. Lab Chip. 2011;11:1484–1490. doi: 10.1039/c0lc00612b. [DOI] [PubMed] [Google Scholar]
- 16.Sieben S, Bergemann C, Lubbe A, Brockmann B, Rescheleit D. J Magn Magn Mater. 2001;225:175–179. [Google Scholar]
- 17.He W, Wang H, Hartmann LC, Cheng JX, Low PS. Proc Natl Acad Sci USA. 2007;104:11760–11765. doi: 10.1073/pnas.0703875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asghar W, Wan Y, Ilyas A, Bachoo R, Kim Y, Iqbal SM. Lab Chip. 2012;12:2345–2352. doi: 10.1039/c2lc21012f. [DOI] [PubMed] [Google Scholar]
- 19.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. J Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geckil H, Xu F, Zhang X, Moon SJ, Demirci U. Nanomedicine. 2010;5:469–484. doi: 10.2217/nnm.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abbott A. Nature. 2003;424:870–872. doi: 10.1038/424870a. [DOI] [PubMed] [Google Scholar]
- 22.Jacquemart I, Pamula E, De Cupere VM, Rouxhet PG, Dupont-Gillain CC. J Colloid Interface Sci. 2004;278:63–70. doi: 10.1016/j.jcis.2004.05.040. [DOI] [PubMed] [Google Scholar]
- 23.Smith IO, Liu XH, Smith LA, Ma PX. Wiley Interdiscipl Rev: Nanomed Nanobiotechnol. 2009;1:226–236. doi: 10.1002/wnan.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Z, Kotaki M, Inai R, Ramakrishna S. Tissue Eng. 2005;11:101–109. doi: 10.1089/ten.2005.11.101. [DOI] [PubMed] [Google Scholar]
- 25.Chen W, Weng S, Zhang F, Allen S, Li X, Bao L, Lam RHW, Macoska JA, Merajver SD, Fu J. ACS Nano. 2012;7:566–575. doi: 10.1021/nn304719q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu B, Lu Q, Yin J, Hu J, Wang Z. Tissue Eng. 2005;11:825–834. doi: 10.1089/ten.2005.11.825. [DOI] [PubMed] [Google Scholar]
- 27.Teixeira AI, Nealey PF, Murphy CJ. J Biomed Mater Res Pt A. 2004;71:369–376. doi: 10.1002/jbm.a.30089. [DOI] [PubMed] [Google Scholar]
- 28.Lee KB, Park SJ, Mirkin CA, Smith JC, Mrksich M. Science. 2002;295:1702–1705. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- 29.Kim DH, Kim P, Song I, Cha JM, Lee SH, Kim B, Suh KY. Langmuir. 2006;22:5419–5426. doi: 10.1021/la060283u. [DOI] [PubMed] [Google Scholar]
- 30.Karuri NW, Liliensiek S, Teixeira AI, Abrams G, Campbell S, Nealey PF, Murphy CJ. J Cell Sci. 2004;117:3153–3164. doi: 10.1242/jcs.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark P, Connolly P, Curtis A, Dow J, Wilkinson C. J Cell Sci. 1991;99:73–77. doi: 10.1242/jcs.99.1.73. [DOI] [PubMed] [Google Scholar]
- 32.Baac H, Lee JH, Seo JM, Park TH, Chung H, Lee SD, Kim SJ. Mater Sci Eng C. 2004;24:209–212. [Google Scholar]
- 33.Gurkan UA, Fan Y, Xu F, Erkmen B, Urkac ES, Parlakgul G, Bernstein J, Xing W, Boyden ES, Demirci U. Adv Mater. 2013;25:1192–1198. doi: 10.1002/adma.201203261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HS, Kim DS, Kwon TH. Microsyst Technol. 2007;13:593–599. [Google Scholar]
- 35.Hutmacher D, Woodfield T, Dalton PD, Lewis JA. Tissue Eng. 2008:403–454. [Google Scholar]
- 36.Turner S, Kam L, Isaacson M, Craighead H, Shain W, Turner J. J Vac Sci Technol B: Microelectron Nanomet Struct. 1997;15:2848–2854. [Google Scholar]
- 37.Sapelkin AV, Bayliss SC, Unal B, Charalambou A. Biomaterials. 2006;27:842–846. doi: 10.1016/j.biomaterials.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 38.Das B, McGinnis SP. Appl Phys A: Mater Sci Process. 2000;71:681–688. [Google Scholar]
- 39.Lee C, Bae SY, Mobasser S, Manohara H. Nano Lett. 2005;5:2438–2442. doi: 10.1021/nl0517161. [DOI] [PubMed] [Google Scholar]
- 40.Gurkan UA, Tasoglu S, Kavaz D, Demirel MC, Demirci U. Adv Healthcare Mater. 2012;1:149–158. doi: 10.1002/adhm.201200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tasoglu S, Kavaz D, Gurkan UA, Guven S, Chen P, Zheng R, Demirci U. Adv Mater. 2013;25:1137–1143. doi: 10.1002/adma.201200285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu F, Inci F, Mullick O, Gurkan UA, Sung Y, Kavaz D, Li B, Denkbas EB, Demirci U. ACS Nano. 2012;6:6640–6649. doi: 10.1021/nn300902w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tasoglu S, Demirci U. Trends Biotechnol. 2013;31:10–19. doi: 10.1016/j.tibtech.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doshi J, Reneker DH. J Electrostat. 1995;35:151–160. [Google Scholar]
- 45.Price RL, Ellison K, Haberstroh KM, Webster TJ. J Biomed Mater Res Pt A. 2004;70:129–138. doi: 10.1002/jbm.a.30073. [DOI] [PubMed] [Google Scholar]
- 46.Zanello LP, Zhao B, Hu H, Haddon RC. Nano Lett. 2006;6:562–567. doi: 10.1021/nl051861e. [DOI] [PubMed] [Google Scholar]
- 47.Liopo AV, Stewart MP, Hudson J, Tour JM, Pappas TC. J Nanosci Nanotechnol. 2006;6:1365–1374. doi: 10.1166/jnn.2006.155. [DOI] [PubMed] [Google Scholar]
- 48.MacDonald RA, Laurenzi BF, Viswanathan G, Ajayan PM, Stegemann JP. J Biomed Mater Res Pt A. 2005;74:489–496. doi: 10.1002/jbm.a.30386. [DOI] [PubMed] [Google Scholar]
- 49.Vidyala SD, Asghar W, Iqbal SM. J Nanobiotechnol. 2011;9:18. doi: 10.1186/1477-3155-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee LJ, Zeng C, Cao X, Han X, Shen J, Xu G. Compos Sci Technol. 2005;65:2344–2363. [Google Scholar]
- 51.Nam YS, Park TG. J Biomed Mater Res. 1999;47:8–17. doi: 10.1002/(sici)1097-4636(199910)47:1<8::aid-jbm2>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 52.Gates BD, Xu Q, Stewart M, Ryan D, Willson CG, Whitesides GM. Chem Rev Columbus. 2005;105:1171–1196. doi: 10.1021/cr030076o. [DOI] [PubMed] [Google Scholar]
- 53.Asghar W, Ramachandran PP, Adewumi A, Noor MR, Iqbal SM. J Manuf Sci Eng. 2010;132:030911. [Google Scholar]
- 54.Chen W, Ahmed H. Appl Phys Lett. 1993;62:1499–1501. [Google Scholar]
- 55.Ilyas A, Asghar W, Allen PB, Duhon H, Ellington AD, Iqbal SM. Nanotechnology. 2012;23:275502. doi: 10.1088/0957-4484/23/27/275502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang HP, Lai KY, Lin YR, Lin CA, He JH. Langmuir. 2010;26:12855–12858. doi: 10.1021/la1012507. [DOI] [PubMed] [Google Scholar]
- 57.Puckett SD, Lee PP, Ciombor DM, Aaron RK, Webster TJ. Acta Biomater. 2010;6:2352–2362. doi: 10.1016/j.actbio.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 58.Park JH, Chung BG, Lee WG, Kim J, Brigham MD, Shim J, Lee S, Hwang CM, Durmus NG, Demirci U, Khademhosseini A. Biotechnol Bioeng. 2010;106:138–148. doi: 10.1002/bit.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee WG, Demirci U, Khademhosseini A. Integr Biol. 2009;1:242–251. doi: 10.1039/b819201d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song YS, Lin RL, Montesano G, Durmus NG, Lee G, Yoo SS, Kayaalp E, Haeggstrom E, Khademhosseini A, Demirci U. Anal Bioanal Chem. 2009;395:185–193. doi: 10.1007/s00216-009-2935-1. [DOI] [PubMed] [Google Scholar]
- 61.Lu J, Rao MP, MacDonald NC, Khang D, Webster TJ. Acta Biomater. 2008;4:192–201. doi: 10.1016/j.actbio.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 62.Xu F, Sridharan BP, Durmus NG, Wang SQ, Yavuz AS, Gurkan UA, Demirci U. PLoS ONE. 2011;6:19344. doi: 10.1371/journal.pone.0019344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moon SJ, Hasan SK, Song YS, Xu F, Keles HO, Manzur F, Mikkilineni S, Hong JW, Nagatomi J, Haeggstrom E. Tissue Eng Pt C: Methods. 2009;16:157–166. doi: 10.1089/ten.TEC.2009.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu F, Sridharan BP, Wang SQ, Gurkan UA, Syverud B, Demirci U. Biomicrofluidics. 2011;5 doi: 10.1063/1.3580752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Durmus NG, Taylor EN, Inci F, Kummer KM, Tarquinio KM, Webster TJ. Int J Nanomed. 2012;7:537. doi: 10.2147/IJN.S27957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stevens MM, George JH. Science. 2005;310:1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 69.Curtis A, Wilkinson C. Biomaterials. 1997;18:1573–1583. doi: 10.1016/s0142-9612(97)00144-0. [DOI] [PubMed] [Google Scholar]
- 70.Yang L, Sheldon BW, Webster TJ. J Biomed Mater Res Pt A. 2008;91:548–556. doi: 10.1002/jbm.a.32227. [DOI] [PubMed] [Google Scholar]
- 71.Khang D, Kim SY, Liu-Snyder P, Palmore GTR, Durbin SM, Webster TJ. Biomaterials. 2007;28:4756–4768. doi: 10.1016/j.biomaterials.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 72.Biggs MJP, Richards RG, Gadegaard N, Wilkinson CDW, Oreffo ROC, Dalby MJ. Biomaterials. 2009;30:5094–5103. doi: 10.1016/j.biomaterials.2009.05.049. [DOI] [PubMed] [Google Scholar]
- 73.Anselme K, Davidson P, Popa A, Giazzon M, Liley M, Ploux L. Acta Biomater. 2010;6:3824–3846. doi: 10.1016/j.actbio.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 74.Flemming R, Murphy C, Abrams G, Goodman S, Nealey P. Biomaterials. 1999;20:573–588. doi: 10.1016/s0142-9612(98)00209-9. [DOI] [PubMed] [Google Scholar]
- 75.Lamers E, van Horssen R, Te Riet J, van Delft F, Luttge R, Walboomers X, Jansen J. Eur Cells Mater. 2010;20:329–343. doi: 10.22203/ecm.v020a27. [DOI] [PubMed] [Google Scholar]
- 76.Zorlutuna P, Rong Z, Vadgama P, Hasirci V. Acta Biomater. 2009;5:2451–2459. doi: 10.1016/j.actbio.2009.03.027. [DOI] [PubMed] [Google Scholar]
- 77.Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Biochim Biophys Acta: Mol Cell Res. 2004;1692:103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 78.Fischer KE, Alemán BJ, Tao SL, Daniels RH, Li EM, Bunger MD, Nagaraj G, Singh P, Zettl A, Desai TA. Nano Lett. 2009;9:716–720. doi: 10.1021/nl803219f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sitti M, Fearing RS. J Adhes Sci Technol. 2003;17:1055–1073. [Google Scholar]
- 80.Spolenak R, Gorb S, Gao H, Arzt E. Proc R Soc A: Math Phys Eng Sci. 2005;461:305–319. [Google Scholar]
- 81.Autumn K, Sitti M, Liang YA, Peattie AM, Hansen WR, Sponberg S, Kenny TW, Fearing R, Israelachvili JN, Full RJ. Proc Natl Acad Sci USA. 2002;99:12252–12256. doi: 10.1073/pnas.192252799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Menguc Y, Yang SY, Kim S, Rogers JA, Sitti M. Adv Funct Mater. 2012;22:1246–1254. [Google Scholar]
- 83.Murphy MP, Aksak B, Sitti M. Small. 2008;5:170–175. doi: 10.1002/smll.200801161. [DOI] [PubMed] [Google Scholar]
- 84.Murphy MP, Kim S, Sitti M. ACS Appl Mater Interfaces. 2009;1:849–855. doi: 10.1021/am8002439. [DOI] [PubMed] [Google Scholar]
- 85.Yurdumakan B, Raravikar NR, Ajayan PM, Dhinojwala A. Chem Commun. 2005:3799–3801. doi: 10.1039/b506047h. [DOI] [PubMed] [Google Scholar]
- 87.Chen L, Liu X, Su B, Li J, Jiang L, Han D, Wang S. Adv Mater. 2011;23:4376–4380. doi: 10.1002/adma.201102435. [DOI] [PubMed] [Google Scholar]
- 88.Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei K, Sun J, Sherman DJ, Behrenbruch CP, Wu H. Angew Chem. 2009;121:9132–9135. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang S, Liu K, Liu J, Yu ZTF, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LWK, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CKF, Tseng HR. Angew Chem Int Ed. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim ST, Kim DJ, Kim TJ, Seo DW, Kim TH, Lee SY, Kim K, Lee KM, Lee SK. Nano Lett. 2010;10:2877–2883. doi: 10.1021/nl100942p. [DOI] [PubMed] [Google Scholar]
- 91.Park GS, Kwon H, Kwak DW, Park SY, Kim M, Lee JH, Han H, Heo S, Li XS, Lee JH, Kim YH, Lee JG, Yang W, Cho HY, Kim SK, Kim K. Nano Lett. 2012;12:1638–1642. doi: 10.1021/nl2045759. [DOI] [PubMed] [Google Scholar]
- 92.Lee SK, Kim GS, Wu Y, Kim DJ, Lu Y, Kwak M, Han L, Hyung JH, Seol JK, Sander C, Gonzalez A, Li J, Fan R. Nano Lett. 2012;12:2697–2704. doi: 10.1021/nl2041707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liotta LA, Tryggvason K, Garbisa S, Hart I, Foltz CM, Shafie S. Nature. 1980;284:67–68. doi: 10.1038/284067a0. [DOI] [PubMed] [Google Scholar]
- 94.Liotta LA, Nageswara Rao C, Wewer UM. Annu Rev Biochem. 1986;55:1037–1057. doi: 10.1146/annurev.bi.55.070186.005133. [DOI] [PubMed] [Google Scholar]
- 95.Wan Y, Mahmood M, Li N, Allen PB, Kim Y, Bachoo R, Ellington AD, Iqbal SM. Cancer. 2011;118:1145–1154. doi: 10.1002/cncr.26349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hughes AD, King MR. Langmuir. 2010;26:12155–12164. doi: 10.1021/la101179y. [DOI] [PubMed] [Google Scholar]
- 97.Han W, Allio BA, Foster DG, King MR. ACS Nano. 2009;4:174–180. doi: 10.1021/nn900442c. [DOI] [PubMed] [Google Scholar]
- 98.Sekine J, Luo S-C, Wang S, Zhu B, Tseng H-R, Yu H-h. Adv Mater. 2011;23:4788–4792. doi: 10.1002/adma.201102151. [DOI] [PubMed] [Google Scholar]
- 99.Wang B, Weldon AL, Kumnorkaew P, Xu B, Gilchrist JF, Cheng X. Langmuir. 2011;27:11229–11237. doi: 10.1021/la2015868. [DOI] [PubMed] [Google Scholar]
- 100.Zhang N, Deng Y, Tai Q, Cheng B, Zhao L, Shen Q, He R, Hong L, Liu W, Guo S, Liu K, Tseng HR, Xiong B, Zhao XZ. Adv Mater. 2012;24:2756–2760. doi: 10.1002/adma.201200155. [DOI] [PubMed] [Google Scholar]
- 101.Asghar W, Kim YT, Ilyas A, Sankaran J, Wan Y, Iqbal SM. Nanotechnology. 2012;23:475601. doi: 10.1088/0957-4484/23/47/475601. [DOI] [PubMed] [Google Scholar]
- 102.Phillips JA, Xu Y, Xia Z, Fan ZH, Tan W. Anal Chem. 2008;81:1033–1039. doi: 10.1021/ac802092j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krasik EF, Hammer DA. Biophys J. 2004;87:2919–2930. doi: 10.1529/biophysj.104.039693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang S, Inci F, Chaunzwa TL, Ramanujam A, Vasudevan A, Subramanian S, Ip AC, Sridharan B, Gurkan UA, Demirci U. Int J Nanomed. 2012;7:2591–2600. doi: 10.2147/IJN.S29629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McEver RP, Zhu C. Annu Rev Cell Dev Biol. 2010;26:363–396. doi: 10.1146/annurev.cellbio.042308.113238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sundd P, Pospieszalska MK, Ley K. Mol Immunol. 2013;55:59–69. doi: 10.1016/j.molimm.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Coghill PA, Kesselhuth EK, Shimp EA, Khismatullin DB, Schmidtke DW. Biomed Microdev. 2013;15:183–193. doi: 10.1007/s10544-012-9715-y. [DOI] [PubMed] [Google Scholar]
- 108.Kim DJ, Seol JK, Lee G, Kim GS, Lee SK. Nanotechnology. 2012;23:395102. doi: 10.1088/0957-4484/23/39/395102. [DOI] [PubMed] [Google Scholar]
- 109.Wan Y, Kim Y-t, Li N, Cho SK, Bachoo R, Ellington AD, Iqbal SM. Cancer Res. 2010;70:9371–9380. doi: 10.1158/0008-5472.CAN-10-0568. [DOI] [PubMed] [Google Scholar]
- 110.Wan Y, Tan J, Asghar W, Kim Y-t, Liu Y, Iqbal SM. J Phys Chem B. 2011;115:13891–13896. doi: 10.1021/jp205511m. [DOI] [PubMed] [Google Scholar]
- 111.Qi S, Yi C, Ji S, Fong CC, Yang M. ACS Appl Mater Interfaces. 2008;1:30–34. doi: 10.1021/am800027d. [DOI] [PubMed] [Google Scholar]
- 112.Zhang Z, Chen N, Li S, Batting MR, Wang Y. J Am Chem Soc. 2012;134:15716–15719. doi: 10.1021/ja307717w. [DOI] [PubMed] [Google Scholar]
- 113.Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Janicke F. Clin Cancer Res. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 114.Sia SK, Whitesides GM. Electrophoresis. 2003;24:3563–3576. doi: 10.1002/elps.200305584. [DOI] [PubMed] [Google Scholar]
- 115.Bhattacharya S, Singh RK, Mandal S, Ghosh A, Bok S, Korampally V, Gangopadhyay K, Gangopadhyay S. J Adhes Sci Technol. 2010;24:2707–2739. [Google Scholar]
- 116.van de Stolpe A, Pantel K, Sleijfer S, Terstappen LW, den Toonder JMJ. Cancer Res. 2011;71:5955–5960. doi: 10.1158/0008-5472.CAN-11-1254. [DOI] [PubMed] [Google Scholar]
- 117.Sieuwerts AM, Kraan J, Bolt J, van der Spoel P, Elstrodt F, Schutte M, Martens JWM, Gratama JW, Sleijfer S, Foekens JA. J Natl Cancer Inst. 2009;101:61–66. doi: 10.1093/jnci/djn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Erdbruegger U, Haubitz M, Woywodt A. Clin Chim Acta. 2006;373:17–26. doi: 10.1016/j.cca.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 119.Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Göttert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. J Am Chem Soc. 2008;130:8633–8641. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ernst O, Lieske A, Jager M, Lankenau A, Duschl C. Lab Chip. 2007;7:1322–1329. doi: 10.1039/b708619a. [DOI] [PubMed] [Google Scholar]
- 121.Zhu H, Yan J, Revzin A. Colloids Surf B: Biointerfaces. 2008;64:260–268. doi: 10.1016/j.colsurfb.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 122.Canavan HE, Cheng X, Graham DJ, Ratner BD, Castner DG. Langmuir. 2005;21:1949–1955. doi: 10.1021/la048546c. [DOI] [PubMed] [Google Scholar]
- 123.Hatch A, Pesko DM, Murthy SK. Anal Chem. 2012;84:4618–4621. doi: 10.1021/ac300496q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li S, Chen N, Zhang Z, Wang Y. Biomaterials. 2013;34:460–469. doi: 10.1016/j.biomaterials.2012.09.040. [DOI] [PubMed] [Google Scholar]
- 125.Plouffe BD, Brown MA, Iyer RK, Radisic M, Murthy SK. Lab Chip. 2009;9:1507–1510. doi: 10.1039/b823523f. [DOI] [PubMed] [Google Scholar]
- 126.Hatch A, Hansmann G, Murthy SK. Langmuir: ACS J Surf Coll. 2011;27:4257. doi: 10.1021/la105016a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gurkan UA, Anand T, Tas H, Elkan D, Akay A, Keles HO, Demirci U. Lab Chip. 2011;11:3979–3989. doi: 10.1039/c1lc20487d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gurkan UA, Tasoglu S, Akkaynak D, Avci O, Unluisler S, Canikyan S, MacCallum N, Demirci U. Adv Healthcare Mater. 2012;1:661–668. doi: 10.1002/adhm.201200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dalby MJ, Gadegaard N, Riehle MO, Wilkinson CDW, Curtis ASG. Int J Biochem Cell Biol. 2004;36:2005–2015. doi: 10.1016/j.biocel.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 130.Gallagher J, McGhee K, Wilkinson C, Riehle M. IEEE Trans Nanobiosci. 2002;1:24–28. doi: 10.1109/tnb.2002.806918. [DOI] [PubMed] [Google Scholar]
- 131.Curtis A, Gadegaard N, Dalby M, Riehle M, Wilkinson C, Aitchison G. IEEE Trans Nanobiosci. 2004;3:61–65. doi: 10.1109/tnb.2004.824276. [DOI] [PubMed] [Google Scholar]
- 132.Andersson AS, Bäckhed F, von Euler A, Richter-Dahlfors A, Sutherland D, Kasemo B. Biomaterials. 2003;24:3427–3436. doi: 10.1016/s0142-9612(03)00208-4. [DOI] [PubMed] [Google Scholar]
- 133.Dalby MJ, Riehle MO, Sutherland DS, Agheli H, Curtis ASG. Biomaterials. 2004;25:5415–5422. doi: 10.1016/j.biomaterials.2003.12.049. [DOI] [PubMed] [Google Scholar]
- 134.Liu Z, Fusi A, Klopocki E, Schmittel A, Tinhofer I, Non-nenmacher A, Keilholz U. J Transl Med. 2011;9:70. doi: 10.1186/1479-5876-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Langb JM, Beebea DJ. Lab Chip. 2013;13:391–396. doi: 10.1039/c2lc41136a. [DOI] [PubMed] [Google Scholar]
- 136.Attard G, de Bono JS. Curr Opin Genet Dev. 2011;21:50–58. doi: 10.1016/j.gde.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 137.Danova M, Torchio M, Mazzini G. Expert Rev Mol Diagn. 2011;11:473–485. doi: 10.1586/erm.11.33. [DOI] [PubMed] [Google Scholar]
- 138.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AGJ, Uhr JW, Terstappen LWMM. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 139.Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI, Fleisher M, Stadler W, Datar RH, Tai YC, Cote RJ. Clin Cancer Res. 2010;16:5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cohen SJ, Punt CJA, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LWMM, Meropol NJ. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 141.Fischer AH. Arch Pathol Lab Med. 2009;133:1367–1369. doi: 10.5858/133.9.1367. [DOI] [PubMed] [Google Scholar]
- 142.Beveridge R. Commun Oncol. 2007;4:79–82. [Google Scholar]
- 143.De Giorgi U, Valero V, Rohren E, Dawood S, Ueno NT, Miller MC, Doyle GV, Jackson S, Andreopoulou E, Handy BC, Reuben JM, Fritsche HA, Macapinlac HA, Hortobagyi GN, Cristofanilli M. J Clin Oncol. 2009;27:3303–3311. doi: 10.1200/JCO.2008.19.4423. [DOI] [PubMed] [Google Scholar]
- 144.Eifler RL, Lind J, Falkenhagen D, Weber V, Fischer MB, Zeillinger R. Cytomet Pt B: Clin Cytomet. 2011;80B:100–111. doi: 10.1002/cyto.b.20560. [DOI] [PubMed] [Google Scholar]
- 145.Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, Yu W, Xiao W, Davis MM, Pease RF, Mindrinos MN, Jeffrey SS, Davis RW. Proc Natl Acad Sci USA. 2009;106:3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hofman VJ, Ilie MI, Bonnetaud C, Selva E, Long E, Molina T, Vignaud JM, Fléjou JF, Lantuejoul S, Piaton E, Butori C, Mourad N, Poudenx M, Bahadoran P, Sibon S, Guevara N, Santini J, Vénissac N, Mouroux J, Vielh P, Hofman PM. Am J Clin Pathol. 2011;135:146–156. doi: 10.1309/AJCP9X8OZBEIQVVI. [DOI] [PubMed] [Google Scholar]
- 147.Lim LS, Hu M, Huang MC, Cheong WC, Gan ATL, Looi XL, Leong SM, Koay ESC, Li MH. Lab Chip. 2012;12:4388–4396. doi: 10.1039/c2lc20750h. [DOI] [PubMed] [Google Scholar]
- 148.Tan SJ, Lakshmi RL, Chen P, Lim WT, Yobas L, Lim CT. Biosens Bioelectron. 2010;26:1701–1705. doi: 10.1016/j.bios.2010.07.054. [DOI] [PubMed] [Google Scholar]
- 149.Kuntaegowdanahalli SS, Bhagat AAS, Kumar G, Papautsky I. Lab Chip. 2009;9:2973–2980. doi: 10.1039/b908271a. [DOI] [PubMed] [Google Scholar]
- 150.Tanaka T, Ishikawa T, Numayama-Tsuruta K, Imai Y, Ueno H, Matsuki N, Yamaguchi T. Lab Chip. 2012;12:4336–4343. doi: 10.1039/c2lc40354d. [DOI] [PubMed] [Google Scholar]
- 151.Lee WC, Bhagat AAS, Huang S, Van Vliet KJ, Han J, Lim CT. Lab Chip. 2011;11:1359–1367. doi: 10.1039/c0lc00579g. [DOI] [PubMed] [Google Scholar]
- 152.Gascoyne PRC, Noshari J, Anderson TJ, Becker FF. Electrophoresis. 2009;30:1388–1398. doi: 10.1002/elps.200800373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen P-i, Morgan B, Trautwein J, Kimura A, Sengupta S, Stott SL, Karabacak NM, Barber TA, Walsh JR, Smith K, Spuhler PS, Sullivan JP, Lee RJ, Ting DT, Luo X, Shaw AT, Bardia A, Sequist LV, Louis DN, Maheswaran S, Kapur R, Haber DA, Toner M. Sci Transl Med. 2013;5:179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Proc Natl Acad Sci USA. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST, Nanus DM, Giannakakou PA, Kirby BJ. Lab Chip. 2010;10:27–29. doi: 10.1039/b917959c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Helzer KT, Barnes HE, Day L, Harvey J, Billings PR, Forsyth A. Cancer Res. 2009;69:7860–7866. doi: 10.1158/0008-5472.CAN-09-0801. [DOI] [PubMed] [Google Scholar]
- 157.Schiro PG, Zhao M, Kuo JS, Koehler KM, Sabath DE, Chiu DT. Angew Chem Int Ed. 2012;51:4618–4622. doi: 10.1002/anie.201108695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rizvi I, Gurkan UA, Tasoglu S, Alagic N, Celli JP, Mensah LB, Mai Z, Demirci U, Hasan T. PNAS. 2013;110:1974–1983. doi: 10.1073/pnas.1216989110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Inci F, Tokel O, Wang S, Gurkan UA, Tasoglu S, Kuritzkes DR, Demirci U. ACS Nano. 2013;7:4733–4745. doi: 10.1021/nn3036232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang S, Inci F, Libero GD, Singhal A, Demirci U. Biotech Adv. 2013;31:438–449. doi: 10.1016/j.biotechadv.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]